Affiliation:
Developmental Biology and Cancer Research, IMRIC, Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel
Email: shulamitk@ekmd.huji.ac.il
ORCID: https://orcid.org/0000-0002-6878-2139
Explor Immunol. 2023;3:158–173 DOI: https://doi.org/10.37349/ei.2023.00095
Received: November 27, 2022 Accepted: February 06, 2023 Published: April 28, 2023
Academic Editor: Noah Isakov, Ben Gurion University of the Negev, Israel; Dominique J. Charron, Hospital Saint Louis, France
The article belongs to the special issue The Role of Adaptor Proteins in Lymphoid Cell Signaling
The growth and differentiation of normal cells are controlled by protein-tyrosine kinases, which serve as receptors for a wide variety of external signals. Small protein modules called Src homology 2 (SH2) and SH3 domains mediate protein-protein interactions in signaling pathways that are triggered by protein tyrosine kinases. The SH2 domain, a protein module of around 100 amino acids, is present in tyrosine kinase targets within the cell. SH2 domains are recruited to activated and autophosphorylated growth factor receptors by directly recognizing tyrosine phosphorylation sites. Growth factor receptors and other phosphoproteins have short phosphotyrosine (pTyr)-containing sequences that are bound by SH2 domains. The SH3 domain, a distinct element of approximately 50 residues that recognizes proline-rich and hydrophobic-amino-acid-containing regions, is frequently found in SH2-containing proteins. Tyrosine kinases can be coupled to downstream targets with SH3-binding sites by proteins with SH2 and SH3 domains acting as adaptors. These intricate and precise biochemical signaling pathways result in the regulation of gene expression, cytoskeletal architecture, and cell metabolism. The role of SH2/SH3 proteins in T cell signaling will be discussed. A special focus will be on the role of the hematopoietic signal transducer with SH2/SH3 domains, Vav1, in health and cancer.
The identification and characterization of the Src homology 2 (SH2) and SH3 domains established a new model of modular regions that play a crucial role in cellular signaling [1]. The SH2 domain was first discovered by Pawson’s group [2], who showed that Fujinami sarcoma virus protein (v-Fps) contains a conserved noncatalytic region of about 100 amino acids, which was named the SH2 domain since it was found in oncogenic Src. Sadowski et al. [2] showed that the SH2 domain of v-Fps mutants has an important role in controlling both cell transformation and tyrosine kinase activity. Another conserved noncatalytic region of ∼60 amino acids, designated the SH3 domain, was then identified, further highlighting the importance of modularly protein-protein regions [1]. The Src family of tyrosine kinases, the CT10 regulator of kinase (Crk) adaptor protein, and phospholipase C-γ (PLC-γ) were among the first signaling proteins identified to share SH3 domains [3, 4]. Support for the concept of modularity was provided by the discovery by Matsuda et al. [5] that v-Crk, the transforming gene of the CT10 retrovirus, encodes a viral gag protein fused solely to SH2 and SH3 domains, revealing that these domains can function alone as oncogenes. It has been demonstrated that the Crk SH2 domain binds directly to proteins that contain phosphotyrosine (pTyr), suggesting that SH2 domains may detect certain tyrosine-containing areas in a phosphorylation-dependent way [5].
The discovery of the SH2 and SH3 domains provided the foundation for the more comprehensive idea of adaptor proteins, which are composed of several different protein-binding modules that connect protein-binding partners and enable the formation of larger signaling complexes [6].
What was discovered about SH2 and SH3 domains following almost 10,000 papers that describe their structure, nature, and function? The 111 human proteins that contain the SH2 domains include small adaptor molecules like growth factor receptor-bound protein 2 (GRB2), scaffold proteins like SH2 domain-containing leukocyte protein of 76 kDa (SLP-76), and SH2 domain-containing protein (Shc), protein kinases like Src, Fps, and Bruton’s tyrosine kinase (Btk), phosphatases like Src homolog domain-containing phosphatase 2 (Shp2), rat sarcoma virus protein (Ras) signaling proteins like GTPase-activating protein (GAP), transcription factors like signal transducer and activators of transcription (Stat), ubiquitination enzymes like Casitas B-lineage lymphoma (c-Cbl), cytoskeleton regulators like Vav1, Tensin, and Chimerin, signal regulators like a suppressor of cytokine signaling (SOCS) [7]. The enormous variety of proteins that include the SH2 region attests to their crucial role in rewiring cell signaling mechanisms.
Tyrosine-phosphorylated sites are selectively and firmly bound by SH2 domains [8, 9]. The interaction is generally independent of the larger protein having a phosphorylated site since binding is only dependent on short peptide sequences and can be imitated by synthetic peptides. The three (sometimes up to five or six) amino acids C-terminal to the phosphorylated tyrosine were discovered to be crucial for specificity, with the N-terminal residues having little to no impact on binding [8, 9].
The local concentration and the relative affinities of potential binding partners determine which SH2 domains will bind to a specific location in vivo. This is the most noticeable effect in the signaling produced by the receptors for peptide growth factors, such as platelet-derived growth factor (PDGF) [10] and epidermal growth factor (EGF) [11]. Activation of the intrinsic tyrosine kinase regions of the receptors is triggered by growth factor stimulation, resulting in rapid autophosphorylation at several sites. The SH2-containing proteins PLC-γ [12], GAP [12], the 85 kDa subunit (p85) of phosphatidylinositol 3-kinase (PI3K) [13], nonreceptor tyrosine kinases and others bind to the auto-phosphorylated receptors with great affinity [14]. The SH2 domains may serve purposes other than merely mediating growth factor-induced signals. For instance, the SH2 domain seems to be involved in the both positive and negative control of the activity of the Src family of nonreceptor tyrosine kinases [15]. The phosphorylation of a C-terminal tyrosine residue maintains the catalytic inactivity of the c-Src protein and other members of its family. c-Src can be released from its autoinhibited confirmation by binding to SH2-proteins [15].
Numerous proteins that include an SH2 domain also encode an SH3 region. With more than 300 domains contained in more than 200 human proteins, the SH3 domain family is one of the largest and best-defined families. Its members are engaged in several different signaling pathways, including the control of the cytoskeleton, endocytosis, and cell proliferation [16]. Proteins with short, proline-rich binding sites are bound by SH3 domains [17]. Although there is a lot of specificity across different SH3 domains for various binding sites, the differences in affinities between high- and low-affinity sites can often be quite minor, just like with SH2 domains. The SH3 domain’s best-known function is in attracting the Ras exchange factor son of sevenless (Sos) to the membrane, which activates Ras [18]. The majority of the time, these domains are more likely to bind constitutively, acting as an intracellular glue rather than a switch. They frequently co-occur with SH2 domains in proteins; however, this is not due to any structural or functional similarity between the domains; rather, it is most likely because these domains are involved in signal transduction complexes on a regular basis [16].
The involvement of SH2 and SH3-containing proteins in T cell signaling is outlined in this review. Of particular focus on this review is Vav1, a hematopoietic signal transducer, which contains one SH2 and two SH3 domains. The role of Vav1 in T cells in health and disease is discussed.
The activation of the T cell receptor (TCR) initiates a series of signaling cascades that control cytokine production, cell survival, proliferation, and differentiation, eventually determining the fate of the cell. In general, the activation of the extracellular domain of the receptor results in conformational changes or receptor aggregation [19]. Since the TCR does not contain an intrinsic protein tyrosine kinase, recruited cytoplasmic protein tyrosine kinases to phosphorylate various tyrosine residues present in its cytoplasmic tail, thus creating binding sites for several signaling molecules that contain specific SH2 domains [20]. This initiates proximal biochemical signals mediated by the key effector enzymes, such as additional protein kinases, phosphatases, and phospholipases, which culminate in distal signaling by activating numerous transcription factors required to translate these events into specific cellular outcomes [21]. However, the proximal and distal signaling has to be integrated, and this is often the role of various adaptor proteins whose main function is to form multiprotein complexes [22].
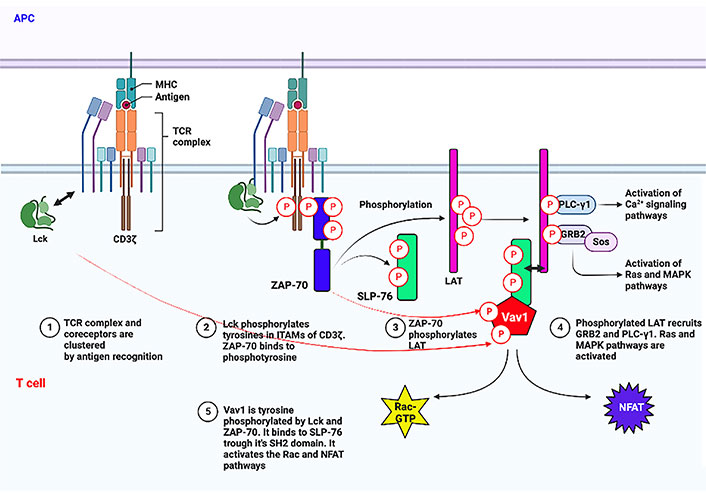
Proximal TCR signaling involving Vav1. The initial signal transduction process following the binding of the TCR to peptide–MHC complexes expressed on antigen-presenting cells [antigen-presenting cell (APC)] is depicted. Especially, tyrosine phosphorylation of Vav1 by Lck and ZAP-70 and its binding to tyrosine phosphorylated SLP-76 through its SH2 region are shown. Arrow with two heads points to association between two proteins. This figure was created with the BioRender.com program. Ca2+: calcium2+; MAPK: mitogen-activated protein kinase; NFAT: nuclear factor of activated T cells; GTP: guanosine triphosphate; MHC: major histocompatibility complex; Lck: lymphocyte-specific protein tyrosine kinase; ITAMs: immunoreceptor tyrosine activation motifs; ZAP-70: zeta-chain-associated protein kinase 70; LAT: linker for activation of T cells
T cell stimulation results from peptides from the MHC or cancer neo-antigens binding to the TCR. The Lck, a member of the Src kinase family, then starts a tyrosine phosphorylation cascade [23, 24]. Lck contains an N-terminus that is myristoylated and palmitoylated enabling its binding to the plasma membrane. It also contains SH3 and SH2 domains, a catalytic tyrosine kinase domain, and a short C-terminal tail [25]. Following conformational changes, activated Lck phosphorylates ITAMs on the TCR-associated CD3 [26]. This tyrosine phosphorylation creates docking sites for ZAP-70 interaction through the SH2 domain facilitating tyrosine phosphorylation on other residues on ITAMs [27, 28]. There is a clear hierarchy between Lck and ZAP-70, with Lck functioning prior to ZAP-70 [29].
The transmembrane adaptor protein LAT [30, 31] and the cytosolically localized SLP-76 [32] are then phosphorylated by activated ZAP-70. LAT and SLP-76 are two of the most important adaptors and they play a critical role in positively regulating TCR signaling. LAT then acts as a hub of the proximal signaling complex (proximal signalosome) by recruiting various other effector proteins, along with PLC-γ1, which links the proximal with several distal signaling pathways upon TCR engagement [30, 33, 34]. The tyrosine phosphorylation sites on SLP-76 serve as binding sites for various Shcs, such as the protein involved in cytoskeletal rearrangements, Vav1 [35], non-catalytic region of tyrosine kinase (NCK) [36], and the protein tyrosine kinase, inducible T cell kinase (ITK) [37]. Vav1 is then tyrosine phosphorylated by Lck and ZAP-70 [38] (illustrated in Figure 1). Concomitantly, phosphorylated tyrosine residues of a 130 kDa multidomain adaptor protein, named SLP-76-associated phosphoprotein (SLAP)/FYN-binding protein (FYB)/adhesion and degranulation promoting adaptor protein (ADAP) associate with the SH2 domain of SLP-76 [39, 40]. By association of SLP-76 with Vav1, NCK, and ADAP, it leads to optimal activation required for T cell activation. Additional layers of complexity of Vav1 tyrosine phosphorylation in T cells other than the standard and canonical Lck-ZAP-70 kinase cascade were recently suggested [41].
Vav1, a hematopoietic specific signal transduction molecule, was identified during a screen for new oncogenes capable of transforming NIH3T3 fibroblasts [42]. Since it was the sixth oncogene detected in Dr. Barbacid’s laboratory, it was termed Vav after the sixth letter of the Hebrew alphabet [42]. Later two other members of the family were isolated, Vav2 and Vav3, which are ubiquitously expressed, and sequence analysis revealed an identical arrangement of domains [43, 44]. A role for Vav1 in T cell signalling was first acknowledged when Vav1 was shown to be tyrosine phosphorylated following TCR stimulation [45, 46]. Moreover, phosphorylation of Vav1 has been detected in a variety of cell types and downstream of many different receptors of the hematopoietic system, such as B-cell receptor (BCR) [45], high-affinity receptor for immunoglobulin E (FcɛRI) [47], cytokine receptors [48], chemokine receptors [49], and integrins [50]. Vav1 is also activated following growth factor tyrosine kinase receptor stimulation when ectopically expressed in non-hematopoietic tissues [45, 46, 51].
To understand the roles and regulation mode of Vav1 activity, it is imperative to know its molecular structure. Vav1 contains the following regions: (1) a calponin homology (CH) domain at its amino terminus, which is involved in Ca2+ mobilization, yet functions in other proteins as an actin binding domain; (2) an acidic (Ac) domain with at least three sites of tyrosine phosphorylation; (3) a Dbl homology (DH) domain that has catalytic guanosine nucleotide exchange factor (GEF) activity towards Rho/Rac GTPases; (4) a pleckstrin homology (PH) domain that was proposed to bind to phospholipids phosphatidylinositol 4,5-bisphosphate (PIP2) and phosphatidylinositol 3,4,5-trisphosphate (PIP3) [52], however, since the PH domain of Vav1 does not fit the consensus motif for PIP3 binding to PH domain, the possibility that it binds to other phosphatidylinositol 5 phosphate (PI5P) and, possibly, other mono-protease inhibitors (PIs) has been raised [53, 54]; (5) a C1 (zinc finger) domain and a proline-rich region (–PPPP–) that mediates binding of Vav1 to SH3 containing proteins; (6) two SH3 regions [N-terminal SH3 (NSH3) and C-terminal SH3 (CSH3)] that can interact with proteins containing proline rich sequences and one SH2 domain located between the SH3 regions that interacts with proteins containing phosphorylated tyrosines [55–57]. By nucleating the ubiquitin ligase Cbl-b and the intracellular domain of Notch1 (ICN1) via its CSH3 domains, Vav1 can inhibit tumorigenesis. This reduces Notch signaling, which is necessary for T cell acute lymphoblastic leukemia (T-ALL) development and results in ICN1 being ubiquitinated and degraded [58]. Potential binding partners for each domain are detailed in several comprehensive reviews [55–57]. Vav proteins are unique in encoding a DH-PH cassette followed by an SH3-SH2-SH3 cassette. This protein structure allows them to participate in multiple signaling pathways in a tightly controlled manner. Thus, in addition to their major function as GEFs for Rho GTPases, Vav proteins also act in many processes as adaptor proteins (Figure 2). Moreover, Vav proteins are the only known GEFs that contain an SH2 domain and conserved tyrosine motifs. This unique structure enables the catalytic activity of Vav proteins to be regulated by tyrosine phosphorylation [59].
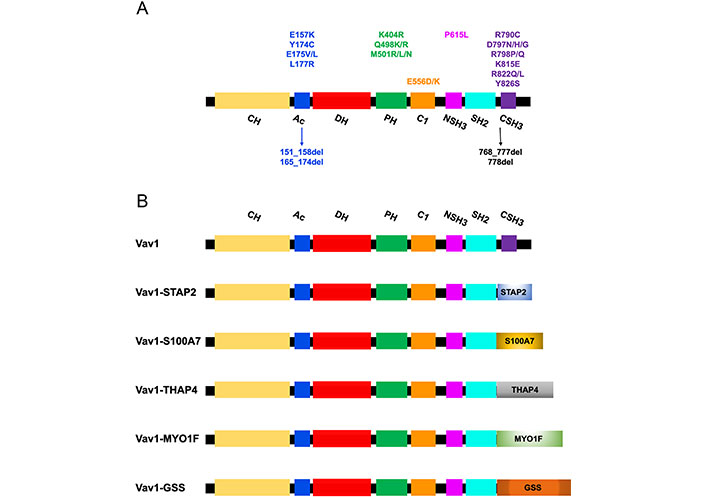
Mutations/deletions and fusions of Vav1 identified in T cell malignancies. (A) The structure of the Vav1 protein indicates its various domains. The localization of missense and deletion mutations (arrows) identified in Vav1 in T cell malignancies are indicated with relations to specific protein regions; (B) fusion proteins of Vav1 in which the CSH3 was replaced by other proteins identified in peripheral T cell lymphoma (PTCL) are indicated. STAP2: signal transducing adaptor family member 2; S100A7: S100 calcium-binding protein A7; THAP4: THAP domain-containing protein 4; MYO1F: myosin IF; GSS: glutathione synthetase
Biochemical analyses confirmed that the DH domain of Vav proteins confers GEF activity towards Rac1, Rac2, and RhoG [60]. Of particular interest, tyrosine phosphorylation of Vav1 leads to the activation of its GEF activity [59]. This mode of regulation is rather complex and requires several events to reach maximum activation. Structural analyses have shown that in Vav1, Tyr174 located at the Ac region, binds to the DH domain and inhibits GEF activity [61]. Phosphorylation of Tyr174 causes tyrosine to move away from the DH domain, abolishing its inhibition. Deletion of the CH domain leads to a constitutively active GEF indicating that this domain also inhibits DH domain activity [62, 63]. Also, the CH domain of Vav1 can bind to the C1 region, suggesting that this inhibition occurs through an intramolecular interaction that occludes the DH domain and blocks access to its substrate GTPases [64]. Similar to other proteins in this family, Vav1 contains a PH domain adjacent to the DH region, suggesting that this domain may regulate GEF activity. It has been suggested that the PH domain of Vav1 binds to the phospholipids, PIP2 and PIP3 and that this binding either inhibits or activates the in vitro GEF activity of Vav1 [52]. Additional intramolecular interactions that regulate the activity of Vav1 relate to the C1 and the CSH3-catalytic region. These include Tyr541 and Tyr544 at the C1 region and Tyr826, Tyr836, and Tyr841 at the CSH3, leading to the activation of both the catalytic and noncatalytic functions of these proteins [55, 65]. Likewise, the CSH3 appears to remain in contact with the catalytic core when Vav proteins are in the “open” state [65]. These intricate intramolecular interactions that control the catalytic function of Vav1 reflect the high degree of regulation of this protein that has to function only under very precise conditions.
The participation of Vav1 in TCR-mediated processes is usually divided into GEF-dependent and GEF-independent pathways (Figure 3). Although this categorization is somewhat artificial and may not always be precise, it reflects the complex structure and function of the protein. Much of the knowledge of Vav1’s role in T cells stems from an analysis of TCR signalling pathways in Vav1-deficient mice which showed severe impairment of activation of mature T cells, including secretion of cytokines, upregulation of activation markers, TCR-induced proliferation, and cytoskeleton organization [66–69]. Also, immune responses in these mice exhibited impaired primary cytotoxic T cell responses to viral infections and defective T cell help in antibody responses to T-dependent antigens [69, 70].
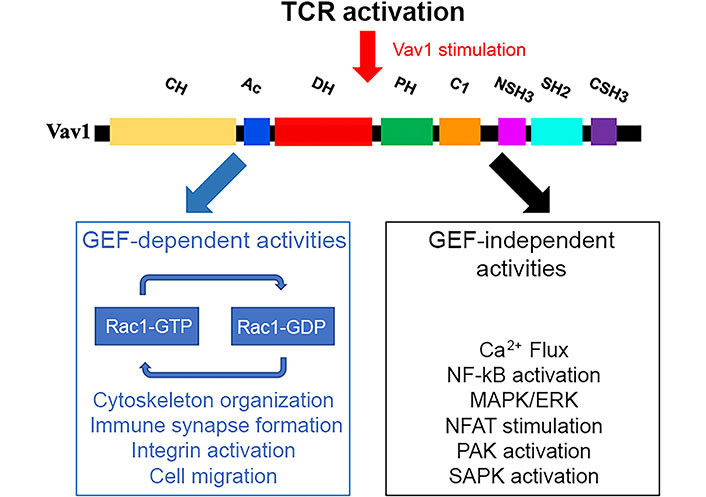
Vav1 functions in GEF-dependent and GEF-independent pathways following TCR stimulation. The various domains of Vav1 described in the legend to Figure 2 enable its participation as a GEF for Rho/Rac GTPases or in pathways that are GEF-independent. These pathways are highlighted in the figure. ERK: extracellular signal-regulated kinase; NF-kB: nuclear factor-kappa B; PAK: p21 activated kinase; SAPK: stress-activated protein kinase; GDP: guanosine diphosphate
One of the hallmarks of Vav1’s activity is its activity as a GEF, determining its participation in cytoskeleton organization in T cells. When a T cell engages an APC or a target cell, an “immunological synapse (IS)” is formed on the surface of the engaged T cell due to the reorganization of key receptors at the cell-cell contact [71–73]. The center of the synapse is composed of two regions: the central supramolecular activation cluster (cSMAC) which is where the TCR and the MHC engage in direct interaction; and the peripheral SMAC (pSMAC), where adhesion molecules and signaling molecules are localized to control the T cell activation and trafficking [74]. Initially, a central zone of integrin lymphocyte function-associated antigen-1 (LFA-1) with a peripheral ring of TCRs forms at the contact site. Within minutes though, this distribution is reversed so there is a concentration of TCRs at the center of the mature IS surrounded by a ring of LFA-1 at the pSMAC [75]. The central area of TCRs is associated with the protein kinases protein kinase C theta (PKCθ), tyrosine kinases: FYN and Lck, whereas LFA-1 becomes clustered as a peripheral ring colocalized with the cytoskeletal protein talin [76]. During this process, the microtubule organizing centre (MTOC) reorients towards the IS [77]. Vav1 is recruited to the IS and is localised predominantly to the periphery of the IS or diffusely across the cell-cell interface, corresponding most closely to the pSMAC distribution [78]. The Vav1 SH2 domain is absolutely necessary for this recruitment to the IS and microclusters, and a functional CSH3 domain is only partially necessary [79]. Several of the aforementioned processes have been discovered to be Vav1-dependent, supporting the idea that Vav1 can regulate the cytoskeleton. Studies of transgenic and knockout mice models identified Vav1 [80, 81], the adaptor proteins ADAP (also known as SLAP-130 or FYB) [39, 40], and Src kinase-associated phosphoprotein of 55 kDa (SKAP-55) [82], and the small GTPase (Ras-related protein 1) Rap1 [83] as major proteins in the cascade between TCR activation and LFA-1 clustering. Following phosphorylation, SKAP-55 binds to ADAP, and ADAP binds to the adaptor protein SLP-76, which is also found in complexes with Vav1. Vav1 then activates Rac, leading to LFA-1-mediated spreading and clustering [84]. Loss of LFA-1 function and failure of the MTOC polarization towards the IS were observed in T cell/APC couples formed from Vav1-deficient T cells [81]. Also, antigen-induced clustering of the TCR and lipid rafts in the middle of the synapse are similarly compromised in Vav1-deficient T cells [85, 86]. Finally, Vav1-deficient DP thymocytes had defective antigen-induced MTOC polarization [81]. While Vav1 is required for inside-out activation of LFA-1, TCR clustering, and cell polarisation, the characteristic changes in cell shape that occur after conjugate formation are not affected by Vav1 deficiency, thus suggesting that Vav1 selectively transduces signals to some, but not all, cytoskeletal rearrangements [81]. Although Vav1 transmits TCR signals to several cell biological processes known to require cytoskeletal rearrangements, the exact pathways by which it does so are not known.
The identification of numerous proteins that bind to the various domains of Vav1 suggests that Vav1 is involved in multiple signaling processes. The most characteristic GEF-independent function of Vav1 proteins is the activation of the NFAT in lymphocytes [57, 87]. These pathways depend on signals stemming from the CH domain of Vav1, the Vav1-dependent activation of PLC-γ and the Ca2+ release from inner reservoirs needed to activate the calcineurin phosphatase, as well as synergistic signals triggered by upstream antigenic receptors [57]. Other GEF-independent roles for Vav1 in T lymphocytes are also associated with biological processes such as the stabilization of SLP-76 microclusters upon TCR stimulation and integrin-mediated spreading [88, 89]. Additionally, the transcription factor PU.1, NFAT, NF-kB, heat shock transcription factor 1, and the androgen receptor have all been implicated in the transcriptional responses mediated by Vav1 [87, 90]. The production of second messengers like diacylglycerol (DAG), inositol triphosphate (IP3), PIP3, phosphatidic acid, arachidonic acid, and cyclic guanosine monophosphate (cGMP) occurs in many of the pathways that Vav1 stimulates. For instance, Vav1 is implicated in the activation of PLC-γ, PI3K, and PKD, in lymphocytes [91]. This results in the stimulation of nonconventional routes for Rho/Rac GEFs through DAG-mediated stimulation of the Ras guanyl releasing protein 1 (RasGRP1)-Ras-Raf-ERK axis [91]. Vav1 might also be activated through mechanisms such as binding to phosphatidylinositol monophosphates and lysine acetylation [54, 92]. Also, the nucleation of the ubiquitin ligase Cbl-b and the ICN1 via Vav1’s CSH3 domain, which leads to reduced Notch signaling, plays a role in NFAT activation [93].
Unexpected expression of Vav1 has been described in numerous studies of a variety of human cancers of various histological origins other than the hematopoietic system, such as neuroblastoma [94], lung [95], pancreas [96], breast [97, 98], ovarian [99], prostate [100], esophageal [101], and brain tumors [102]. In addition, mutations in various domains of the Vav1 protein have been found in human cancers such as hematologic malignancies (detailed below), lung adenocarcinoma, and squamous cell carcinomas [103], as well as other cancers (https://cancer.sanger.ac.uk/cosmic/). Since Vav1 is involved in numerous signaling pathways with various outcomes, it is obvious that these mutations can induce different outcomes on the downstream signaling of the protein.
This implies that the normal regulatory control of cellular proliferation may be overwhelmed if a signal protein like Vav1 is produced, either as a mutant or an overexpressed protein.
The review focused here on the involvement of Vav1 in hematopoietic malignancies, especially T cell malignancies. High expression of Vav1 was shown to predict a poor prognosis of acute myeloid leukemia (AML) [104]. Also, numerous studies reported missense/deletion mutations (Figure 2A) and fusions/translocations (Figure 2B) in Vav1 in T cell lymphoma (TCL) [105–109]. The alterations in Vav1 range from 5% in angioimmunoblastic TCL (AITL), 7–11% in peripheral PTCL-not otherwise specified (NOS), 11% in anaplastic large cell lymphoma (ALCL) [106, 107, 110, 111], and 18% in adult T cell leukemia/lymphoma (ATLL) [112]. Most of the accumulation of missense mutations and localized in-frame deletions of Vav1 were detected in the Ac region (Glu157, Tyr174, Glu175, Leu177, 151_158del, and 165_174del) and the CSH3 domain [Arg790, Asp797, Arg798, Lys815, Arg822, and Tyr826 (Figure 2A)] [108]. Additional mutations are also marked in Figure 2A.
The Vav1 CSH3 domain is also frequently deleted in various subtypes of mature T cell neoplasms (TCNs) as a result of Vav1 fusion, with numerous partner genes, such as Vav1-THAP4, Vav1-MYO1F, Vav1-S100A7, Vav1-STAP2, and Vav1-GSS (Figure 2B) [108].
However, the accumulation of mutations in the Ac and CSH3 regions raises the possibility that such genetic changes lead to the generation of a Vav1 protein that is over-active as a GEF, as these domains were shown to be cardinal for the regulation of its GEF activity. Indeed, alterations in either the Ac or SH3-SH2-SH3 domains impair their interaction with the DH domain, thus enabling activation of the GEF activity [113], leading to hyperactivation of TCR signaling in vitro [106, 107].
The contribution of wild-type Vav1 [114–116] or Vav1 mutants [108, 117] to tumor development has been studied in vivo recently, using genetically engineered mouse models (GEMMs). Expression of wild-type Vav1 in specific organs, such as the lungs [114] or the pancreas [116], indicated that while expression of Vav1 only did not lead to tumor development, co-expression of Vav1 with mutant K-Ras (K-RasG12D/Vav1 mouse) synergistically enhanced tumor development in these organs [114, 116]. Also, of great interest is the fact that the development of lung cancer in K-RasG12D/Vav1 mice was accompanied by distinct changes in the organization of the tumor microenvironment, and especially the emergence of tertiary lymphoid structures (TLSs) [114]. Expression of wild-type Vav1 in transgenic mice using the ubiquitous ROSA26 promoter led to its expression in epithelial tissues of various organs, including the pancreas, liver, and lung [115]. Surprisingly, while no carcinomas developed in these organs, B-cell lymphomas (BCLs) were detected, accompanied by an increase in ERK phosphorylation in the lymphomas [115]. Colony stimulating factor 1 (CSF1), one of the growth factors have investigated as a potential candidate to mediate paracrine stimulation in the lymphocytes, was highly expressed in the epithelial compartment of Rosa Vav1 mice. The discovery that its particular receptor, CSF1 receptor (CSF1R), is strongly expressed in BCLs strongly suggests the possibility of cross-talk between epithelial cells that express Vav1 and release CSF1 and lymphocytes that express CSF1R, which results in the development of BCLs [115]. These findings offer a novel mechanism by which Vav1 contributes to tumor propagation and malignant progression.
Analysis of tumor generation in the immune system indicated that in adoptive T cell transfer experiments, a trivalent subclass Vav1 mutant in CD4+ T cells can directly cause the development of AITL in vivo. This protumourigenic effect engages the GEF-dependent and GEF-independent activities of oncogenic Vav1 mutants [118]. Using a GEMM approach, Fukumoto et al. [108] demonstrated that the ectopic expression of Vav1 gain-of-function mutations, Vav1 (165-174del) and Vav1-STAP2, leads to the development of T cell lymphomagenesis only when tumor protein P53 (TP53), a tumor suppressor gene is deleted [108]. In a similar study, Robles-Valero et al. [117] showed in a gene-edited mouse model that the trivalent functional subclass of Vav1 mutations plays critical roles in the development of AITL, yet it requires the elimination of the suppressor activity of TP53. It is possible that the requirement for loss of TP53 is related to issues such as overcoming the negative selection that is probably imposed on thymocytes expressing active versions of Vav1. Taken together, even potent mutations in Vav1 that deregulate and activate its functions are not sufficient for it to function as an oncogene alone, and additional molecular insults are required.
The complex family of proteins known as adaptors often has several binding domains for pTyr, proline-rich regions, and lipid interactions, and also has sequence motifs that are involved in binding to such domains. Some of the members of this family contain catalytic enzymatic activity, while others do not. Vav1, a hematopoietic signal transducer, possesses adaptor protein domains, such as SH2 and SH3, along with enzymatic GEF activity. This complex protein structure endows Vav1 with a cardinal role in T cell biology. Since Vav1 was shown to contribute to the development and maintenance of various malignancies, including malignancies of the hematopoietic system, it is seemingly an excellent target for therapeutic drug design. However, Vav1 can be a double-edged sword in this regard, as a treatment that is not targeted specifically to the protein in the cancer cells alone may seriously damage the immune system.
Ac: acidic
ADAP: adhesion and degranulation promoting adaptor protein
AITL: angioimmunoblastic T cell lymphoma
APC: antigen-presenting cell
BCLs: B-cell lymphomas
Ca2+: calcium2+
c-Cbl: Casitas B-lineage lymphoma
CH: calponin homology
Crk: CT10 regulator of kinase
CSF1: colony stimulating factor 1
CSH3: C-terminal Src homology 3
DH: Dbl homology
ERK: extracellular signal-regulated kinase
GEF: guanosine nucleotide exchange factor
GRB2: growth factor receptor-bound protein 2
ICN1: intracellular domain of Notch1
IS: immunological synapse
ITAMs: immunoreceptor tyrosine activation motifs
LAT: linker for activation of T cells
Lck: lymphocyte-specific protein tyrosine kinase
LFA-1: lymphocyte function-associated antigen-1
MHC: major histocompatibility complex
MTOC: microtubule organizing centre
NFAT: nuclear factor of activated T cells
NSH3: N-terminal Src homology 3
PH: pleckstrin homology
PIP3: phosphatidylinositol 3,4,5-trisphosphate
PLC-γ: phospholipase C-γ
pSMAC: peripheral supramolecular activation cluster
Ras: rat sarcoma virus protein
SH2: Src homology 2
SLP-76: Src homology 2 domain-containing leukocyte protein of 76 kDa
STAP2: signal transducing adaptor family member 2
TCR: T cell receptor
TP53: tumor protein P53
ZAP-70: zeta-chain-associated protein kinase 70
Thanks are due to Dr. Gillian Kay for editing the manuscript.
SK: Conceptualization, Writing—original draft, Writing—review & editing, Visualization.
The author declares that she has no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
This work was partially supported by grants from the Israel Cancer Association [ICA; 07/20; supported by Ms. Y. Doron] Israel Science Foundation [333/13&654/18] the Hubert H. Humphrey Center for Experimental Medicine and Cancer Research; and the Alex U Soyka Pancreatic Cancer Research Project. The funders had a role in paying for the professional editing of the manuscript and for creating the figures.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Tadashi Matsuda ... Kenji Oritani
Mari Hikosaka Kuniishi ... Takanori So
Laura E. McMillan, Christoph Wülfing
Xiaoyu Jiang, Izidore S. Lossos
