Affiliation:
Department of Microbiology, Immunology and Genetics, Faculty of Health Sciences, Ben Gurion University of the Negev, Beer Sheva 84105, Israel
Email: noah@bgu.ac.il
ORCID: https://orcid.org/0000-0002-1412-0957
Explor Immunol. 2023;3:217–232 DOI: https://doi.org/10.37349/ei.2023.00099
Received: January 13, 2023 Accepted: March 29, 2023 Published: June 30, 2023
Academic Editor: Buyong Ma, Shanghai Jiaotong University, China
The article belongs to the special issue The Role of Adaptor Proteins in Lymphoid Cell Signaling
The Src homology 2 (SH2) and SH3 domain-containing chicken tumor virus number 10 (CT10) regulator of kinase (Crk) adaptor proteins include three cellular members that serve as integral constituents of multiple receptor-linked signal transduction pathways. CrkI and CrkII are products of alternative RNA-splicing which is transcribed from a single gene, while Crk-like (CrkL), which is highly homologous to CrkII, is encoded by a different gene. Thanks to their modular structure, the Crk adaptor proteins can simultaneously interact with activated receptors and a wide range of effector molecules, and orchestrate the assembly of complexes containing enzymes and substrates at the receptor site. They are involved in the regulation of a large number of cellular processes which control cell growth, differentiation, transformation, and apoptosis. Cell activation-dependent tyrosine phosphorylation of CrkII and CrkL serves as a major posttranslational modification mechanism that introduces conformational changes in the proteins by promoting an intramolecular interaction between the phosphotyrosine and the self SH2 domain. The resulting conformational change induces downregulation of CrkII- and CrkL-dependent biological processes. A second type of posttranslational modification mechanism regulates the structure and function of the CrkII adaptor protein by immunophilin-mediated protein isomerization. Two of the most abundant immunophilins in T lymphocytes which function as peptidyl-prolyl cis-trans isomerases (PPIases), namely cyclophilin A (CypA) and FK506-binding proteins (FKBPs), can associate with CrkII and catalyze its reciprocal cis-trans isomerization. This mechanism is of special importance for the regulation of T lymphocyte functions and for T cell-mediated immune responses, since immunophilin inhibitors, such as cyclosporin A (CsA) and FK506, function as immunosuppressive drugs that can prevent allotransplanted graft rejection. The present manuscript focuses on selected functions of Crk adaptor proteins, predominantly in T lymphocytes, and reviews in more detail the current knowledge on the immunophilin-dependent regulation of the structure and function of the CrkII adaptor protein.
The mammalian chicken tumor virus number 10 (CT10) regulator of kinase (Crk) adaptor proteins include three members representing the cellular counterparts of the viral oncogene, v-Crk, which was originally isolated from the avian sarcoma virus CT10 (Figure 1). The protein product of the oncogene was found to be capable of stimulating tyrosine phosphorylation of signaling proteins in chicken fibroblasts, but was missing a catalytic kinase domain and was therefore termed “CT10 regulator of kinase”, and abbreviated as Crk [1, 2].
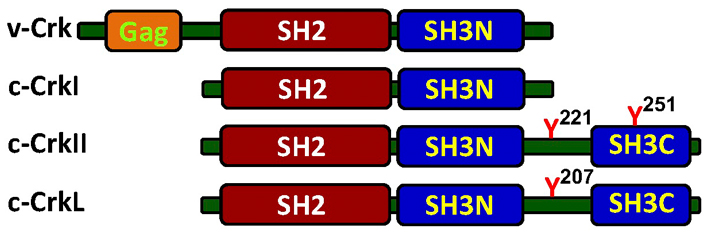
Schematic structure of the Crk adaptor proteins. The cellular Crk adaptor proteins possess a single Src homology 2 (SH2) domain and one or two SH3 domains, while the viral protein, v-Crk, possesses an additional group-specific antigen (Gag) at the N-terminus. The Crk-SH2 domain binding site has a preference towards tyrosine-phosphorylated proteins containing a consensus motif of pY-x-x-P. This motif is found in a number of signaling proteins, including zeta-chain associated protein kinase 70kDa (ZAP70), phosphatidylinositol 3-kinase (PI3K), Casitas B-lineage lymphoma (Cbl), and Crk-associated substrate lymphocyte type (CasL), all of which undergo tyrosine phosphorylation in a cell activation-dependent manner. While CrkI and CrkII are alternative RNA-spliced variants that are transcribed from a single gene on human chromosome 17, band p13 [3], Crk-like (CrkL) is transcribed from chromosome 22, band q11 [4]. A single copy of the phospho-Y-x-x-P motif is also found in the linker region between the two SH3 domains in CrkII and CrkL. Phosphorylation of the tyrosine residues in this motif, namely Y221 and Y207 in the human CrkII and CrkL, respectively, can promote an intramolecular association with the Crk-SH2 domain, which alters the protein folding, blocks the SH2 domain and inhibits CrkII/CrkL association with SH2 binding partners. SH3N: N-terminal SH3; SH3C: C-terminal SH3
Since their discovery, considerable efforts have been invested in order to define the biological roles of the Crk adaptor proteins and clarify their input in signal delivery from various cell surface receptors. Additional studies attempted to identify proteins that constitutively or inducibly associate with individual Crk adaptor proteins and learn about the function of these binding partners in different cellular processes. Furthermore, experiments were also designed to identify the enzymes and clarify the mechanisms that regulate the conformation and function of Crk adaptor proteins by posttranslational modification.
The ubiquitously expressed Crk adaptor proteins have a modular domain architecture consisting of SH2 and SH3 domains, through which they interact with various binding partners and contribute to the regulation of multiple networks of signal transduction pathways. By recruiting enzymes and substrates into the vicinity of activated cell surface receptors, Crk proteins are capable of promoting the activation of effector molecules which are essential for the propagation of signals downstream of the activated receptor. Indeed, Crk proteins have been implicated in cellular processes that regulate cell adhesion, proliferation, differentiation, transformation, and death.
The human CrkI and CrkII proteins are derived from a single gene located on chromosome 17, band p13 [3]. They are products of two alternative transcripts encoding different isoforms with distinct biological activities. In contrast, the human CrkL protein, which exhibits 57% identity with the human CrkII, is a product of a distinct gene which is located on chromosome 22, band q11 [4].
The Crk and CrkL genes are highly conserved through evolution, as indicated by the high degree of identity of nucleic acid sequences of the human and mouse CrkII (99%) and CrkL (97%). Due to the structural similarity between CrkII and CrkL, the SH2 and SH3 domains of the two proteins share many binding partners, and the two proteins show functional similarities and overlapping roles in different biological systems. However, CrkII and CrkL exhibit some specific non-redundant essential functions, as was demonstrated in CrkII and CrkL gene-knockout models in mice. In both of these models, selective knockout of either the Crk or the CrkL gene resulted in multiple developmental defects that led to early death in utero or shortly after birth. The fact that mice in each of the knockout systems suffered from developmental defects and early death suggests that each of the two proteins regulates essential functions that cannot be compensated by the other gene [5–7]. This assumption is supported by the observation that deficiency of either Crk or CrkL does not alter the steady-state level of expression of the other protein.
Due to the Crk adaptor proteins’ modular structure, their critical functions in various signal transduction pathways and their involvement in oncogenic processes, they stand as potential candidates for drug targeting. Indeed, cell-penetrating Crk-SH3 binding peptides were shown to disrupt Crk-dependent signaling pathways and inhibit the growth of chronic myeloid leukemia (CML) cells [8, 9], in which the breakpoint cluster region-Abelson protein tyrosine kinase (Bcr-Abl) onco-protein constitutively phosphorylates CrkL and amplifies CrkL signals that promote cell growth. In addition, a cyclophilin A (CypA) inhibitor which targets CrkII was found to inhibit cell migration and reduce metastasis formation in an in vivo orthotopic model of breast cancer [10]. Future development of highly specific, high affinity cell-penetrating peptides that modulate the structure of CrkII and block Crk-SH2 and SH3 domain interactions with selected binding partners are likely to provide means for therapeutic intervention in various Crk-dependent hyperproliferative diseases.
All three Crk adaptor proteins are expressed in T lymphocytes where they form multimolecular complexes with a wide range of molecules [11]. The Crk-SH3-domain-binding guanine-nucleotide releasing factor (C3G) is the first protein to be identified as a Crk-binding partner [12, 13]. C3G regulates the conversion of Ras-associated protein 1 (RAP1) guanosine triphosphatase (GTPase) to an active guanosine triphosphate (GTP)-bound protein which in turn leads to activation of the lymphocyte function-associated antigen-1 (LFA-1) integrin and increases T cell adhesion [14, 15]. C3G associates with CrkII and CrkL adaptor proteins in T cells [16, 17] and participates in the regulation of lymphocyte intravasation and extravasation, as well as recruitment to sites of inflammation [15, 18].
Another protein that associates with the Crk-SH2 domain is the product of the Cbl protooncogene [19, 20]. Cbl is an E3 ubiquitin-protein ligase that undergoes tyrosine phosphorylation in activated T lymphocytes and posttranslationally modifies proteins by ubiquitination. CrkL- and Cbl-containing complexes were reported in various cell types that were stimulated by different agonists [21–23], as well as in Bcr/Abl-transformed leukemia cells [24].
The CrkII protein was found to constitutively associate with C3G and transiently with Cbl in resting and T cell antigen receptor (TCR)-stimulated T cells, respectively [16, 17, 25]. Stimulation of T cells induces Cbl-mediated ubiquitination of the adjacent C3G protein, which then undergoes proteasomal degradation [25]. The data indicate that CrkII functions as a scaffold that recruits Cbl to the vicinity of C3G in TCR-stimulated T cells, and that the tyrosine phosphorylation and activation of Cbl promote the ubiquitination and degradation of C3G. This CrkII-dependent mechanism appears to play an important role in terminating TCR/CD3-induced activation signals, and thereby regulate the length and intensity of the T cell responses.
Several additional effector molecules were found to interact with CrkII in TCR-stimulated T cells, including the TCRζ chain and ZAP70 protein tyrosine kinase (PTK), although the relevance of these interactions to the regulation of the cell activation process is not fully understood.
It should be mentioned that CrkL, but not CrkI and CrkII, is required for actin polymerization in response of T cells to the integrin ligand, intercellular adhesion molecule (ICAM) [26], and appears to regulate T cell migration to sites of inflammation.
The Crk adaptor proteins possess a modular structure that enables them to simultaneously interact with two or more binding partners and form multiprotein complexes that convey signals from activated cell-surface receptors. They can directly bind an activated receptor and link it to effector molecules that are required for signal delivery from the occupied receptor to the nucleus or to other subcellular compartments.
The SH2 domains are structurally conserved modular components of a wide range of functionally distinct proteins that mediate interactions with specific phosphotyrosyl peptides and participate in signal initiation and propagation downstream of a variety of receptors.
The 118 amino acid-long Crk-SH2 domain is located in the amino-terminus of all members of the Crk family of adaptor proteins (Figure 1), and recognizes specific phosphotyrosine residues in the context of a p-tyrosine (Tyr)-X-X-Pro/Leu motif [27, 28]. The conserved residues within its ligands form direct contact with the SH2 binding surface, while the specificity and affinity of interaction is determined by contextual information, provided by the electric charge and the structure of the residues that extend outside of the canonical binding region.
The Crk-SH2 domain is highly versatile and can transiently interact with multiple tyrosine phosphorylated proteins in TCR-stimulated T cells. Some of the known Crk-SH2 binding partners in T cells include the TCRζ chain [29], the ZAP70 PTK [30, 31], Cbl [19, 20], the p85 protein, which serves as the regulatory subunit of the PI3K [32], and the CasL [33].
The Crk adaptor proteins possess a highly conserved SH3 domain, C-terminal to the SH2 domain, which recognizes polyproline type II (PPII) motifs in the context of Pro-X-X-Pro-X- [lysine (Lys)/arginine (Arg)] [34]. The PPII motif is characterized by a left‐handed helix which makes a full turn every three residues [35]. In contrast to the Crk-SH2-induced transient interactions, Crk-SH3N mediates constitutive interactions with its binding partners in a cell activation-independent manner. In T lymphocytes, the Crk SH3N domain mediates constitutive interaction with the C3G protein and the resulting complex plays an important role in integrin-mediated cell adhesion and migration [16].
Many studies that intended to identify Crk-SH3N domain-binding partners were initially performed using fibroblasts and epithelial cells or cell-free in vitro methods. These studies revealed a list of Crk-SH3N domain-binding partners which participate in the regulation of cell proliferation, adhesion and migration, and include the Abl [36, 37], Abl-related gene (Arg) [38], Jun N-terminal kinase 1 (JNK1) [39] and hematopoietic progenitor kinase 1 (HPK1) [40], as well as the son-of-sevenless (SOS) Ras/Rac guanine nucleotide exchange factor 1 (SOS1) [41], and the DOCK180 (dedicator of cytokinesis) [42] guanine nucleotide exchange factors.
A flexible stretch of 48 amino acids that connects the SH3N and SH3C domains in CrkII and CrkL, termed linker region, plays an important role in regulation of the CrkII/CrkL protein conformation and function. Both Crk proteins are subject to Abl-mediated phosphorylation of a critical, linker region-residing tyrosine residue (Tyr221 and Tyr207 in human CrkII and CrkL, respectively) [36], which can auto-inhibit CrkII/CrkL by promoting an intramolecular interaction with the SH2N domain. In addition, the linker region in CrkII, but not in CrkL, possesses an isomerase-binding motif which enables the induction of conformational changes in CrkII by isomerases, such as CypA and FK506-binding proteins (FKBPs) [17]. The regulatory functions of this motif which determine the accessibility of the CrkII SH2 and SH3N domains to their binding partners [43–46] will be discussed in detail in one of the following chapters.
The CrkI protein is a shorter splice-variant of CrkII which lacks the linker region and the SH3C domain (see Figures 1 and 2). As a result, CrkI is not subject to regulation by tyrosine kinases or isomerases and retains a constitutive single extended structure in which the SH2 and SH3N domains are accessible for interaction with their binding partners.
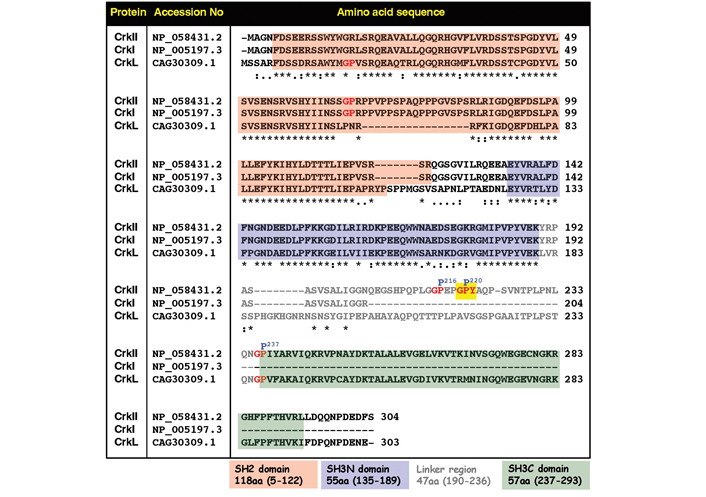
Comparative analysis of amino acid sequences and positions of functional domains and selected regulatory sites in the human CrkII, CrkI, and CrkL proteins. Amino acid sequences which include the SH2 domain (pink background), the SH3N domain (blue background), and the SH3C domain (green background) are indicated. Residues within the linker region are in grey color. The CypA recognition motif possessing the Gly-Pro220-Tyr motif is indicated by red letters on a yellow background. Two other Gly-Pro motifs within the linker region are indicated by red letters. “*” in this figure indicates residues in that column are identical in all sequences; “:” in this figure indicates conserved substitutions; “.” in this figure indicates semi-conserved substitutions
A C-terminal domain in both CrkII and CrkL, termed SH3C, possesses an amino acid sequence and core structural characteristics of a classical SH3 domain, but exhibits a very low affinity to PPII motifs [47–49]. In contrast to the classical SH3 domains, which possess three aromatic residues in the PPII binding pocket, the CrkII- and CrkL-SH3C possess three hydrophobic residues at the putative binding pocket which drastically reduce its affinity to polyproline rich motifs [49]. However, the CrkII-SH3C domain appears to function as an autoregulatory element which, under certain conditions, can stabilize CrkII in a closed conformation that blocks the protein’s biological functions [43, 50]. Due to the lack of SH3C domain in CrkI, this protein is not subject to a negative regulation and is, therefore, more effective than CrkII and CrkL in upregulating signaling cascades that promote cell transformation [51, 52].
T lymphocytes undergo activation following TCR engagement with cognate peptide-bound major histocompatibility complex (MHC) receptors that are displayed on the surface of antigen presenting cells (APC). TCR engagement results in activation of TCR-linked tyrosine kinases which phosphorylate specific effector molecules at the plasma membrane and, with the help of adaptor proteins, create signaling complexes that transduce the signals to various cell compartments, including the nucleus.
One of the early activation events in T lymphocytes is the phosphorylation of CrkII on one or more tyrosine residues [32]. Earlier in vitro studies demonstrated that CrkII undergoes phosphorylation on Tyr221 within the middle of the linker region. Phosphorylation of Tyr221, which is mediated predominantly by the Abl PTK [36, 37], leads to autoinhibition of CrkII due to an intramolecular interaction between phospho-Tyr221 and the self SH2 domain [36, 53]. The resulting conformational change interferes with the SH2 and SH3N domain binding to their endogenous ligands and induces disassembly of CrkII protein complexes, leading to inhibition of CrkII-dependent downstream signaling events [37, 53]. Regulation of CrkII-dependent signal transduction pathways by phosphorylation of Tyr221 was also reported following ligand binding to various receptor tyrosine kinases, including the tropomyosin receptor kinase A (TrkA) [54], the epidermal growth factor receptor (EGFR) [55], and the platelet-derived growth factor receptor (PDGFR) [56].
While the precise biological role of phosphorylation of CrkII Tyr221 is not completely understood, it is assumed that receptor engagement promotes rapid CrkII-SH2-mediated interactions with tyrosine-phosphorylated effector molecules that initiate signaling cascades, and that this process is followed by phosphorylation of CrkII Tyr221 and results in disassembly of the CrkII protein complexes, leading to downregulation of the Crk-dependent signaling pathways [57]. This course of action enables the transient activation of receptor-ligand interaction-induced signal transduction pathway, and a successive signal termination, via a CrkII Tyr221 phosphorylation-dependent mechanism.
A second site of cell stimulation-dependent Abl-mediated phosphorylation of CrkII was reported within the SH3C domain, on Tyr251 [58]. The phospho-Tyr251 residue was found to exhibit high affinity towards a subset of SH2 domains, including Abl-SH2. The resulting interaction with phospho-Tyr251 further propagated the activation signal due to transactivation of Abl [59, 60]. Support for this hypothesis was obtained by replacing CrkII Tyr251 by phenylalanine (Phe), a non-phosphorable mimetic residue, which abrogated Abl transactivation by CrkII [58]. The current findings suggest that tyrosine phosphorylation of CrkII can provide both positive and negative regulatory signals for the activation of the Abl PTK and its downstream signaling events.
The Crk proteins undergo posttranslational modifications that determine their conformation and ability to interact with distinct binding partners. A pioneering study by Sarkar et al. [43] revealed a previously unknown mechanism of regulation of CrkII, which is mediated by peptidyl-prolyl cis-trans isomerases (PPIases). The authors reported that the CrkII linker region is susceptible to conformational transitions from cis to trans, and vice versa, which give rise to two isomers with an overall distinct conformation. While the trans isomer acquires an open conformation in which the SH3N domain is free to interact with other molecules, folding of the cis isomer forms a closed conformation in which the two SH3 domains are in close proximity and the SH3N domain binding pocket is partially obstructed by the neighboring SH3C domain. Using nuclear magnetic resonance (NMR) spectroscopy, the authors studied a truncated chicken CrkII which included the SH3N-linker-SH3C regions (aa135-297), or a shorter version of this peptide, in which the SH3N was excluded. They found that the glycine 237 (Gly237)-proline 238 (Pro238) peptide bond within the linker region (that corresponds to Gly236-Pro237 in the mouse and human CrkII) can undergo cis-trans isomerization and that, in solution, the predominant isomer acquires the cis conformation (~90%). However, addition of an SH3 ligand to the solution increased the proportions of the trans isomers, suggesting that occupation of the SH3N domain by its ligand slows down the rate of the trans-to-cis isomerization of CrkII. Furthermore, Sarkar et al. [43] demonstrated that the cis-trans isomerization of the Gly237-Pro238 peptide bond in chicken CrkII is drastically accelerated in the presence of the PPIase CypA, suggesting that CrkII might serve as a physiological target for CypA [43].
A following study by Sarkar et al. [45] provided additional high resolution data elucidating the structure of the cis and trans conformers of Pro238 chicken CrkII, and demonstrated how interaction between the linker region and the SH3C domain is modulated by Pro isomerization. Further studies suggested that the effect of CypA on chicken CrkII-Pro238 might lead also to the formation of an intermediate state which is dependent on the global organization of the specific domains [61].
Members of the two major groups of immunophilins, which include the cyclophilins and FKBPs, were extensively studied in T lymphocytes, predominantly due to their ability to interact with the potent immunosuppressive drugs, cyclosporin A (CsA) and FK506, respectively [62, 63]. The two structurally distinct complexes of cyclophilin-CsA and FKBP-FK506 exhibit high affinity towards the Ca2+/calmodulin-dependent protein phosphatase calcineurin and, following a physical interaction, inhibit the Ca2+-dependent dephosphorylation of the transcription factor nuclear factor of activated T cell (NFAT) [64, 65]. As a result, nuclear translocation of the phosphorylated NFAT is prohibited and T cell activation and proliferation is repressed due to lack of cytokine transcription. CsA and FK506 are therefore widely used in allotransplanted patients in order to prevent immune-mediated graft rejection.
The ability of CypA and FKBP to interact with human CrkII was demonstrated using glutathione S-transferases (GST)-CrkII fusion protein in a pull-down assay and coimmunoprecipitation studies [17]. Additional results obtained using human Jurkat T cells alluded to the possibility that in vivo binding of CypA and FKBP to CrkII may also affect CrkII-dependent cellular functions. In these studies, the ability of T cell-derived human CrkII-SH3N to interact with C3G was found to be CypA-dependent, since preincubation of CrkII with an enzymatically active recombinant CypA significantly increased C3G binding to CrkII [17]. Furthermore, the CypA-mediated increased interaction between CrkII and C3G was completely abolished by inclusion of CypA inhibitors in the assay system, suggesting that the observed effect of CypA on CrkII-C3G interaction was dependent on the catalytic activity of CypA. These findings led to the hypothesis that, during the preincubation reaction, CypA modulated the conformation of CrkII, and altered the cis/trans steady-state balance of CrkII towards the formation of excess trans conformers that can interact with C3G.
Additional studies in human T lymphocytes provided further support for the in vivo regulatory effects of immunophilins on CrkII. In these studies, a comparison between the expression levels of CrkI and CrkII in Jurkat T cells revealed that CrkII is expressed at ~5-fold higher levels compared to CrkI [46]. In contrast, binding of C3G to CrkI was ~5-fold more efficient than C3G binding to CrkII. Despite the fact that CrkI is fully identical to the first 204 amino acids in the CrkII N-terminal (see Figure 2), these observations are not surprising, since CrkI misses the phosphorylation site at Tyr221, cannot form intramolecular interactions, and, therefore, adopts an extended “open” conformation in which both SH2 and SH3N domains are accessible for interaction with binding partners, including C3G, which bind to the SH3N domain. In contrast, the majority of CrkII proteins (~90% [45]) adopts a cis conformation and a “closed” compact structure in which the SH3N domain is not accessible for interaction with ligands, such as C3G. Furthermore, the addition of CsA and FK506 to the cell culture had no effect on the extent of CrkI interaction with C3G, but further reduced CrkII-binding efficiency of C3G, apparently by blocking the immunophilin-dependent cis-to-trans isomerization of CrkII, which result in higher proportions of CrkII cis conformers which could not interact with C3G [46].
Additional supporting data for the assumption that CypA can isomerize CrkII were obtained using fluorescence resonance energy transfer (FRET)-based in vivo studies in T cells that had been transiently transfected with PicchuX, a modified version of the chimeric plasmid phosphorylation indicator of Crk chimeric unit (Picchu), which includes a carboxy terminal CAAX box derived from the Ki-Ras sequence [66]. The CAAX box (where C is cysteine, A is an aliphatic amino acid, and X is one of several different amino acids) represents a consensus prenylation sequence which restricts protein localization predominantly to the plasma membrane. The PicchuX plasmid encodes human CrkII1-236 with fluorescent tags at its amino- and carboxy-terminals, including a cyan fluorescent protein (CFP) and a yellow fluorescent protein (YFP), respectively. It can, therefore, be utilized as a FRET biosensor. PicchuX was originally designed for monitoring Abl-mediated and CrkII-dependent signal transduction pathways [66, 67]. Monitoring of FRET emission by PicchuX-expressing cells demonstrates increased FRET values when conformational changes in human CrkII occur, due to Abl-mediated phosphorylation of Tyr221 which forms an intramolecular interaction with the self SH2 domain.
Transient expression of PicchuX in human Jurkat T cells followed by confocal microscopy of resting cells revealed a basal high level of FRET emission [17, 46]. Treatment of the cells with CsA and FK506 induced a significant increase in FRET emission values, suggesting that the drugs inhibited the activity of the cellular immunophilins and shifted the cis/trans steady-state balance towards the formation of excess cis conformers. The above results were reconfirmed using an independent analysis on a fluorescence-activated cell sorter (FACS) [17].
Additional studies using the PicchuX-transfected T cells revealed that the effect of immunophilin inhibitors on the FRET emission is concentration and time dependent [46]. Increased FRET was observed within 1 h treatment of the cells with CsA and FK506 and maximal emission was observed at ~8 h post-addition of the drugs [17].
Previous studies demonstrated that CypA binds linear peptides that possess a Gly-Pro motif [68, 69]. It catalyzes the cis-trans isomerization of peptide bonds between the Pro residue and the preceding amino acid [70].
A search of the chicken CrkII linker region amino acid sequence revealed three potential binding motifs for CypA, namely GP217, GP221 and GP238 (see Figure 3), and structural and functional analyses indicated that CypA-mediated isomerization of chicken CrkII occurs at the GP238 motif [45, 43]. While SH3N-ligand interaction can be blocked in both chicken and human CrkII by PPIase-mediated isomerization, structural studies indicated that the two proteins utilize different strategies for autoinhibition of their SH3N domain [45]. In addition, the observations showing that human CrkII is sensitive to isomerization by CypA and FK506 were obtained using a shorter version of human CrkII (1–236) [17, 46] which lacks the isomerization motif (Pro238) that was identified in chicken CrkII [17, 46]. A comparative amino acid sequence analysis of CrkII from multiple species revealed that the chicken CrkII isomerization epitope, which consists of GP238F, is unique among all species analyzed, including other avians (Figure 4). The equivalent epitope, which is completely conserved in all higher organisms analyzed, including mammalians, avians (except for the chicken), reptiles, and amphibians, includes isoleucine (Ile) instead of Phe at position 239.
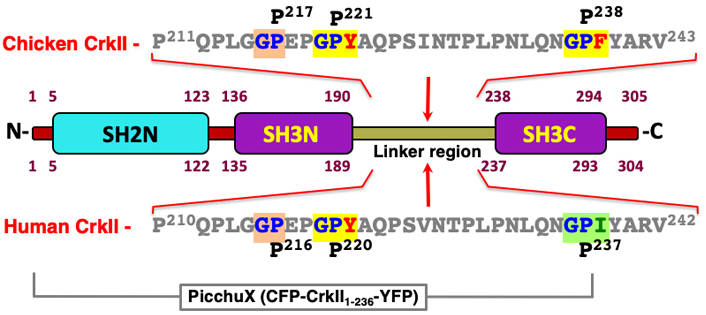
The structure of the CrkII protein and comparison between the CypA-specific recognition and isomerization sites in the human and chicken CrkII. The human and chicken CrkII are highly homologous proteins that possess an N-terminal SH2 domain and two SH3 domains (SH3N and SH3C). A 47 amino acid long region which links the two SH3 domains includes also a site of isomerization by the cyclosporin A (CsA) PPIase. Amino acid sequences of the human and chicken CrkII linker regions are shown, where Gly-Pro sequences representing potential recognition sites for CypA are highlighted [68]. In vitro studies of a truncated protein possessing the chicken CrkII carboxy-terminus revealed that the Gly-Pro238-Phe motif is a site of isomerization by CypA [43]. However, the human CrkII homologous sequence that includes the Gly236-Pro-Ile appeared to be insensitive to the PPIase [52, 71]. The difference between the human and chicken CrkII sensitivity to CypA was attributed to the nature of the third residue in this motif, Ile and Phe, in the human and chicken CrkII, respectively. In vivo studies demonstrated that the full-length human CrkII, as well as a truncated CrkII which includes the first 236 residues are sensitive to isomerization by CypA [17]. On the other hand, the alternative spliced variant of CrkII, termed CrkI, which is identical in its sequence to the first 204 residues of CrkII, is insensitive to CypA [17]. Thus, the PPIase isomerization site in the human CrkII resides in the linker region between amino acids 205-236. This region possesses a Gly-Pro220-Tyr motif, which appears to be the preferred CypA binding and isomerization site. This motif is fully conserved among evolutionary distant taxa, including all mammalian, avian, reptile and amphibian species tested (see Figure 2). In addition, the third residue in this motif (Tyr221) is highly similar to the third residue found in the chicken isomerization motif (Phe239), since both are structurally related aromatic residues with a hydrophobic side chain that can be interchangeable in proteins with no effect on their biological activities. A second Gly-Pro motif in the human CrkII linker region includes a structurally non-related negatively charged glutamic acid (E) residue at the third position which is likely to exhibit very low affinity to CypA
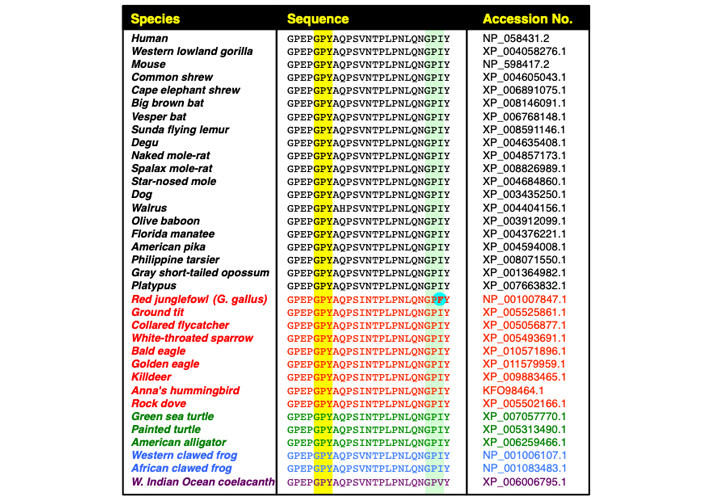
Evolutionary conservation of the immunophilin-binding motif, GP220Y, in the CrkII linker region. The figure includes sequence alignment of the C-terminal half of the CrkII linker region, corresponding to residues 215-239 in the human CrkII protein derived from 35 organisms, including mammalians (black), avians (red), reptiles (green), amphibians (blue) and fish (purple). Alignment was performed using Clustal Omega Multiple Sequence Alignment program (https://www.ebi.ac.uk/Tools/msa/clustalo/). The conserved GP220Y motif is highlighted (above the yellow column). A second motif within the chicken CrkII, which was reported to serve as a substrate for immunophilins (GP238F), and the aligned sequences in other organisms are also highlighted (above the light green column). The phenylalanine residue within the chicken CrkII GP238F motif (in bold letters above a light blue background), which was reported to serve as a site of isomerization by CypA [43], is found only in the red junglefowl [Gallus (G.) gallus], the ancestor of the domestic chicken
A comparative amino acid sequence analysis of the three members of the Crk family of proteins demonstrates that the Gly-Pro-Tyr motif is found in the linker region of CrkII, but not in CrkI or CrkL, providing an explanation for the lack of sensitivity of CrkI and CrkL to regulation by PPIases (Figure 2).
Sequence alignment of the N-terminal and C-terminal portions of the CrkII linker region demonstrated that a high degree of sequence conservation is found predominantly in the C-terminal portion of the linker region (Figure 5).
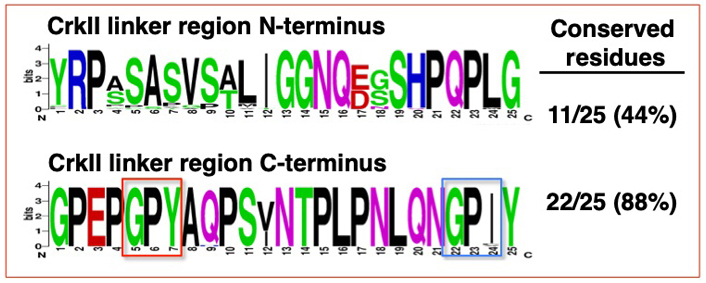
Sequence alignment of the N-terminal and C-terminal portions of the CrkII linker region. Sequence alignment was performed on the N-terminal and C-terminal half of the CrkII linker region, corresponding to residues 190-214 and 215-239 in the human CrkII protein, respectively. The alignment was performed on 35 organisms (see Figure 4), using WebLogo (http://weblogo.berkeley.edu/logo.cgi). The conserved GP220Y motif in the C-terminus is marked (red frame). A second motif within the chicken CrkII, which was reported to serve as a substrate for immunophilins (G237PF), and the aligned sequences in the 35 organisms is also marked (blue frame)
The properties of the specific amino acid at position 239 were found to be directly related to the distinct folding characteristics of the chicken and the human CrkII [72]. Thus, reciprocal point mutations revealed that Phe239 replacement by Ile stabilized the chicken CrkII and decreased its ability to bind C3G, while substitution of Ile239 by Phe in the human CrkII had an opposite effect; it destabilized the protein and increased its C3G binding ability [72].
Analysis of a molecular surface model of the human CrkII (Figure 6) further demonstrated that the GP220Y sequence, which serves as a CypA binding motif in human CrkII, is found on an outer surface area highly accessible for interaction with CsA. In contrast, the GP238I sequence, which is homologous to the CypA-binding site in the chicken CrkII, appears to be buried within the human CrkII protein and is, therefore, less accessible for interaction with CypA.
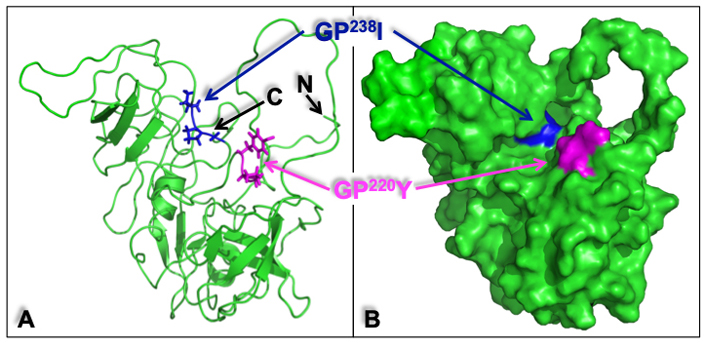
Structure of the human CrkII represented in a ribbon diagram and molecular surface model. The ribbon diagram (A) and the molecular surface model (B) of human CrkII [protein data bank identification (PDB ID): 2EYZ] provide a structural interpretation of some of the key experimental observations and demonstrate the potential positions of the CrkII-binding sites for CypA. The position of GP220Y, the CypA binding motif in human CrkII (in magenta), and the position of GP237I (in blue), which corresponds to the CypA-binding motif in the chicken CrkII (GP238F), are indicated. The GP220Y motif is located on an accessible surface area and can easily support the interaction with CsA. In contrast, the GP238I motif appears to be buried within the protein and is, therefore, less accessible for interaction with other proteins. The amino (N)- and carboxy (C)-termini of the protein are also indicated. Structural alignment and visualization were performed using PyMOL
The above studies indicate that human CrkII is sensitive to isomerases and that in contrast to chicken CrkII, which undergoes isomerization at the GP238F motif, human CrkII possesses a distinct isomerization site. Human CrkI possesses an extended conformation which is insensitive to isomerases and is identical to the N-terminal 204 residues of CrkII. In contrast, the PicchuX-encoded human CrkII includes the amino-terminal 236 residues and is sensitive to isomerases. Thus, the isomerization site in human CrkII occurs at a short segment within the linker region, between amino acids 205 and 236. A search of this region showed that human CrkII possesses two copies of a Gly-Pro motif, namely GP216E and GP220Y.
The GP220Y-containing motif in human CrkII is highly similar to the GP238F-containing motif in chicken CrkII, since Tyr (Y) and Phe (F) are similar aromatic residues and Phe replacement by Tyr represents a conservative substitution [73]. Phe and Tyr can be interchangeable in a variety of proteins with no significant effect on the protein’s structure or function. In contrast, the Gly residue in the GP216E-containing motif in chicken CrkII is a non-aromatic negatively charged acidic residue which represents a non-conserved replacement of Phe. Mutating the PicchuX-Tyr221 to Ala, using site-directed mutagenesis, revoked the sensitivity of PicchuX to CsA and FK506, further indicating that the GP220Y-containing motif in human CrkII is the site of isomerization (unpublished data). Amino acid sequence comparison of CrkII from 35 different species demonstrate that the human CrkII GP220Y motif is fully conserved throughout the evolution and that together with CypA, is found in cells of most vertebrate species. Thus, the conformation-dependent mode of regulation of CrkII by CypA appears to be a wide-spread mechanism which is likely to regulate signal transduction from a large number of cell surface receptors.
Despite their relatively small size and lack of ability to catalyze biochemical reactions, the ubiquitously expressed Crk adaptor proteins play extremely important roles in transducing signals from a large variety of receptors. In T lymphocytes, Crk adaptor proteins were found to interact with the antigen receptor ζ chain and the antigen receptor-linked signal transducing ZAP70 PTK, which are critical for the induction of T cell-mediated immune responses. In addition, the Crk adaptor proteins are essential for the regulation of T cell adhesion and recruitment to sites of inflammation. The involvement of the Crk adaptor proteins in a myriad of regulatory mechanisms and biological processes is made possible by several different factors. First, the modular structure of Crk enables it to associate via its SH2 and SH3 domains with many different binding partners and form distinct protein complexes. The repertoire of Crk binding partners differs from one cell type to another, in accordance with the distinct cell type-specific proteins. In addition, differential expression of the three individual Crk proteins in different cell types at distinct developmental stages, and the non-redundant functions of the three Crk proteins, further increase the flexibility and variety of Crk-regulated mechanisms.
Finally, cell activation-induced posttranslational modifications of Crk and of its binding partners provide additional opportunities for Crk-dependent assembly and disassembly of qualitatively and quantitatively different complexes at distinct subcellular compartments. While some of the mechanisms that regulate Crk function by posttranslational modifications were identified and partially characterized, additional studies are required in order to understand the full spectrum of mechanisms that regulate the conformation and function of Crk under different cell activation conditions and in distinct cell types. Deciphering the mode of regulation of Crk adaptor proteins and revealing the mechanisms by which Crk regulates different cellular process are extremely important. Such information will undoubtedly lead to the identification of new molecules and mechanisms that can be targeted in order to intervene with physiological and pathological signal transduction pathways that regulate distinct cell functions, as well as cell growth, differentiation and transformation.
Abl: Abelson protein tyrosine kinase
C3G: chicken tumor virus number 10 (CT10) regulator of kinase-Src homology 3-domain-binding guanine-nucleotide releasing factor
Cbl: Casitas B-lineage lymphoma
Crk: chicken tumor virus number 10 (CT10) regulator of kinase
CrkL: Crk-like
CsA: cyclosporin A
CypA: cyclophilin A
FKBP: FK506-binding protein
FRET: fluorescence resonance energy transfer
Gly237: glycine 237
Ile: isoleucine
Phe: phenylalanine
Picchu: phosphorylation indicator of Crk chimeric unit
PPIase: peptidyl-prolyl cis-trans isomerase
PPII: polyproline type II
PTK: protein tyrosine kinase
SH2: Src homology 2
SH3C: C-terminal SH3
SH3N: N-terminal SH3
TCR: T cell antigen receptor
Tyr: tyrosine
ZAP70: zeta-chain associated protein kinase 70kDa
We thank Ms. Margalit Krup for technical assistance and Drs. Michiyuki Matsuda and Carmen Guerrero for the gifts of reagents.
NI: Conceptualization, Writing—original draft, Writing—review & editing.
The author declares that he has no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
This work was funded in part by the
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 2457
Download: 20
Times Cited: 0
Tadashi Matsuda ... Kenji Oritani
Mari Hikosaka Kuniishi ... Takanori So
Laura E. McMillan, Christoph Wülfing
Shulamit Katzav
Xiaoyu Jiang, Izidore S. Lossos
