Affiliation:
1School of Life Science, Institute for Immunology, Tsinghua University, Beijing 100084, China
Email: Kabeerhaneef16@gmail.com
ORCID: https://orcid.org/0000-0002-7638-7358
Affiliation:
2Department of Pediatric Haematology, Oncology and BMT, The Children’s Hospital and Institute of Child Health, Lahore 53700, Pakistan
Email: Kabeerhaneef16@gmail.com
Affiliation:
3Department of Biochemistry, Cancer Institute of the second affiliated hospital, School of Medicine, Zhejiang University, Hangzhou 310058, Zhejiang, China
ORCID: https://orcid.org/0000-0002-0503-2947
Affiliation:
4Centre of Excellence in Molecular Biology, University of the Punjab, Lahore 53700, Pakistan
ORCID: https://orcid.org/0000-0003-1430-9538
Affiliation:
5School of Agriculture Sciences and Food Technology, Shandong University of Technology, Zibo 255000, Shandong, China
Affiliation:
6National Institute for Biotechnology and Genetic engineering (NIBGE), Faisalabad 38000, Pakistan
7Pakistan Institute of Engineering and Applied Sciences (PIEAS), Nilore, Islamabad 45650, Pakistan
Affiliation:
4Centre of Excellence in Molecular Biology, University of the Punjab, Lahore 53700, Pakistan
ORCID: https://orcid.org/0000-0002-5190-6844
Explor Immunol. 2021;1:140–154 DOI: https://doi.org/10.37349/ei.2021.00011
Received: April 29, 2021 Accepted: July 11, 2021 Published: August 31, 2021
Academic Editor: Wangxue Chen, Human Health Therapeutics Research Center, Canada
The article belongs to the special issue Vaccine-induced Immune Responses Against SARS-CoV-2 Infections
Coronavirus disease-2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus (SARS-CoV)-2 spread globally and creates an alarming situation. Following the SARS-CoV-2 paradigm, therapeutic efficacy is achieved via repurposing several antiviral, antibacterial, and antimalarial drugs. Innate and adaptive immune cells work close to combat infection through the intricate production of antibodies (Abs) and inflammatory cytokines. As an essential component of the immune system, Abs play an important role in eliminating viruses and maintaining homeostasis. B lymphocytes (B cells) are effector cells, stringent to produce neutralizing Abs to combat infection. After recognizing SARS-CoV-2 antigens by a surface receptor called B cell receptors (BCRs) on the plasma membrane, the BCRs transmembrane signal transduction and immune activation results in Ab production and development of immune memory. Thus, it ensures that plasma B cells can quickly start an intricate immune response to generate efficient protective Abs to clear the pathogen. Nevertheless, considering therapeutic challenges in the context of the new coronavirus pandemic, this review addresses the molecular mechanism of the immune activation and function of novel SARS-CoV-2 specific B cells in the production of SARS-CoV-2 specific Abs. Additionally, these studies highlighted the Ab-mediated pathogenesis, the intriguing role of nano-scale signaling subunits, non-structural proteins during COVID-19 infection, and structural insights of SARS-CoV-2 specific Abs.
Undoubtedly, the whole world lives with the reality of coronavirus disease-2019 (COVID-19) disease, a pandemic that has claimed thousands of lives. The novel coronavirus disease (nCOVID-19) caused by severe acute respiratory syndrome coronavirus (SARS-CoV)-2 has been reported from Wuhan with remarkable consequences and has spread worldwide, creating alarming situations [1–3]. Phylogenetic analysis of SARS-CoV-2 reveals proximity with β subgroup [4], placed in Coronaviridae family [5]. Interestingly, SARS-CoV-2 shows a higher tendency to bind human angiotensin-converting enzyme-2 (hACE2) in contrast to severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome (MERS) to induce lungs infection [6]. Genetic annotations revealed that the SARS-CoV-2 genome potentially encodes four structural [membrane (M), nucleocapsid (N), spike (S), envelope (E)] and six accessory proteins (7, 8, 7a, 7b, 3a, 6) [1]. Receptor-binding domain (RBD), a subunit of surface S glycoprotein, triggers attachment with host hACE2 [7]. Intriguing genome mapping reveals 79.6% identity features between SARS-CoV-2 and SARS-CoV [8]. Following the tremendous increase in death toll and severity of the disease, healthcare emergencies have posed an urgent call for developing vaccines, suitable drug candidates, and high-affinity antibodies (Abs) to combat the SARS-CoV-2 pandemic [5, 9–12]. At present, several antiviral drugs (remdesivir, umifenovir, lponavir, ritonavir) alone or in combination with antimalarial (chloroquine) and antibacterial are repurposed to achieve therapeutic efficacy [1, 13–15]. Additionally, convalescent plasma (CP) therapy is beneficial in the treatment of SARS-CoV-2 infected patients [11, 12, 15]. However, severe threats and devastation in lung pathologies are caused by SARS-CoV-2 in contrast to SARS-CoV and MERS coronavirus (MERS-CoV) [16, 17].
Following infection, innate (macrophage, dendritic cells) and adaptive (B and T) immune cells coordinate efficiently to neutralize the pathogens [18, 19]. In vivo studies have revealed that following interaction of external invading Ags with a surface-anchored receptor called the B cell receptor (BCR), a series of signal transduction events occurs via phosphorylation of the immunoreceptors tyrosine-based activation motif (ITAM) interlinked immunoglobulin (Ig)α, Igβ domains via underlying kinase phosphorylated spleen tyrosine kinase (pSyk) [2]. These events led to the subsequent recruitment of proximal signaling molecules and adaptor proteins to produce neutralizing Abs (NAbs) and develop immune memory [20, 21]. B cell receptors (BCRs) configuration reveals noncovalent meticulous interactions of membrane-tethered with Igα, Igβ [21–23]. Interestingly BCRs transduced proximal signaling further develop immune memory to combat reoccurrence of infection [24]. Several studies have reported that B lymphocytes (B cells) producing effective SARS-CoV-2 specific NAbs (IgG, IgM, IgE) have been successfully isolated and identified in convalescent patient’s plasma [25]. Multiple technical platforms, including immunology, high-speed, high-resolution live-cell single-molecule fluorescence imaging technologies, have illustrated potential mechanisms to uncover a series of events showing how B cells undergo Ab production [26]. Here we review the literature, highlighting how nanoscale signaling immune (B and T) cells contribute to COVID-19 mediated infection. Furthermore, we illustrate some Ab-mediated parameters against SARS-CoV-2 concerning some cross-sectional studies. These studies could provide detailed insights in the future for the development of therapeutic entanglements for SARS-CoV-2 infection (Figure 1).
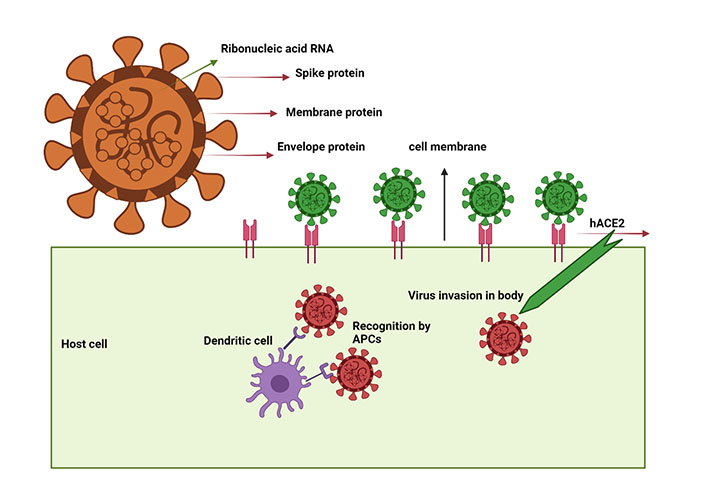
SARS-CoV-2 virus schematic illustration showing the target proteins and mechanism of entry into the host cell. SARS-CoV-2 virus structural proteins include S, M, and E protein. When SARS-CoV-2 enters host cells, it is recognized by the hACE2 and processed by APCs, particularly dendritic cells, which initiate the signaling cascade events
Antiviral immunity emerged through the intricate association between the Ag and APCs through delicate interactions confronted through external projections and dendrites of in vivo APCs [5]. Following viral infection, bridge cells (macrophages and dendritic cells) are infiltrated at the infection tropism to combat infection [27]. Major histocompatibility complex class (MHC)I molecules are required for the efficient processing and presentation of exogenous invading Ags to B cells [5], which results in humoral immune responses and activation of CD8+ T cells [5]. However, sophisticated mechanisms achieved through collective contributions of interferons (IFNs) and interleukins (IL), including IFNs, IL-2, IL-12, and IL-33, constrain viral replication and further assassinate virus-infected cells [28, 29]. Besides, T cell interventions contribute a significant role to clear infection through the production of inflammatory cytokines, thus offering a comprehensive strategy to prevent the reoccurrence of COVID-19 disease [29]. Similar studies have shown how T cell subset counts fluctuate during COVID infection [30]. Typically, T cell counts in COVID-19 patients with severe and moderate symptoms seem diminished (541.5/μL) than the normal range (955.0–2,860.0/μL) [31], in contrast to B cell and NKT cells [32, 33].
Similarly, the key difference in CD4+ and CD8+ T cells reveal the onset of differential cytokine expressions during SARS-CoV-2 infection [30]. T cell subset contributions during SARS-CoV-2 infection have already been investigated, offering new insights to designing a vaccine against SARS-CoV-2. Similar interventions have demonstrated the influence of T cell subsets such as T helper (TH)1, TH2, TH17, and regulatory T cell (Treg) on IL and cytokine secretion mechanisms during SARS-CoV-2 infection, with TH1 cells secreting significantly less IL-6 [31, 34]. Interestingly, TH2 cells during COVID-19 infection triggered IL-4 and IL-3 cytokine production [35]. Corresponding studies reported that T cells, especially naïve regulatory and induced Treg cells (iTreg) [CD45RO+CD3+CD4+CD25+CD127low+, iTreg], seem diminished in severe COVID-19 infected patients, thereby exaggerating the immune response to trigger disease severity [36]. Besides the activation of costimulatory receptors like—CD28, OX40 (CD134), and 4-1BB (CD137) were observed during activation of CD4+ T cells [37]. Co-stimulatory receptor molecules increase in patients with SARS-CoV-2 infection [37]. A considerable increase in OX40 and 4-1BB expression was identified in COVID-19 infected individuals compared to healthy controls [38]. Inflammatory cytokines [tumor necrosis factor (TNF)-α, IL-8, 1L-2, IL-10] are mostly co-related to disease.
Several cellular activities, such as cellular exhaustion, have been investigated during the chronic phase of COVID-19 infection, affecting disease progression [39]. Infected cells mainly express programmed cell death protein 1 (PD-1), cytotoxic T-lymphocyte-associated Ag 4 (CTLA-4), T cell Ig and mucin domain 3 (Tim-3) Ags fluctuating COVID-19 infection [40]. To better decipher the lungs pathology during SARS-CoV-2 infection, several interventions revealed the collective contributions of C-X-C motif (CXC) family receptors, including CXC ligand (CXCL) 10 (CXCL-10), C-C chemokine ligand (CCL)2, CCL5 stringent to trigger the site-specific aggregation and recruitment of neutrophils in lungs. Potentially, these mechanisms trigger lung injury due to the release of antimicrobial factors such as reactive oxygen species (ROS), proteases, and neutrophil extracellular traps (NETs) [41]. Other studies have revealed the expression of CXCL8 (IL-8), a major neutrophil-attracting chemokine in patients with severe COVID-19 infection [42]. Cytotoxic T lymphocytes induce apoptosis by various means such as the production of cytotoxic granules (perforin and granzyme), expression of cytokines, such as TNF, Fas ligand (FasL), and TNF-related apoptosis-inducing ligand (TRAIL) in virus-infected cells [43].
A large proportion of biomarkers have been identified from humans. To investigate mRNA and DNA, PCR-directed amplification techniques and protein assays are broadly undergoing [44]. Similarly, Ab production against specific SARS-CoV-2 viral infections, including NAbs against N, RBD, and S proteins, could be detected to confirm the persistence of the virus [10]. Patients suffering from SARS-CoV-2 infection have been reported positive for Ab production [10]. Several studies have reported the onset of triggered cytokine (IL-18, IL-8, IL-6) production in SARS-CoV-2 infected patients, suggesting positive biomarkers to trigger diagnostic levels [45]. Similar investigations have reported diminished levels, especially IL-1, IL-4, IL-12, and TNF-a during the SARS-CoV-2 diseased [46].
Following the interaction of Ags, BCR undergoes signal transduction events through phosphorylation of ITAMs to produce plasma cells to generate effective Abs [22]. BCRs delicate configuration possessed membrane-tethered Ig, non-covalently associated with Iga, Igβ domains (Figure 2) [20]. Following BCR interactions with SARS-CoV-2 structural Ags, BCRs undergo series of signal transduction events [20]. These events lead to several well-ordered molecular events, including isotype class switching, affinity maturation, and Ab production against SARS-CoV-2 [5, 20, 21]. BCR’s intricate, methodical configuration reveals efficient capability to translate the external physical, chemical, and antigenic cues and harbor sophisticated mechanisms of generating chemical signal entities to reshape the immune mechanism [22, 26]. However, potential investigations on how BCRs of SARS-CoV-2 specific B cells isolated from CP of infected people respond to BCR signaling, spatiotemporal dynamics, and Ab production remains obscure.
Potential contributions of B cells response during SARS-CoV-2 pandemic are crucial during vaccine production. Similar interventions reported that Igs (IgG, IgM) concentrations were disturbed after SARS-CoV-2 infection [47]. Similar studies indicated that SARS-CoV-2 exposed patients showed positivity for SARS-CoV-2 specific IgG and IgA at some time frames [47–50]. Patients experiencing the active phase of SARS-CoV-2 infection generally showed peak amounts of Abs in contrast to patients [31, 48, 51], with relative IgG (amounts of 6.3 compared to 2.1) in a time of 30–45 days after the first symptoms surfaced [2]. Similar speculations have reported that patients suffering from active, mild, or severe SARS-CoV-2 infection develop Abs despite disturbances in B cell counts and IgA, IgM, and IgG expression profiles [3].
As an adaptive immune system, it’s vital to elaborate on B cell counts and their role during the SARS-CoV-2 infection trajectory [52, 53]. Several studies have reported that following SARS-CoV-2 infection, the magnitude of Ab-secreting B cells was significantly higher than in healthy individuals, implying that they could serve as a prognostic indicator of disease [4]. Furthermore, these studies highlighted a reduced number of marginal zone B (MZB) cells in SARS-CoV-2 infected people compared to healthy individuals [54]. Similar studies have demonstrated that the triggered population of Ab-secreting plasma cells in patients correlates with disease severity; however, determining whether the Abs secreted are protective or pro-inflammatory requires additional attention [55]. B cell types, especially MZB and follicular B (FBC) cells were reported altered in SARS-CoV-2 infection, suggesting efficient interplay of B cells between innate and adaptive immune cells [56]. In comparison between SARS-CoV-2 infected and healthy people, naïve B cell count frequencies were found similar, irrespective of the diseased condition [57]. SARS-CoV-2 patients exhibit a reduction of CXC receptor (CXCR)5 expression in major B cell subsets, suggesting a hallmark of disease [58, 59]. However, the changes in the B cell subsets were not associated with co-infection, immune suppression, or treatment with steroids or other clinical features (Figure 2).
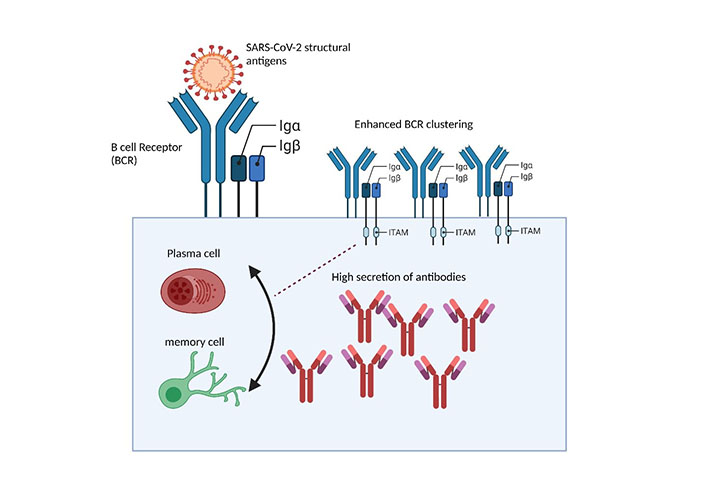
BCRs surface interaction with SARS-CoV-2 structural glycoprotein antigens triggers humoral immune response. Surface interaction of SARS-CoV-2 structural antigens with BCR initiates the aggregation of BCRs into BCR microclusters, through the phosphorylation of ITAMs interlinked Igα, Igβ domains which are subsequently carried through underlying enzymes resulting in triggered series of humoral immune responses to combat infection
Concerning COVID-19 pathogenesis, several other significant factors play a contributing role [5]. However, considering the immune system during SARS-CoV-2 infection, an intricate decisive step in the process of SARS-CoV-2 induced pathogenesis is cellular receptor recognition [59]. S protein having RBD and S2 on viral envelop, recognize angiotensin-converting enzyme (ACE)2 ligand on bronchioles epithelial cells [60]. This recognition process is arbitrated by transmembrane serine protease 2 (TMPRSS2) [61], disintegrin metalloproteinase 17 (ADAM17) [62], membrane proteins (vimentin and clathrin), and angiotensin II type 1a receptor (AT1a R). All these facilitate the coalition of the virus envelope following the viral RNA entrance into the cytoplasm of the target cells [63]. Upon viral entry into the host cell, this mechanism induces autophagy and eventually hypoxia which may lead to SARS-CoV-2 induced pneumonia acute respiratory distress syndrome (ARDS), and even death [64]. Additionally, COVID-19 could target and infect various cells having ACE2+ receptors [65]. Many studies isolate the SARS-CoV-2 from the 67% of patients who develop diarrhea during the infection, indicating that the disease may spread to other organs (heart, kidney, and small intestine) with high ACE2 expression [66]. However, ACE2+ receptors on other cells like intestine enterocytes directly correlated with age [58], indicating that children have less susceptibility to infection than old-age persons [67].
A recent study confirms that SARS-CoV-2 can use host cell novel receptor CD147 to gain entry into the host cells besides ACE2 [68, 69]. People having asthmatic and diabetic complications lead to the escalation of CD147 expression levels [70]. SARS-CoV-19 virus RNA behaves as a toxin due to Ag consequences with toll-like receptor (TLR)7 [71], via myeloid differentiation factor 88 (MyD88)-dependent and TIR-domain-containing adapter-inducing interferon-beta (TRIF) dependent pathways [72]. Pro-inflammatory cytokines (IL-1, IL-6, TNF-α) and type I IFN-α/β expression can trigger the cytokine storm phenomena caused by vascular leakage and severe SARS-CoV-2 infection [73, 74]. Corresponding studies showed that genetic inactivation of the TLR4 gene might reduce lung injury in SARS-CoV-2 infected patients [75]. Many studies revealed that TLR4 activation induces the most substantial immune effect in cytokine release [75]. However, SARS-CoV-2 is unlikely to activate TLR4 directly, as TLR4 responds to bacteria and viruses like the dengue virus [non-structural protein (NSP) 1].
To deeply investigate the mechanism of SARS-CoV-2 mediated pathogenesis, NSPs appear necessary for RNA transcription, translation, and viral replication in SARS-CoV-2 infection [76]. The structural dogma of SARS-CoV-2 reveals the intricate involvement of NSPs to ameliorate lung pathology. Mechanistically, NSPs proteins trigger lung pathology through triggered replication and evasive immune mechanisms [76]. SARS-CoV-2 genome is supplemented with NSP3 and NSP5 regions having proteolysis activity [papain-like protease (PLpro), 3C-like protease (3CLpro)] which helps the cleavage of polyproteins into NSPs 1–16 [5]. NSPs are encoded through Orf1a/Orf1ab particles [76]. The viral attachment and host response are dependent on the numbers of NSPs (NSP3, NSP5, NSP8, NSP9, NSP10) [76]. NSP3, having 1945 amino acids, is the prime NSPs in the SARS-CoV-2 genome and subsumes many functional domains like MHC-I and MHC-II T cell and B cell activation [61]. Besides, that could be a safe target for vaccine development [77]. NSP3 and NSP9-SARS-CoV-2 exhibit a high sequence similarity and a higher degree of functional conservation with SARS-CoV-NSP3 and NSP9 regions [78]. NSP9 plays a significant role in the SARS-CoV-2 infection trajectory [79]. The NSP9 intervenes in viral replication, virulence, and viral genomic RNA multiplication [79]. The NSPs proteins can target therapeutic agents due to their precise biological function and vibrant enzyme active sites. The cocktail vaccine containing a structural protein (SP) and an NSP would stimulate effective complementary immune responses [80, 81].
Anti-SARS-CoV-2 specific Abs and fraction analysis of Abs seroconversion (IgM, IgG, IgE) were investigated among infected patients, suggesting different disease severity outcomes [44]. Structural features of SARS-CoV-2 specific Abs and detailed insights for binding with high-affinity functional epitopes are fundamental to achieving better therapeutic efficacy [10]. Potential illustrations regarding the characterization of structural and neutralizing parameters of SARS-CoV-2 specific Abs have provided detailed insights to better decipher the mechanism of Ab-mediated pathogenies during different stages of development [82]. Healthy individuals reveal the persistence of fucosylated IgG [83], while non-fucosylation has been observed during pregnancy, increasing the risk of disease severity [84]. Considering structural parameters of Abs, several experimental inculcations have developed anti-SARS-CoV-2 Abs, harboring the stringent potential to devastate infected cells through Ab-dependent cellular cytotoxicity (ADCC) mechanism [85]. Persistence of fucosylated Abs in serum of severely ill patients target glycoprotein (S) of SARS-COV-2 in contrast to mild and asymptomatic patients, suggesting efficient potential to neutralize the infection [10].
Overall, magnitude, efficacy, and quantity consideration of Ab responses are fundamental to achieve better therapeutic efficacy [86]. The potential capability of binding with specific epitopes triggers the neutralization response [10]. For example, the crystallizable fragment (Fc) mediated region in the case of influenza virus is required to achieve neutralization [87, 88], while in terms of SARS-CoV, NAbs blocked hACE2 [6, 89]. Following SARS-CoV-2 infection, collaborative immune mechanism amplified through Abs, complement proteins, bridge APCs (macrophages, dendritic cells) trigger immune cascade activation resulting in clearance of pathogens [56, 90]. Interestingly Ab-dependent enhancement (ADE) phenomenon emerged during specific frames of anti-pathogen-associated molecular patterns (PAMPs), lead to the devastation of the immune response [10, 91]. However, in-depth investigations illustrating how the ADE phenomenon could provide deep insights to target specific molecules to prevent autoimmune pathologies is still matter of considerations. The persistence of Fc receptors (FcRs) on immune cells (monocytes and macrophages) triggers immunopathology during dengue virus infection [92]. In terms of SARS-CoV and ACE2, endosomal pH and protease independent pre-existing FcRs interactions facilitate virus entry mechanism [93].
Pre-existing investigations have identified several factors (concentration, specificity, affinity, and isotype switching) stringent to check interplay, whether anti-SARS-CoV-2 Abs are neutralizing, host protective, damaging, and cause ADE or inflammation [10, 94]. Similar speculations have revealed that mice immunized with SARS-CoV-2 vaccines encoding S and N portions produced (anti-N, S) NAbs with the same magnitude. At the same time, re-exposure triggered the onset of anti-SARS-CoV acquired N, proteins Ab response with elevated expression of cytokine and site-specific aggregation of neutrophils leading to the devastation of lungs pathology [95]. The low volume of Ab production in FcRs expression triggers ADE-mediated pathology [48]. Higher Ab concentrations direct anti-SARS-CoV-2 mediated targeting, suggesting Ab magnitude as a potential candidate to trigger ADE-mediated pathologies [48].
Deciphering the dynamics parameters and optimizing NAbs production sequelae of SARS-CoV-2 infection is fundamental for efficient formulations of intricate strategies for developing therapeutic options and vaccines against SARS-CoV-2 coronavirus [96]. Several corresponding investigations have showed that virus magnitude seems triggered during the first week, followed by a gradual temporal decrease during the second week [4]. Enhanced infection load might be crucial to transmit the virus during disease severity [97–99]. Several investigations have reported the significant correlation of SARS-CoV-2 specif Abs with age and sex, exhibiting an increased Abs titers during severe infection in contrast to mild and asymptomatic infections [48, 49]. Additionally, corresponding speculations have reported the persistence of IgG and IgM after ten days of SARS-CoV-2 disease and the onset of seroconversion [100, 101]. Studies conducted by Chen et al. [65] reported the augmented persistence of the hACE2 receptor in older patients in contrast to young people, posing severe threats of carrying an infection.
NAbs milieu induced through passive/active immunization or virus infection is crucial to combat disease [9, 56, 102]. Currently, several monoclonal NAbs, including Ag-binding fragments (Fab), variable region, and single-chain variable fragment (scFv) purified through cutting multidisciplinary approaches against MERS and SARS-CoV-2, are undergoing to treat COVID-19 infection [9]. The specified monoclonal region reveals specificity to bind respective SARS-CoV-2 S and RBD regions to further inhibit S2 mediated entry and attachment mechanisms, thus likely efficient to treat infections [103–105]. NAbs show cross-reactivity against SARS-CoV and MERS as summarized in Table 1. S230.15 and m386 NAbs targeting SARS-CoV showed a cross-neutralizing potential against SARS-CoV-2, interacting RBD surface proteins, and inhibiting binding potential with hACE2. Likewise, the humanized 80R NAbs have revealed the functional capability to disrupt the RBD-hACE2 mechanism [106–107]. However, its anti-SARS-CoV-2 potential is obscure yet. Experimental investigations using HEK293T cells to analyze the antiviral efficacy of mouse-derived NAbs against SARS-CoV viruses highlight the powerful role of blocking RBD-hACE2 interactions [92, 108]. Currently, polyclonal Abs from CP of infected patients are considered comprehensive therapeutic modalities for treating patients suffering from SARS-CoV-2 infection [10–12]. Despite the emergence of NAbs, there is a desperate need for technologies to develop functional monoclonal Abs and determinants to treat SARS-CoV-2 patients directly [94].
Considering phylogenetic and genome homology features of SARS-CoV-2 with SARS-CoV and MERS, researchers are trying to figure out the cross-neutralizing potential of SARS-CoV Abs against SARS-CoV-2 [109–111]. The CR3022, humanized monoclonal Ab harbor neutralization efficacy against SARS-CoV, has been evaluated to show cross-neutralization against SARS-CoV-2, with stringent potential to inhibit RBD-hAEC2 binding interactions [50, 112] (Table 1). S1 specific serum NAbs from recovered animals and humans have shown cross-neutralizing potential against SARS-CoV [91], showing the intricate capability to mediate S protein-specific entry mechanisms. Moreover, it’s imperative to apply SARS-CoV specific NAbs against the current SARS-CoV-2 infection wave after robust efficacy and trial studies [113]. In the future, these cross-reactive applications could help to design specific monoclonal Abs (MoAbs) with potential efficacy to bind SARS-CoV-2 specific PAMPs regions (Table 1) [10].
Overview of Abs undergoing to treat SARS-CoV-2
| Ab | Isolating tropism | Potential efficacy (µg/mL) | Binding affinity (nmol/L, Kd) | Potential binding domain | In vivo activity | Cross-reactivity against SARS-CoV | References |
|---|---|---|---|---|---|---|---|
| CR3022 | Humanized | 0.114 | 6.3–115 | RBD | N/A | Present | [4] |
| CV-30 | Humanized | 0.003 | 3.6 | RBD | N/A | absent | [114, 115] |
| COVA1-12 | Humanized | 1.4 | 2.7 | RBD | N/A | absent | [10, 116] |
| CB6 | Humanized | 0.026 | 2.49 | RBD | N/A | absent | [117] |
| CCL12.1 | Humanized | 0.0222 | 5.83 | RBD | N/A | absent | [10] |
| 4A8 | Humanized | 0.61 | 91.7 | RBD | NTD | absent | [9, 103] |
List of Abs undergoing to treat SARS-CoV-2, revealing potential isolating source tropism, binding specificity, affinity, and cross-reactivity against SARS-CoV strain
We summarized, the intricate subunits’ during SARS-CoV-2 infection and the potential possible parameters of Abs undergoing treating COVID-19 patients. Furthermore, we illustrate a brief overview of adaptive immune cell (B and T) interventions and immunopathology during COVID-19. There are so many Abs mediated parameters against SARS-CoV-2, but we can not list them all. Thus, anti-SARS-CoV-2 parameters should be paid explicit attention in the future to identify the epitope targets for the developments of monoclonal NAbs against SARS-CoV-2. Likewise, it is significant to investigate the role of BCR signaling and meticulous interventions of BCR interlinked signaling subunit response during BCR signaling in results to SARS-CoV-2 Ags. These studies could provide detailed insights for developing therapeutic entanglements against a cohort of pathogens and viral diseases.
Abs: antibodies
ACE: angiotensin-converting enzyme
ADE: antibody-dependent enhancement
Ag: antigen
APCs: antigen-presenting cells
B cells: B lymphocytes
BCRs: B cell receptors
CCL: C-C chemokine ligand
COVID-19: coronavirus disease-2019
CP: convalescent plasma
CXCL: C-X-C motif ligand
FcRs: crystallizable fragment receptors
hACE2: human angiotensin-converting enzyme-2
Ig: immunoglobulin
IL: interleukins
ITAM: immunoreceptor tyrosine-based activation motif
MERS: Middle East respiratory syndrome
MHC: major histocompatibility complex class
N: nucleocapsid
NAbs: neutralizing antibodies
NSP: non-structural protein
RBD: receptor-binding domain
S: spike
SARS-CoV: severe acute respiratory syndrome coronavirus
TH: T helper
TLR: toll-like receptor
TNF: tumor necrosis factor
The author is thankful to Tsinghua University, School of Life Science (SLS) for providing all kind of facilities to prepare the manuscript.
KH and RS are the main authors of this manuscript who designed the manuscript and content outlines; MUA, MSIK, and ZUR provide technical assistance and revise the manuscript critically; ZF and OSA provide critical overview and support in figure drafting. SB carefully overviewed and revised the manuscript.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2021.
Copyright: © The Author(s) 2021. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 4891
Download: 75
Times Cited: 0
Eduardo Tosta
Krupanidhi Sreerama
Eknath D. Ahire, Sanjay J. Kshirsagar
Kenneth Lundstrom
Sevilay Hintistan, Hatice Demirağ
Mandeep Garg ... Suruchi Garg
Swapan K. Chatterjee ... Maria Nilda M. Munoz
Marileia Chaves Andrade ... Hellen Oliveira Rosa
Devlina Ghosh ... Abhishek Saxena
Yoanna Slabakova ... Tsvetelina Velikova
Balram Ji Omar ... Manju O. Pai
Basista Rabina Sharma, P. Veeranna Ravindra
