Affiliation:
1Division of Hematology, Department of Medicine, University of Miami Miller School of Medicine, Sylvester Comprehensive Cancer Center, Miami, FL 33136, USA
ORCID: https://orcid.org/0000-0002-3517-9843
Affiliation:
1Division of Hematology, Department of Medicine, University of Miami Miller School of Medicine, Sylvester Comprehensive Cancer Center, Miami, FL 33136, USA
2Department of Molecular and Cellular Pharmacology, University of Miami, Miami, FL 33146, USA
Email: ilossos@med.miami.edu
ORCID: https://orcid.org/0000-0002-9346-9013
Explor Immunol. 2023;3:186–206 DOI: https://doi.org/10.37349/ei.2023.00097
Received: October 30, 2022 Accepted: January 10, 2023 Published: June 28, 2023
Academic Editor: Noah Isakov, Ben Gurion University of the Negev, Israel
The article belongs to the special issue The Role of Adaptor Proteins in Lymphoid Cell Signaling
Human germinal center (GC)-associated lymphoma (HGAL) is a multi-domain adaptor protein expressed in GC B lymphocytes, T follicular helper (Tfh) cells and lymphomas derived from these cells. HGAL expression is an independent predictor of longer survival of diffuse large B-cell lymphoma (DLBCL) and classical Hodgkin’s lymphoma (HL) patients. HGAL regulates B cell receptor (BCR) signaling and immunological synapse formation by binding to either the downstream effectors [e.g., spleen tyrosine kinase (Syk)] or other signaling regulators [e.g., growth factor receptor-bound protein 2 (Grb2)]. HGAL regulates the cytoskeleton that reshapes B cell morphology during BCR signaling and cell motility by at least two molecular mechanisms: enhanced Ras homolog gene family member A (RhoA) signaling and inhibition of myosin-actin translocation. These effects on the cytoskeleton decrease lymphoma dissemination in animal models and contribute to decreased lymphoma dissemination in patients. The latter may contribute to the association of HGAL protein expression with longer survival of patients with DLBCL and HL tumors. The ability to regulate multiple and distinct functions simultaneously in B cells implies that the HGAL protein level is tightly regulated. It was demonstrated that HGAL can be regulated by PR/SET domain 1 (PRDM1)/B lymphocyte-induced maturation protein-1 (BLIMP1) and interleukin-4 (IL-4) at the transcription level, by microRNA-155 (miR-155) at the post-transcriptional level, and by F-box protein 10 (FBXO10) at the post-translational level. Constitutive enforced expression of HGAL at physiological levels leads to lymphoid hyperplasia and DLBCL in mice. Future studies need to focus on identifying HGAL interactome, dissecting its interaction network, and understanding HGAL spatiotemporal signaling in live cells in physiological conditions. Further, the recent demonstration of HGAL expression in Tfh cells requires the determination of its function in these cells. These studies will contribute to new insights into the biology of these cellular subsets and how immune dysregulation contributes to lymphomagenesis.
In recent years, it has become increasingly evident that the functions of many proteins can only be fully understood in the context of protein interaction networks. The description of such networks provides the keys to our understanding of disease processes [1–3]. Adaptor proteins play key roles in regulating and integrating physiological responses [4–6], such as cell migration [7–9], division [10–12], and differentiation [13–15]. Adaptor proteins contain a variety of protein-binding modules facilitating protein interactions [16–18]. The sequence of binding motifs of adaptor proteins, as well as their subcellular localization and the proximity of binding partners, determine their signaling specificity [19–21].
For effective humoral immunity, mature B cells must respond to foreign antigens and generate antigen-specific effector cells [22]. The germinal center (GC) is an important site for the generation and selection of B cells bearing high-affinity antibodies. A comprehensive understanding of the regulation of B-cell receptor (BCR) signaling and antigen processing is crucial for vaccine development and therapeutic intervention for immunodeficiency and autoimmune disease [23–26]. The American Society of Hematology proposed a “Roadmap for Discovery and Translation in Lymphoma” to define molecules necessary for GC response and to identify key protein-protein interactions and post-translational modifications that regulate BCR signaling [27].
One of the proteins expressed specifically in GC B [28–30] and T follicular helper (Tfh) [31] cells is human GC-associated lymphoma (HGAL), also known as GC B-cell expressed-expressed transcript 2 (GCET2) [32], or GC associated signaling and motility (GCSAM) [33]. HGAL was cloned by us following its identification during gene expression profiling studies of lymphoma tissues and purified normal B cell subsets [28, 29, 34]. HGAL is an adaptor protein expressed in GC-derived B cell lymphomas such as a subset of diffuse large B-cell lymphoma (DLBCL) [29] and classic Hodgkin lymphoma (cHL) [35, 36], and its expression is associated with improved survival and thus may serve as a prognostic biomarker [34–36].
The HGAL gene is located on chromosome 3q13 and encodes a 178-amino acid (aa) protein with 51% identity and 62% similarity to the murine GC-specific protein M17 [37]. It was demonstrated that in M17 knockout mice, this protein is dispensable for GC formation, class-switch recombination, immunoglobulin somatic hypermutation, and mounting of T-cell-dependent antibody responses [37]. However, in contrast to their wild-type littermates, M17 deficient mice exhibited reduced-sized Peyer’s patches [37]. Tightly regulated expression of the HGAL and M17 proteins, restricted to B lymphocytes and Tfh cells in the GC compartment, and their multiple binding motifs including an immunoreceptor tyrosine-based activation motif (ITAM) [38–40], implicated in signal transduction in B and T lymphocytes, suggest they have specific signaling functions.
Initial studies demonstrated that HGAL is specifically expressed in normal GC lymphocytes and GC-derived lymphomas [29, 34–36]. This restricted HGAL expression points to the existence of specific and tightly controlled mechanisms regulating its expression. It was have shown that HGAL expression is increased following B-cell stimulation with interleukin-4 (IL-4) and IL-13 [34, 36], which are secreted by cells in the GC microenvironment, and is down-regulated by BCR and CD40 stimulations [28, 41, 42] (Figure 1) controlling GC B cell activation and differentiation to later B cell ontogeny stages [28]. Concordantly, it was shown that PR/SET domain 1 (PRDM1)/B lymphocyte-induced maturation protein-1 (BLIMP1) that is functioning as a master regulator of terminal B cell differentiation into plasma cells and mediates termination of GC reaction, transcriptionally downregulates HGAL messenger RNA (mRNA), and consequently protein expressions [43]. PRDM1 is a direct DNA-binding transcription repressor recognizing the sequence 5’-MAGYGAAAYK-3’ [44, 45]. A bioinformatic search of the HGAL promoter revealed two potential homologies to this sequence located at positions –1,608 and –1,383 upstream of the transcription initiation site. Chromatin immunoprecipitation (ChIP) and reporter assays using wild-type and mutated constructs confirmed direct repression of HGAL expression by PRDM1 [43]. This effect may mediate the loss of HGAL expression upon terminal differentiation of GCBs to plasma cells and may contribute to the absence of HGAL expression in post-GC lymphoid tumors. Further, PRDM1 which is frequently inactivated in DLBCL tumors by mutations and deletions was shown to function as a tumor suppressor [46, 47]. Thus, it is possible that the PRDM1 tumor suppressor effect is at least partially mediated by repression of HGAL, which is highly expressed in a subset of DLBCL and plays a role in the pathogenesis of this malignancy (see below “Conditional expression of HGAL leads to the development of DLBCL in mice”).
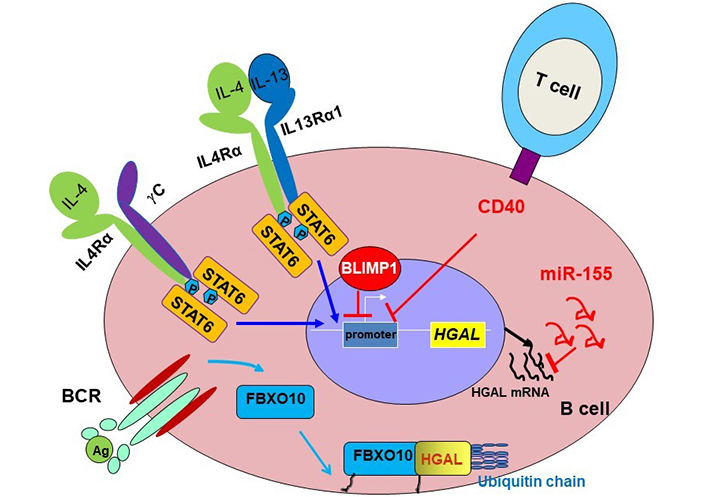
Schematic diagram of HGAL expression regulation. HGAL protein is tightly regulated at multiple levels. It was demonstrated that HGAL expression is increased following B-cell stimulation with IL-4 and IL-13 [34, 36], which are secreted by cells in the GC microenvironment, and is down-regulated by BCR [48] and CD40 stimulations controlling GCB activation and differentiation to later B cell ontogeny stages [28]. PRDM1/BLIMP1 transcriptionally downregulates HGAL mRNA and consequently HGAL protein expression [43]. MicroRNA-155 (miR-155), implicated in the control of GC reaction and lymphomagenesis, also directly down-regulates HGAL expression by binding to its 3’-untranslated region [49]. BCR stimulation induces rapid and reversible palmitoylation of the stem cell factor-F-box protein 10 (SCF-FBXO10) ubiquitin E3 ligase, resulting in F-box protein 10 (FBXO10) relocation to the cell membrane, where it binds and targets HGAL protein for ubiquitylation and subsequent proteasomal degradation [48]. IL4Rα: IL-4 receptor-alpha; STAT6: signal transducer and activator of transcription 6; γC: gamma chain; P: phosphorylation; Ag: antigen
miR-155, implicated in the control of GC reaction [50–52] and lymphomagenesis [53, 54], also directly down-regulates HGAL expression by binding to its 3’-untranslated region [49]. Since miR-155 is more commonly expressed in non-GC DLBCL, it may also contribute to the loss of HGAL expression in these tumors.
HGAL expression is also regulated at the post-translational level [48]. BCR stimulation induces rapid and reversible palmitoylation of the SCF-FBXO10 ubiquitin E3 ligase, leading to FBXO10 relocation to the cell membrane, where it targets HGAL protein for ubiquitylation and degradation [48]. FBXO10 recognition and degradation of HGAL is phosphorylation independent and relies on an evolutionarily conserved aa residue (H91) of HGAL and FBXO10 re-localization to the cytoplasmic membrane [48].
More recently it was demonstrated that HGAL is also expressed in normal Tfh cells within the GC and lymphomas of Tfh derivation [31]. These studies demonstrated that HGAL is a reliable marker of the Tfh phenotype, showing comparable and sometimes even more robust staining compared with that of currently used Tfh markers, and can be used for routine diagnostic workup of these tumors [31]. However, mechanisms controlling HGAL expression in these cells and tumors are unknown and will need to be elucidated in future studies.
HGAL is a relatively small adaptor protein harboring specific domains/motifs facilitating interactions with other proteins or subjected to post-translational modifications resulting in altered HGAL functions (Figure 2).
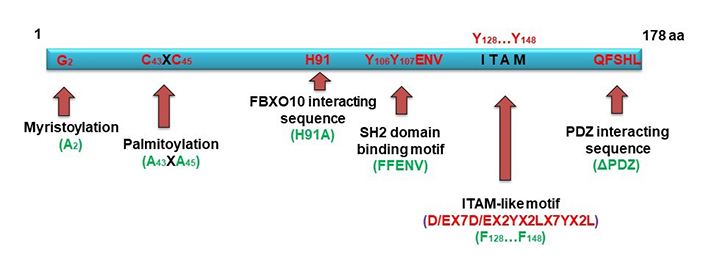
Schematic diagram of HGAL structural features (in red) and its mutations (in green). HGAL is a multi-domains adaptor protein. N-terminal lipid myristoylation and palmitoylation by covalent attachment of a myristoyl moiety to an N-terminal glycine residue and of a palmitoyl moiety to cysteine (S-palmitoylation), respectively, lead HGAL to the inner membrane and facilitate delivery of extracellular cues into the cells [55]. C-terminal post-synaptic density-95, disks-large and zonula occludens-1 (PDZ) domain interacting sequence can directly interact with Ras homolog gene family member A (RhoA)-guanine nucleotide exchange factor (GEF; such as RhoPDZ-GEF and LARG) and enhances RhoA signaling [56]. FBXO10 recognition and degradation of HGAL is phosphorylation independent and relies on a single evolutionarily conserved HGAL amino acid residue (H91) and FBXO10 re-localization to the cytoplasmic membrane [48]. Src homology domain 2 (SH2) domain binding motif and ITAM-like motif are subjected to post-translational phosphorylation [55, 57, 58], thus mediating specific protein interactions [55, 57, 58]. Mutations used to interrogate HGAL functions are shown in green
The major structural characteristic of HGAL is the presence of an ITAM with the consensus sequence YxxI/Lx6-12YxxI/L. Phosphorylated ITAM motifs, found in many receptors or adaptor proteins in the immune system [39, 40], serve as docking sites for tandem SH2 domains of spleen tyrosine kinase (Syk) family kinases and are involved in cell activation [59, 60]. It was indeed demonstrated that HGAL directly binds to Syk, enhancing BCR signaling [57]. HGAL harbors YYENV, which when phosphorylated can also bind the SH2 domain contained within the Src oncoproteins and many adaptor proteins participating in signal transduction [e.g., growth factor receptor-bound protein 2 (Grb2)] [61, 62]. Among its multiple functions, Grb2 also controls lymphoid follicle organization and GC reaction [63] and was shown by us and others to bind to HGAL via it’s phosphorylated YYENV motif [58, 64]. We showed that this interaction also regulates BCR signaling and can be eliminated by HGAL (FFENV) mutation [58].
Another motif located in the HGAL C-terminus is a PDZ interacting sequence: PDZ is an acronym combining the first letters of the first three proteins discovered to share the same structural domain—post synaptic density protein 95 (PSD95) [65, 66], Drosophila disc large tumor suppressor 1 (Dlg1) [66], and zonula occludens-1 (ZO-1) protein [66]. Proteins containing PDZ domains play a key role in anchoring receptor proteins in the membrane to cytoskeletal components by organizing signaling complexes. We demonstrated that the PDZ interacting sequencing at the C-terminal of HGAL mediates interaction with PDZ-Rho guanine nucleotide exchange factor (RhoGEF), regulating RhoA activity [56]. RhoA is a member of the Rho family of guanosine triphosphate hydrolases (GTPases)—a family of small (~21 kDa) signaling G-proteins, playing a key role in regulating cytoskeletal dynamics in a wide variety of morphogenetic events, such as cell migration and vesicle trafficking [67]. This interaction implicated HGAL function in motility and cytoskeleton regulation.
HGAL is a cytoplasmic protein that can be anchored to cell membranes via post-translational modifications—myristoylation and palmitoylation [55]. Myristoylation is a protein modification by the covalent attachment of a myristoyl moiety to an N-terminal glycine residue, which allows for protein-lipid interactions. Myristoylation plays an essential role in membrane targeting and functions in a variety of signal transduction pathways [68]. Palmitoylation is a protein modification by covalent attachment of a palmitoyl moiety to cysteine (S-palmitoylation) residues, usually in their C-termini. Palmitoylation enhances the hydrophobicity of proteins, contributes to their membrane association, and plays a significant role in the subcellular trafficking of proteins between membrane compartments [69]. Using [3H]myristic acid and [3H]palmitic acid labeling, we showed that HGAL was myristoylated and palmitoylated. Mutagenesis confirmed that G2 at the N-terminal of HGAL is the myristoylation site, and C43XC45 is the palmitoylation site [55]. Lipid modifications lead HGAL to the cell membrane. Elimination of either myristoylation or palmitoylation alone results in partial loss of membrane location; however, elimination of both abolishes the HGAL membrane localization, restricting it to the cytosol [55].
Based on these structural characteristics, it is not surprising that HGAL interacts with multiple proteins and simultaneously regulates diverse biological activities.
B cells are an integral part of the adaptive immune response [22, 26, 70]. They participate in multiple immunological processes including affinity maturation of antibodies, antigen presentation, immunological memory, and regulatory cytokine production [22, 26, 70]. B cells express surface BCR that is pivotal for their function and survival [71, 72]. The BCR is composed of membrane immunoglobulin (mIg) molecules that are associated with immunoglobulin alpha (Igα)/Igβ (also named CD79a/CD79b) heterodimers, facilitating intracellular signaling [73, 74]. Upon binding of an antigen, there is an aggregation of BCRs in lipid rafts leading to rapid activation of multiple intracellular kinases, including Src family kinases such as Lck/Yes-related novel protein (Lyn), the Syk kinase, and the tyrosine kinase expressed in hepatocellular carcinoma (Tec) family kinase Bruton’s tyrosine kinase (Btk) that initiate and propagate multiple signaling cascades regulating intracellular Ca2+, gene expression, cell metabolism, and cytoskeleton organization [73, 74]. The complexity and fine tuning of BCR signaling permit a wide variety of biological outcomes, including cell survival and proliferation or apoptosis, immune activation or tolerance (anergy), and differentiation into antibody-producing cells or memory B cells [70, 73–76]. This variability is at least partially achieved by tight regulation of magnitude and duration of BCR signaling via the involvement of proteins enhancing BCR biochemical signaling (e.g., CD19 complex facilitating recruitment of protein kinases), limiting activation by negative feedback loops [e.g., Lyn/CD22/SH2-containing tyrosine phosphatase-1 (SHP-1)/SH2-containing inositol 5’-phosphatase-1 (SHIP-1) activation] or regulation of BCR internalization [73, 74]. The nature of the antigens also defines the activation outcome [73, 74, 77]. Although B cells can strongly respond to soluble antigens, the predominant form of antigen in vivo is bound to the surface of antigen presenting cells (APC) [77]. Binding to membrane-bound antigens leads to the formation of an immunological synapse (IS), also known as the supermolecular activation cluster (SMAC) [77]. This structure is composed of concentric rings: central SMAC (cSMAC) and peripheral SMAC (pSMAC), each containing segregated clusters of distinct proteins. cSMAC includes BCR and protein tyrosine kinases but excludes tyrosine phosphatases CD45, and SHP-1. pSMAC is comprised of actin and adhesion molecules such as lymphocyte function-associated antigen-1 (LFA-1), and very late activation antigen-4 (VLA-4), which can provide stability and support to the synapse [77]. The synapse serves as a platform for BCR signaling transduction and antigen gathering for internalization, both of which are essential for full B cell activation and differentiation into antibody producing effector cells or memory cells [77].
HGAL markedly increases ligand-induced and mildly increases tonic BCR signaling [55]. HGAL regulates and enhances BCR signaling by several mechanisms controlling both BCR-induced biochemical signaling and BCR clustering in signalosomes [55, 57, 58]. The enhanced BCR-induced biochemical signaling is achieved via direct HGAL binding to Syk that enhances its kinase activity [55, 57]. This function of HGAL is crucially dependent on myristoylation, palmitoylation, and lipid raft localization, as determined by HGAL colocalization with Lyn in cells fractionated following a detergent-free method [55]. In resting B cells, a proportion of HGAL resides in lipid rafts together with the Lyn kinase while BCR, for the most part, is located outside rafts [55] (Figure 3A). Syk may associate with HGAL in these rafts, but such an association is likely nonproductive in terms of Syk kinase activity [55]. These changes during BCR engagement, leading to BCR recruitment to lipid rafts and Lyn-mediated phosphorylation of ITAMs within HGAL and CD79a/CD79b. The latter attracts and activates Syk, which, through association with phosphorylated HGAL acquires intensified kinase activity leading to enhanced induction of the BCR signaling pathway [55]. Activated Syk then phosphorylates HGAL that moves out of the lipid raft/BCR signalosome. BCR stimulation also induces rapid and reversible SCF-FBXO10 ubiquitin E3 ligase palmitoylation resulting in relocalization to the cell membrane, where it targets HGAL for ubiquitylation and proteasomal degradation in the cytoplasm. In vivo, FBXO10 binding to HGAL is dependent on its membrane relocalization following BCR-induced palmitoylation, recognition of a single evolutionarily conserved HGAL aa residue (H91), and is independent of HGAL phosphorylation (Figure 3B). Using HGAL myristoylation and palmitoylation mutants we showed that cytosolic HGAL does not associate with Syk, nor does it enhance BCR signaling. Whether HGAL acyl modifications are lost in the process of cytoplasmic relocalization and proteasomal degradation is unknown. While S-palmitoylation is a dynamic and reversible modification process, N-myristoylation is irreversible [78]. However, myristoylated proteins bind transiently to the membrane owing to their weak hydrophobic nature with a half-life in the order of minutes, which contrasts with hours for palmitoylated or myristoylated and palmitoylated proteins [79]. Overall, these findings demonstrate the importance of temporal-spatial HGAL localization in lipid rafts for the regulation of BCR signaling [55].
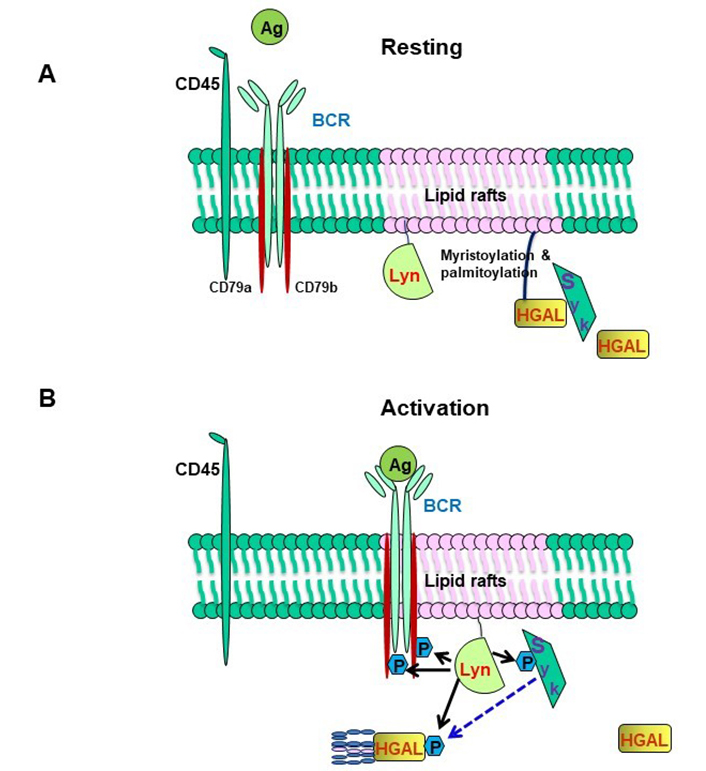
HGAL within lipid rafts enhances BCR signaling. (A) In resting B cells, HGAL is mainly located within lipid rafts together with Lyn; (B) in BCR-stimulated B cells, Lyn phosphorylates tyrosine residues within the ITAMs of CD79a/b, and this attracts and activates Syk. Coassociation of Syk with HGAL enhances Syk kinase activity and BCR signaling strength [55]. BCR stimulation also induces FBXO10 palmitoylation, relocation to the cell membrane leading to HGAL ubiquitylation (shown in Figure 1). Following BCR engagement, HGAL is shunted from lipid rafts to ultimately end up in the cell cytoplasm for destruction by the proteasome [48]
HGAL also regulates BCR signaling via interaction with Grb2 [58]. Grb2 is a ubiquitously expressed adaptor protein containing 2 flanking SH3 domains and a central SH2 domain preferentially binding to the pYXN motif [80, 81]. Grb2 negatively regulates BCR signaling [82]. Grb2 localizes to BCR microclusters and decreases BCR-induced Ca2+ influx [82]. Upon BCR activation and HGAL phosphorylation, yielding a pYEN sequence that binds to the SH2 domain of Grb2, HGAL negates Grb2 inhibitory effects on BCR-induced intracellular signaling [58]. Using multiple Grb2 and HGAL mutants it was demonstrated that Grb2 and HGAL adaptor proteins directly modulate each other’s effects on BCR-induced intracellular signaling [58]. However, Grb2 does not impact HGAL-enhanced Syk kinase activity following BCR stimulation [58].
In addition to the Grb2 effects on the BCR-induced intracellular signaling, it is also involved in B-cell synapse formation by enhancing the movement of BCR microclusters to gather antigens in the cSMAC [58]. It was previously demonstrated, that upon BCR stimulation, Grb2 initially localizes to microsignalosomes, indirectly recruits dynein, and subsequently moves together with antigen to gather in the cSMAC [83]. Using B cell lymphoma cell lines it was shown that BCR activation leads to Grb2 colocalization with BCR and HGAL in cSMACs [58]. The HGAL and Grb2 interaction is not required for Grb2’s localization to the cSMAC [58] but enhances the rate of cSMAC formation and increases BCR accumulation in the cSMAC. Concomitant knockout of both Grb2 and HGAL prevents cSMAC formation to a larger extent than individual knockout of these proteins [58].
cSMAC formation is also dependent on Syk [84]. Both Grb2 and HGAL interact with Syk suggesting that these 3 proteins may be part of the same multi-protein complex regulating BCR signaling. Grb2 interacts with Syk attenuating its activation by Lyn and leading to decreased BCR biochemical signaling while promoting cSMAC formation (Figure 4, left) [84]. HGAL directly binds to Syk, increasing its kinase activity and leading to enhanced BCR biochemical signaling and accelerating cSMAC formation by increasing BCR accumulation in cSMACs (Figure 4, right) [58]. Grb2-HGAL interaction counteracts each other effects on BCR biochemical signaling while cooperating in accelerated and larger cSMAC formation (Figure 4, middle) [58]. These interactions may play an important role in regulating the magnitude and duration of BCR signaling and antigen presentation.
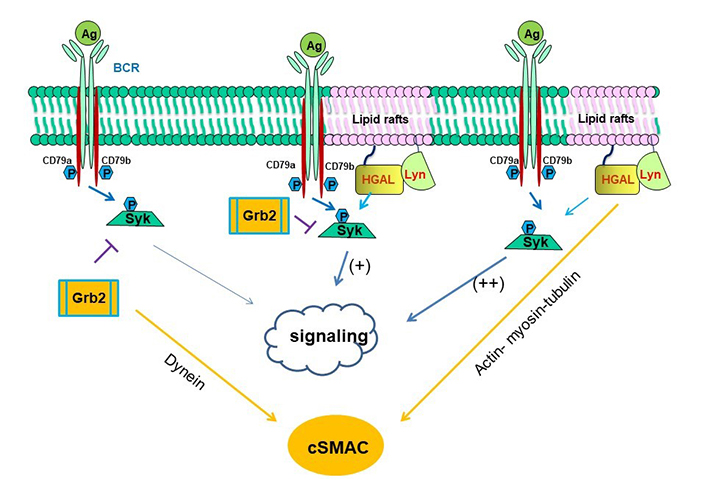
Schematic diagram of biological effects attributed to interaction among HGAL, Syk and Grb2 proteins [58]. On the left side of the figure, it is shown that Grb2 directly interacts with Syk attenuating its activation by Lyn and leading to decreased BCR-induced intracellular signaling, while promoting cSMAC and immunological synapse formation by indirectly recruiting dynein, promoting its own and BCR bound to antigen centripetal movement to the cSMAC. On the right side of the figure, it is shown that HGAL directly binds to Syk, increasing its kinase activity and leading to enhanced BCR signaling. HGAL also contributes to the faster dynamic of cSMAC formation and increases BCR accumulation in the cSMAC by interacting with actin, myosin, and tubulin, which mediate centripetal movement of BCR to c SMAC. In the middle of the figure, it is shown that Grb2-HGAL binding facilitates their interaction with Syk, while distinctively affecting Syk function: attenuated by Grb2 and enhanced by HGAL. Both Grb2 and HGAL facilitate the formation of cSMAC [58, 83]
Lymphocyte motility is critical for various physiological and pathological processes, including immune response and lymphoma dissemination [84–88]. Cytoskeleton proteins play a major role in these processes [89–93]. Cytoskeletal regulatory proteins that have been implicated in lymphocyte migration and may also lead to immune dysfunction syndromes include Wiskott-Aldrich syndrome protein (WASP), WASP-interacting protein (WIP), actin-related protein 2/3 complex subunit 1B (Arpc1B), WD40 repeat protein 1 (Wdr1), hematopoietic protein 1(Hem1), dedicator of cytokinesis 2 (DOCK2), DOCK8, and Rac family small GTPase 2 (Rac2) [94]. However, the identity of intracellular proteins exclusively expressed in normal or malignant lymphocytes that by modulating cytoskeleton activity regulate migration and dissemination of lymphoma is largely unknown. Identifying the functions of these proteins is important since deregulated lymphocyte motility may predispose to lymphoma development by targeting cells to microenvironments that inhibit cellular differentiation and promote malignant transformation [95, 96]. These proteins, by controlling lymphoma dissemination, may also affect lymphoma aggressiveness and patients’ prognosis.
In the normal GC, B-lymphocytes are functionally and spatially segregated from extra-GC compartments [97]. Some of the GC B-lymphocytes demonstrate limited motility between the GC light and dark zones, whereas other B cells are stationary throughout these zones [97]. In contrast to the usually spherical shape of naive and memory B lymphocytes, GC B-cells commonly exhibit irregular contours with prominent asymmetric cytoplasmic processes, resulting in polarized shapes [97]. The mechanisms regulating the motility of GC lymphocytes and lymphoma cells are only partially elucidated [98–102]. It was demonstrated that HGAL, in addition to regulating BCR signaling [55, 57, 58], also decreases spontaneous and stimuli induced [e.g., stromal cell derived factor 1 (SDF-1), IL-6, fibronectin] motility by several molecular mechanisms [56, 103–105].
Lymphocyte motility is a multistep process resulting from coordinated cytoskeletal remodeling mediated by cyclic interactions between the myosin head domains projecting from the myosin filaments and F-actin in the presence of ATP. Hydrolysis of the ATP by myosin releases free energy to produce a mechanical force for directed actin-based movement [106, 107]. HGAL interacts directly and independently with both actin and myosin [103, 104]. HGAL protein interacts with two myosin regions: the head (S1) and tail [104]. Although the tail domain generally mediates interaction with the ‘cargo’ and/or other myosin molecules, the head domain is also involved in binding to F-actin, which is coupled to steps in the hydrolysis of ATP and the transduction of free energy into directed actin-based movement. HGAL is not affecting the actomyosin adenosine triphosphatase (ATPase) activity, although it increases the strength of actin-myosin binding [104]. This leads to the prolongation of the myosin-actin interaction, contributing to a decreased ability of myosin to translocate actin. An HGAL-induced decrease in actin filament sliding velocity, as demonstrated by the in vitro motility assays, supports this idea [104]. Alternatively, but not mutually exclusive, HGAL binding to myosin and/or actin may serve as a ‘load’ that resists actin filament motion, hence leading to the same effects [104]. It is possible to hypothesize that HGAL may function as a molecular tether between the thick myosin-containing filaments and filamentous actin, adjusting the kinetics of acto-myosin interactions and thus cellular motility. The HGAL-mediated bridging properties may be like those imposed by another myosin-binding protein, myosin-binding protein C. It was demonstrated that myosin-binding protein C forms a linkage between the thick and thin filaments of cardiac muscle affecting cardiac contractility [108]. Thus, both HGAL and myosin-binding protein C can bind to two different regions in the myosin molecule and affect actomyosin motility. The findings of no change in actomyosin ATPase activity suggest that HGAL does not affect the number or rate of myosin cross-bridges undergoing a transition from the weakly to strongly bound state and the generation of force. In general, these findings show that HGAL interaction with actin and myosin may contribute to the decreased motility of GC lymphocytes expressing HGAL protein. Further, it was have previously shown that HGAL colocalized with actin in the pSMAC [58]. Recently, Wang et al. [109] demonstrated the formation of an actomyosin arc network in the pSMAC that promotes immunological synapse formation by mechanistically moving antigen microclusters into the cSMAC. The combination of these observations raise a hypothesis that previously reported HGAL-mediated enhancement in the rate of cSMAC formation and increased BCR accumulation in the cSMAC may be mediated by HGAL effects on these actomyosin arc network. This will need to be investigated in future studies.
Rho GTPases are molecular switches transmitting extracellular cues to intracellular signaling pathways to regulate the cytoskeletal dynamics in a wide variety of morphogenetic events, such as cell migration, cytokinesis, vesicle trafficking, and endocytosis [110, 111]. RhoA is one of the key members of the Rho GTPases family. RhoA cycles between active guanosine triphosphate (GTP)-bound and inactive guanosine diphosphate (GDP)-bound states. This cycling is controlled by their interaction with GEFs and GTPase-activating proteins (GAPs). In the activated form, they can bind to numerous effector proteins, leading to the activation a multitude of downstream signaling cascades [112, 113].
HGAL regulates the RhoA signaling pathway in B-cell lymphoma cells and may have a similar role in GC lymphocytes [56]. HGAL enhances RhoA activation, leading to inhibition of lymphocyte motility and chemotaxis by activation of its downstream effector Rho-associated kinase (ROCK), which phosphorylates myosin regulatory light chain (MRLC), myosin phosphopantetheinyl transferase (PPTase) subunit myosin phosphatase target subunit 1 (MYPT1) and cofilin, which mediate cytoskeletal reorganization, as reflected by increased formation of focal adhesions and stress fibers. Increased RhoA activity is contributing to the inhibitory effects of HGAL on lymphocyte motility. It may be attributed to the firmer attachment to the extracellular matrix due to increased focal adhesions formation. Alternatively, HGAL-mediated RhoA-induced phosphorylation of MRLC and myosin II activation may result in cell motility inhibition, since recent studies demonstrated that myosin II ablation leads to increased cell migration while myosin activation decreased cell motility [114–116].
HGAL enhances the activation of RhoA by binding and stimulating the guanidine nucleotide exchange activity of the RhoA-specific RhoGEFs, PDZ-RhoGEF, and LARG, leading to the inhibition of lymphoma cell motility [56]. This binding is mediated by the HGAL C-terminal PDZ binding motif (Figure 2) [56]. Knockdown of PDZ-RhoGEF and LARG increased lymphoma cell chemotaxis, similarly to the reported migration increase of marginal zone B-lymphocytes in p115-RhoGEF−/−mice [117]. It was demonstrated that HGAL activates PDZ-RhoGEF by directly binding to its catalytic Dbl homology (DH)-domain. This binding may result in conformational changes in the PDZ-RhoGEF protein that either enhance the intrinsic guanidine nucleotide exchange activity of the DH-domain or alleviates auto-inhibitory effects of other protein domains [56].
It was further demonstrated that HGAL regulates BCR-mediated cell motility. BCR stimulation in vitro increased RhoA activity leading to decreased both spontaneous and chemotaxis induced DLBCL cell motility. HGAL expression further increased BCR-induced RhoA activation and reduced DLBCL cell motility following BCR stimulation. Further, HGAL knockout ameliorated BCR-induced RhoA activation and BCR-mediated inhibitory effects on DLBCL cell motility [105].
HGAL, as most adaptor proteins, likely interacts with many other unknown proteins involved in the regulation of cell motility. To comprehensively examine the global cellular HGAL interactome, we employed the recently published proximity biotinylation (BioID) method that relies on proximity-dependent protein biotinylation [118, 119]. In this method, an Escherichia coli (E. coli) biotin protein ligase harboring an R118G mutation [referred to escherichia coli biotin ligase* (BirA*)] is fused in frame to the HGAL protein. The fused BirA* mutant catalyzes the formation of activated biotin (biotinoyl-5’-adenosine monophosphate), which can covalently react with the epsilon amine of lysine residues in directly interacting and neighboring proteins and permanently tag them, allowing their purification with streptavidin coupled to mass spectrometric analysis. A total of 96 individual proteins potentially interacting with HGAL-BirA were identified. These included 30 unique proteins (besides HGAL itself) that were considered part of the cytoskeleton, to be interacting with the cytoskeleton, or to be regulating cellular motility (Table 1) [105]. Of note, actin was among these proteins, consistent with our previous observations [103, 104]. Gene set enrichment analysis revealed statistically significant enrichment for proteins belonging to the cytoskeleton, cell junction, cell projection, tumor invasiveness, and microtubule cytoskeleton signatures (Table 2) [105]. The direct interaction between HGAL and tubulin α and β, was confirmed using coimmunoprecipitation and co-sedimentation of recombinant HGAL tagged with thioredoxin (TRX) with isolated microtubules. These findings suggest that HGAL interacts with multiple components of the cytoskeleton [105], thus affecting cellular motility through multiple complementary mechanisms (Figure 5).
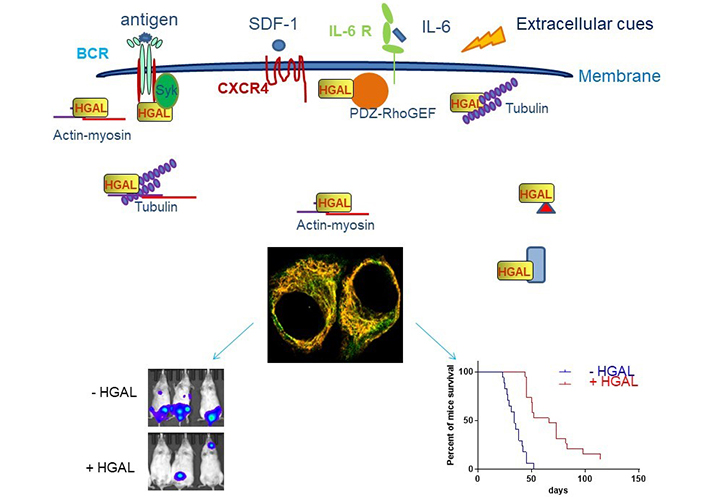
HGAL inhibits cell motility. HGAL inhibits cell motility by directly binding to multiple cytoskeleton proteins [103–105] or interacting with effectors or regulators of distinct signaling pathways which regulate cytoskeleton (actin, myosin, tubulin, and PDZ-RhoGEF are shown as indicated and red triangle and blue rectangle represent other multiple interacting proteins) [56–58]. This allows HGAL to affect multiple cell motility mechanisms in response to distinct extracellular cues, such as antigen [105], integrins [56], chemo-attractants [56, 105], and cytokines [56, 103, 105]. The confocal microscopy image of unstimulated Raji cells stably expressing HGAL-green fluorescent proteins (GFP; green) and stained with Alexa Fluor 555-phalloidin (yellow) demonstrate colocalization of HGAL and actin (orange). In vivo experiments demonstrate that HGAL expression in lymphoma decreases tumor dissemination and prolongs animal survival [105]. Mice were injected intravenously with 5 × 106 luciferase-transduced U2932 human lymphoma cells. D-luciferin was injected intraperitoneally (150 mg/kg body weight) 15 min before acquiring images. Bioluminescence images were taken once per week. Representative bioluminescent images of mice with U2932 Mock and U2932 HGAL lymphoma and Kaplan-Meier plots of mouse survival demonstrated a significant difference between the U2932 Mock and U2932 HGAL mice. CXCR4: C-X-C chemokine receptor type 4; IL-6R: IL-6 receptor
Note. Adapted with permission from “HGAL inhibits lymphoma dissemination by interacting with multiple cytoskeletal proteins,” by Jiang X, Lu X, Gentles AJ, Zhao D, Wander SA, Zhang Y, et al. Modified from Blood Adv. 2021;5:5072–85 (https://doi.org/10.1182/bloodadvances.2021004304). CC BY NC ND.
Cytoskeletal proteins interacting with HGAL
| Protein symbol | Protein name |
|---|---|
| ACTG1 | Actin, cytoplasmic 2 |
| ADD3 | Gamma-adducin |
| AFDN | Afadin |
| ALDH18A1 | Delta-1-pyrroline-5-carboxylate synthase |
| ARHGAP25 | Rho GTPase-activating protein 25 |
| CCT2 | T-complex protein 1 subunit beta |
| CCT7 | T-complex protein 1 subunit eta |
| CLINT1 | Clathrin interactor |
| CRKL | Crk-like protein |
| EHD1 | EH domain-containing protein 1 |
| EPB41L5 | Band 4.1-like protein 5 |
| EPS15 | Epidermal growth factor receptor substrate 15 |
| ERBIN | Erbin |
| EZR | Ezrin |
| GCSAM (HGAL) | GCSAM protein |
| MARK2 | Serine/threonine-protein kinase MARK2 |
| MSN | Moesin |
| PICALM | Phosphatidylinositol-binding clathrin assembly protein |
| PLEKHA5 | Pleckstrin homology domain-containing family A member 5 |
| PLS3 | Plastin-3 |
| PPFIBP1 | Liprin-beta-1 |
| SLC3A2 | 4F2 cell-surface antigen heavy chain |
| SNX1 | Sorting nexin-1 |
| SWAP70 | Switch associated protein 70 |
| TNS3 | Tensin-3 |
| TUBA1C | Tubulin alpha-1C chain |
| TUBA4A | Tubulin alpha-4A chain |
| TUBB | Tubulin beta chain |
| TUBB2B | Tubulin beta-2B chain |
| TUBB4B | Tubulin beta-4B chain |
| WASF2 | Wiskott-Aldrich syndrome protein family member 2 |
Gene set enrichment analysis of HGAL interacting proteins
| Gene set | No. of genes in set | Overlap with HGAL interactome | –log10 (FDR) |
|---|---|---|---|
| GO_CYTOSKELETON | 1967 | 30 | 14.91 |
| GO_CELL_JUNCTION | 1151 | 22 | 12.19 |
| GO_CELL_LEADING_EDGE | 350 | 14 | 11.19 |
| WANG_TUMOR_INVASIVENESS_UP | 374 | 14 | 10.86 |
| GO_CYTOSKELETAL_PART | 1436 | 21 | 9.64 |
| GO_CELL_PROJECTION | 1786 | 22 | 8.85 |
| GO_IMMUNE_SYSTEM_PROCESS | 1984 | 22 | 8.10 |
| GO_CYTOSKELETAL_PROTEIN_BINDING | 819 | 15 | 7.79 |
| GO_ACTIN_CYTOSKELETON | 444 | 12 | 7.76 |
| GO_ACTIN_BINDING | 393 | 11 | 7.15 |
| GO_CYTOSKELETON_ORGANIZATION | 838 | 14 | 6.72 |
| GO_LAMELLIPODIUM | 172 | 8 | 6.52 |
| GO_MICROTUBULE_CYTOSKELETON | 1068 | 15 | 6.37 |
FDR: false discovery rate; GO: gene ontology
Note. Reprinted from “HGAL inhibits lymphoma dissemination by interacting with multiple cytoskeletal proteins,” by. Jiang XY, Lu XQ, Gentles AJ, Zhao DK, Wander SA, Zhang Y, et al. Blood Adv. 2021;5:5072–85 (https://doi.org/10.1182/bloodadvances.2021004304). CC BY NC ND.
We previously observed that in humans, HGAL expression in DLBCL tumors is associated with a limited, less disseminated stage of disease [120]. To examine if this observation is a direct result of HGAL expression in these tumors, several novel animal models for DLBCL dissemination were established. By using these models based on HGAL expression and knockdown, it was showned that mice harboring HGAL expressing DLBCL tumors exhibit decreased lymphoma dissemination and longer survival [105]. Further, decreased in vivo motility of DLBCL cells expressing HGAL [105]. These findings recapitulate observations in humans, establishing the role of HGAL in lymphoma dissemination and explaining the longer survival of patients with HGAL-expressing lymphomas.
Overall, these studies revealed a novel molecular mechanism of inhibitory effects of HGAL on the motility of GC-derived lymphoma cells that may contribute to the favorable outcome of patients with HGAL expressing DLBCL and cHL tumors [34, 36].
HGAL enhances BCR signaling [55, 57] and reduces lymphocyte motility [56, 104, 105]. Deregulation of these processes was shown to contribute to lymphomagenesis. Mutations and aberrations in BCR signaling [e.g., caspase recruitment domain-containing protein 11 (CARD11), CD79, myeloid differentiation primary response 88 (MYD88)] [38] and cellular motility proteins [e.g., sphingosine-1-phosphate receptor-2 (S1PR2), guanine nucleotide-binding protein subunit alpha-13 (GNA13), RhoA, and actin] were implicated in lymphoma pathogenesis [95, 96, 121, 122] via enhancement of BCR signaling and targeting of cells to microenvironments preventing cellular differentiation and promoting malignant transformation, respectively. The concomitant regulation of lymphocyte motility and BCR signaling by HGAL may represent a unique mechanism for coordination between antigen stimulation and lymphocyte motility. While HGAL is not mutated in DLBCL, its specific upregulation in GC lymphocytes with concomitant loss of downregulation due to, for example, PRDM1/BLIMP1 mutations commonly detected in DLBCL [46, 123, 124], may contribute to lymphomagenesis. To examine the effects of HGAL constitutive expression in vivo, mice that conditionally express human HGAL at different stages of hematopoietic development using 3 restricted Cre-mediated approaches to initiate the expression of HGAL in hematopoietic stem cells (Sca1-Cre), pro-B cells (Mb1-Cre), and GCBs [activation-induced cytidine deaminase (Aid)-Cre] were regarded (Figure 6) [125]. Immune stimulation resulted in larger GCs in mice in which HGAL expression was initiated in GCBs. All three mouse strains developed lymphoma that resembles human GCB-type DLBCL (morphologically, immunophenotypically, and genetically) at a frequency of 12% to 30% starting at age 13 months, leading to shorter survival [125]. In these tumors, we also observed mutations in multiple genes [e.g., proviral integration site for Moloney murine leukemia virus 1 (PIM1), GNA13, fatty acid synthase (FAS), phosphodiesterase-4D-interacting-protein (PDE4DIP), NFKBIA, and protein tyrosine phosphatase non-receptor type 6 (PTPN6)] that were implicated in the pathogenesis of human DLBCL. We also observed increased BCR signaling and ennhanced BCR synapse intensity and density following BCR stimulation in Rosa26HGAL/Mb1-Cre lymphocytes in comparison with lymphocytes not expressing HGAL originating from WT control littermates [125]. These data suggest that constitutive expression of HGAL, irrespective of the differential stage at which HGAL is conditionally expressed, leads to a block in B-cell differentiation at the GC reaction, as indicated by the expression of peanut agglutinin (PNA) and enriched expression of other GC genes, as well as the presence of mutated immunoglobulin genes in all the analyzed tumors. These observations are in line with data from human lymphoma patients, in whom HGAL is expressed in 90% of GCB-type DLBCLs [36]. The findings that it takes about 13 months to develop lymphoma suggest that secondary genetic aberrations or external triggers are likely required for lymphomagenesis, and these were observed as mentioned above.
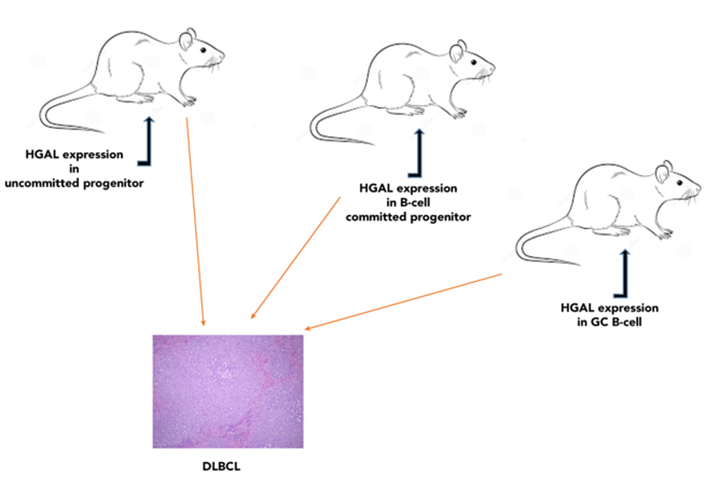
Conditional expression of HGAL in mice results in lymphoma that resembles human GCB-type DLBCL. Mice conditionally expressing human HGAL at different stages of hematopoietic development (hematopoietic stem cells, pro-B cells, or GCBs) developed lymphoma that resembles human GCB-type DLBCL [125]. Representative images of large B-cell lymphoma in the spleen showing multiple splenic nodules of large pleomorphic cells with effacement of normal spleen structure [hematoxylin and eosin (HE); original magnification ×400)]
Note. Reprinted from “Conditional expression of HGAL leads to the development of diffuse large B-cell lymphoma in mice,” by Raboso-Gallego J, Casado-García A, Jiang XY, Isidro-Hernández M, Gentles AJ, Zhao SC, et al. Blood. 2021;137:1741–53 (https://doi.org/10.1182/blood.2020004996). © 2021 American Society of Hematology.
In summary, by using mouse genetics, it was demonstrated that enforced constitutive HGAL expression drives lymphoma formation that phenocopies human GCB-type DLBCL. These results implicate HGAL in human lymphomagenesis.
While major advances in our knowledge of HGAL functions were obtained over the last 20 years since we cloned this gene [34], there are still major gaps. The precise and comprehensive analyses of the regulation of HGAL expression in B cells, as well as in Tfh cells [31], where it was recently shown to be expressed, are needed. As an adaptor protein, HGAL interacts with multiple proteins, and the functions of these interactions need to be identified. Furthermore, the HGAL interactome was examined in non-stimulated cells, while the interactome of activated/phosphorylated HGAL protein is largely unknown. The recent demonstration of HGAL expression in Tfh cells [31] opened a new area of research on the function of HGAL in these cells. While extensive knowledge on HGAL function in B cells and B cell lymphomas was accumulated, there are still gaps in knowledge of HGAL function in antigen internalization, processing, and presentation, which are controlled by the cytoskeleton that is regulated by HGAL. We have previously demonstrated that HGAL localized to cell membrane principally regulates BCR signaling, while cytoplasmic HGAL is involved in motility regulation. However, the control and interplay between these HGAL intracellular pools need further studies. For example, we demonstrated that upon BCR stimulation HGAL translocated form the membrane to the cytoplasm, and whether this re-localization is playing a major role in mediating BCR-induced inhibition of lymphocyte motility is unknown. While we showed that constitutive HGAL expression leads to DLBCL, the interplay with other genes regulating lymphocyte motility and involved in lymphomagenesis (e.g., GNA13) is needed. We hope that over the next decade, at least some of these questions will be addressed.
aa: amino acid
BCR: B cell receptor
BirA*: escherichia coli biotin ligase*
BLIMP1: B lymphocyte-induced maturation protein 1
Cre: Cre/lox system
cSMAC: central supramolecular activation cluster
DLBCL: diffuse large B-cell lymphoma
FBXO10: F-box protein 10
GC: germinal center
GCB: germinal center B cell
GEF: guanine nucleotide exchange factor
GNA13: G protein subunit alpha 13
Grb2: growth factor receptor-bound protein 2
HGAL: human germinal center-associated lymphoma
HL: Hodgkin’s lymphoma
IL-4: interleukin-4
ITAM: immunoreceptor tyrosine-based activation motif
LARG: leukemia-associated Rho guanine nucleotide exchange factor
Lyn: Lck/Yes-related novel protein
miR-155: microRNA-155
PDZ: C-terminal post-synaptic density-95, disks-large and zonula occludens-1
PRDM1: PR/SET domain 1
pSMAC: peripheral supramolecular activation cluster
RhoA: Ras homolog gene family member A
RhoGEF: Rho guanine nucleotide exchange factor
SCF: stem cell factor
SH2: Src homology domain 2
Syk: spleen tyrosine kinase
Tfh: T follicular helper
We thank to Sylvester Comprehensive Cancer Center for its continuous support over the years that included access to facilities and funding.
XJ and ISL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing—original draft. ISL: Funding acquisition, Project administration, Resources, Supervision, Writing—review & editing.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
ISL was approved by National Cancer Institute [1R01CA233945, U01CA195568], University of Miami SCCC (the Intramural Funding Program), Dwoskin and Anthony Rizzo Families Foundations and Jaime Erin Follicular Lymphoma Research Consortium. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 3451
Download: 28
Times Cited: 0
Tadashi Matsuda ... Kenji Oritani
Mari Hikosaka Kuniishi ... Takanori So
Laura E. McMillan, Christoph Wülfing
