Affiliation:
1Biochem123 Education, NW7 4AU London, UK
Email: abrownbscmsc@gmail.com
ORCID: http://orcid.org/0000-0001-5238-6943
Affiliation:
2Independent researcher, 31195 Lamspringe, Germany
ORCID: http://orcid.org/0000-0001-7638-3181
Affiliation:
3Independent researcher, Biochem123 Education, NW7 4AU London, UK
ORCID: http://orcid.org/0000-0002-8200-0885
Affiliation:
4Rocket Program Education Business School, W1B 3HH London, UK
ORCID: http://orcid.org/0000-0001-9176-8565
Affiliation:
5Medical University of Holguin, Holguin, HOG 80 100, Cuba
ORCID: http://orcid.org/0000-0002-7817-0494
Affiliation:
7Think Vaccines LLC, Houston, TX 77005, USA
ORCID: http://orcid.org/0000-0003-2466-4920
Explor Immunol. 2023;3:525–553 DOI: https://doi.org/10.37349/ei.2023.00119
Received: June 16, 2023 Accepted: October 11, 2023 Published: November 21, 2023
Academic Editor: Noah Isakov, Ben Gurion University of the Negev, Israel; Vladimir N. Uversky, University of South Florida, USA
The article belongs to the special issue Old and New Paradigms in Viral Vaccinology
Since 2019, notable global viral outbreaks have occurred necessitating further research and healthcare system investigations. Following the coronavirus disease 2019 (COVID-19) pandemic, in 2022, whilst severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strains evolved, monkeypox virus (MPXV) infections became more evident. MPXV is of the Orthopoxviridae genus, belonging to the family Poxviridae. Zoonotic transmission (animal-to-human transmission) may occur. The Orthopoxviridae genus includes other orthopoxviruses (OPXVs) present in animal host reservoirs that include cowpox viruses (CPXVs), vaccinia virus (VACV), and variola virus (VARV), with the latter being a causal agent of smallpox and excessive mortality. This review aims to present facts about MPXV-specific pathogenesis, epidemiology, and immunology alongside historical perspectives. MPXV was rarely reported outside Africa before April 2000. Early research since 1796 contributed towards the eradication of VARV leading to immunisation strategies. The World Health Organisation (WHO) announcement that VARV had been eradicated was confirmed in 1980. On the 23rd of July 2022, the WHO announced MPXV as a health emergency. Therefore, concern due to the propagation of MPXV causing monkeypox (mpox) disease requires clarity. Infected hosts display symptoms like extensive cellular-initiated rashes and lesions. Infection with MPXV makes it difficult to differentiate from other diseases or skin conditions. Antiviral therapeutic drugs were typically prescribed for smallpox and mpox disease; however, the molecular and immunological mechanisms with cellular changes remain of interest. Furthermore, no official authorized treatment exists for mpox disease. Some humans across the globe may be considered at risk. Historically, presenting symptoms of mpox resemble other viral diseases. Symptoms include rashes or lesions like Streptococcus, but also human herpes viruses (HHVs), including Varicella zoster virus (VZV).
Monkeypox virus (MPXV), the causal virion of monkeypox (mpox) disease, affects living animal populations by causing systemic disease, but mainly in specific tissues and cells through host cell receptors and intracellular propagation. Historically, research into variola virus (VARV) occurs under World Health Organisation (WHO) rules due to such virulence seen by the original VARV eradicated pathogen. Recently research development utilises lesser virulent orthopoxviruses (OPXVs), like those derived from vaccinia virus (VACV), as well as cowpox virus (CPXV), although horsepox virus (HPXV) was examined recently. Considerably less was known in 1980 when VARV (major/minor) was eradicated from the molecular and cellular interactions involved in OPXV propagation [1]. In 2017, reports appeared analysing a 1902 VARV vaccine suggestive of high (99.7%) levels of similarity with HPXV, although the origins of Poxviridae remain unknown [2, 3]. Below is shown some historical perspectives of Orthopoxviridae (see Figure 1).
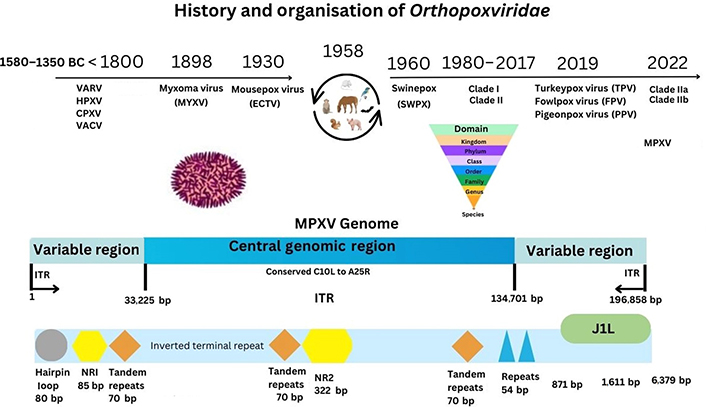
MPXV genome. ITR: inverted terminal repeat; NR1: non-repeat 1; ECTV: ectromelia virus
The MPXV particle size is 200–450 nm in length, with an approximate width of 160–260 nm, and a genome size of approximately 197 kb [4, 5]. Viral components are a linear double-stranded deoxyribonucleic acid (dsDNA) genome, a nucleoprotein core containing transcription factors (TFs), surface tubules, and two lateral bodies [6]. MPXV encodes structural proteins with virions that are ovoid/brick-shaped, together with a lipoprotein outer membrane. The MPXV genome is composed of approximately 190 open reading frames (ORFs). Approximately 90 ORFs are considered to encode MPXV proteins required for replication. Some of these may undergo gene mutations affecting biological function [7, 8]. There are an additional two conserved gene sequences (NR1 and NR2) that are known to flank ITR sequences in OPXV genomes. Therefore, NR1 and NR2 may act as regulatory sequences. Currently classified utilising genomic surveillance (www.nextstrain.org), MPXV is known by clade I (Congo Basin) or clade II (West Africa) nomenclature [9, 10]. When clades were classified, disruptions to ORFs were noted that may explain differences in virulence [11, 12].
MPXV infects a host through intradermal, oropharynx, and nasopharyngeal routes. Infection accompanied by rashes and lesions that resemble those of MPXV can be lesions similar to other viral or bacterial infections. For example, exanthem skin rashes or lesions are associated with epithelial and mucosal cell membrane disruption during infection with echovirus, coxsackievirus, cytomegalovirus (CMV), or Neisseria meningitidis (meningococcal disease) [13–16]. In addition, chickenpox lesions caused by human herpesvirus 3 (HHV3)/Varicella zoster virus (VZV) have similarities to those caused by mpox disease. Viral gene DNA transcription into messenger RNA (mRNA) occurs with translation into proteins intracellularly following infection.
In brief, Poxviridae use cell permeation, adsorption, membrane fusion, and lysis, which are four key stages to propagate while interacting with cellular proteins [13]. Globally, mpox disease has occurred in more than 110 previously non-endemic countries reaching more than 90,656 laboratory-confirmed cases worldwide as of 11 October, 2023 [17]. It is possible that increased usage of diagnostic testing during the COVID-19 pandemic thereby allows greater scientific analysis into mpox cellular disease mechanisms [18]. Different generations born after 1980, or potentially earlier could have variations in immune responses against OPXV that include MPXV infections [19]. Historical studies estimating mutations in dsDNA OPXVs, like VARV now extinct, indicate that before eradication OPXVs may have evolved or mutated at between 1 × 10–5 to 1 × 10–6 substitutions/site per year [20, 21]. The current review focuses on existing contributory MPXV proteins and will attempt to clarify immunological responses since the VARV pandemic more than 43 years ago.
Initially, MPXV was isolated in the laboratory from lesions found on imported primates in Copenhagen in 1958, but also subsequently in monkeys and squirrels (1985), together with dormice (Graphiurus), rodents (Cricetomys), and prairie dogs (2003). These are known to be the primary MPXV hosts [22–25].
Other OPXVs exist in different animal species, for example, isolation of Eptesipoxvirus occurred (2017) in a microbat (Eptesicus fuscus), while some are known to infect cattle (Capripoxviruses) causing lumpy skin disease (LSD) [26–28]. On May 25, 2022, the WHO issued a public health statement due to increases in MPXV cases detected. The risk of transmission in vulnerable populations also includes both elderly and children. The information available suggests that children in the endemic area are affected and vulnerable populations in non-endemic could be affected [29]. There is some data on whether MPXV infection affects pregnancy and gestation [30]. From early 2022, the median age of MPXV cases is 34 (range 29–41). Like other OPXVs, MPXV may affect pregnant women more severely than healthy non-pregnant females [31]. Some reports suggest a link between congenital infection and fetal death from virus propagation [31].
Since 2017, surveillance confirmed the existence of subclade IIb B.1. Moreover, subclade IIb A.2 is considered to possess 42 nucleotide mutations, including those of apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3) that play a role in outbreaks seen in 2022 [5, 10, 32, 33]. All systems put in place to monitor SARS-CoV-2 can therefore be useful in the context of MPXV surveillance and other pathogens from the OPXV family given the severity of both. At the time of the WHO announcement, it was noted that SARS-CoV-2 prevalence was transitioning between BA2/4/5 strains at that time. Details on MPXV co-infection remain limited even now [34]. The recent outbreak in 2022 began in the United Kingdom (UK) in a patient who had traveled to Nigeria in April 2022 (see Supplementary materials). On May 21, 2022, the WHO announced that 92 confirmed cases of mpox had occurred in 12 countries outside the endemic areas in Central and West Africa (see Supplementary materials). On July 23, 2022, the WHO Director-General declared the current mpox outbreak a public health emergency of international concern. As of October 11, 2023, 90,656 confirmed mpox cases had been reported from 115 locations worldwide (108 countries with non-historically reported cases; see Supplementary materials).
Clades of MPXV can vary by infection fatality rate (IFR), as indicated by clade I (Congo Basin and Nigeria) at up to 10.0%, with the latter clade IIa (West Africa clade), and more recent clade IIb suggested to have an IFR of 1.0–3.7% [35]. Transmission rates are indicated by R0 and a recent study by Du et al. [36] indicated that MPXV R0 is around 1.39 (95% confidence interval: 1.37, 1.42). R0 remains a parameter used to define transmissibility in human populations [37, 38]. In comparison with 2009 influenza (H1N1) and 2020 SARS-CoV-2, R0 was quantified at 1.7 and 2.4 respectively, with other Paramyxoviridae (e.g., measles) R0 estimated between 12 and 18 [39–41]. Between 1980 and 2000, MPXV remained detected in mostly African countries with cases reported in the Democratic Republic of Congo (DRC), Gabon, Ivory Coast, and the Central African Republic until 2003 in the United States of America (USA) [17]. Notably, research during the DRC outbreak quantified incidence risk between outbreaks of 1981–1986 and 2005–2007. During the 1980s, this is suggested at 0.72/10,000 individuals, with latter outbreaks at 14.42/10,000 individuals, respectively [42]. Prior articles indicate occurrences in individuals between the age of 21 and 40 that may be indicative of a lack of immunity to OPXVs, like VARV, unknown to date [22]. Between 2009 and 2014 (n = 645) it was confirmed that 80% of prior OPXV outbreaks occurred in children under 15. Analysis indicated either 93% or 98% met the criteria for MPXV infection; however low specificity to the available serological analysis of between 9% and 26% respectively was indicated [6]. Serological diagnostic testing is validated before use as a diagnostic for the specificity of reagents (e.g., monoclonal antibodies) and was indicated problematic with OPXVs before and after 2000 due to antibody cross-reactivity with other OPXVs [43–46]. However, more recently, 119 antigen, DNA, and antibody tests are now available subject to specificity and licensing approval (see Table S1).
Lymphadenopathy reactions occur when the lymphatic system, tissues, and cells react as with other viral lesions that occur during chickenpox (VZV) infection generating an immune response [6]. More recent surveillance of serology in animals is feasible and has occurred during a 2017 CPXV outbreak. For example, in four alpaca herds, seroprevalence was established (range 16.2–81.6%) alongside monitoring of OPXV (CPXV) spread between rodents and the vole (Microtus arvalis) between 2007 and 2017 [47, 48]. In December 2019, as the COVID-19 pandemic was recognized by the WHO, investigations examined real-time quantitative polymerase chain reaction (PCR) cycle thresholds (CT-range 22.6–38.1). This was identified in a case study to find that 88.1% (n = 42) of MPXV-infected individual sample types obtained from residential surfaces were positive for MPXV DNA. The likelihood of finding DNA was greater on surfaces such as bed linen and tablet screens [49]. Moreover, in 2021, confirmation occurred (CT-range 16.1–35.7) that MPXV was isolated from porous surfaces with viability of up to 15 days (about 2 weeks) [50]. Indeed, analysis confirmed that the amount of infectious MPXV viral load necessary to instigate mpox disease remains unknown from these investigations [51–53].
mpox disease has an incubation period of 7 to 14 days and starts with a 1–4 days prodromal stage of fever, headache, and myalgia with/or without swelling that may occur within lymph nodes (LNs), indicative of an immune response occurring in germinal centres (GCs). Lymphadenopathy can be accompanied by exhaustion, chills, back pain, sore throat, and malaise [54]. The appearance of lesions together with rashes and/or pustules disturbs skin epithelial cell layers [55, 56]. These occur as macular, papular, vesicular, or pustular and may also start in the mouth [57]. Initially, a rash begins as macules, progressing to papules, vesicles, and pustules before scab appearance after 7–14 days [28, 29]. The eschar falls off with epithelial cells and surrounding tissue healing over 2–4 weeks later and renewing. An individual is no longer considered infectious from this time onward [56]. During 2022, it was noted that lesions in certain areas were less or more common [58]. Such clinical signs and symptoms could be mistaken for other viral diseases (e.g., VZV, HHV3), but also bacterial infections like Treponema pallidum (syphilis) or lesser virulent poxviruses causing skin lesions like molluscum contagiosum. There is a recent study across 16 country reports in which 647 mpox cases were compared [55]. It was noted that of these, 524 (80.9%) individuals affected were middle-aged men who have sex with men (MSM), of which 483 (92.2%) reported sexual contact, a unique clinical feature of this mpox outbreak [55]. The authors found that lymphadenopathy was significantly lower in smallpox-vaccinated cases compared to unvaccinated cases [55]. In addition, the median duration between exposure and onset of symptoms was significantly shorter in human immunodeficiency virus (HIV)-infected patients with T cell [cluster of differentiation 4 (CD4+)] counts below 200 than in those with higher levels [55]. However, in half of the cases, no prodromal symptoms were noticed before the rash, and one-fifth of those who reported prodromal symptoms did not have a fever. Therefore, regardless of whether prodromal symptoms preceded the rash, caution is needed in patients with a contact history of rash consistent with mpox. Furthermore, rash was detected in the genital area in more than half of the cases. All of the latter should be considered when clinically suspecting mpox nowadays, different from previous cases and outbreaks [55].
Diagnostics began testing for MPXV with real-time PCR for generic OPXV genes or others (see Tables S1–5). Assays also utilised include enzyme-linked immunosorbent assay (ELISA), western blot, or tissue protein immunohistochemistry. More recently, clustered regularly interspaced short palindromic repeats (CRISPRs), and other biosensors (e.g., CRISPR/Cas12b) are in development that could potentially be suitable for viral detection. These could be more cost-effective than traditional diagnostics and potentially more accurate than lateral flow device (LFD) testing [59–63].
During acute infection, real-time PCR is the preferred test as per current WHO guidelines [17]. For example, usage in a variety of cohort studies (n = 3) quantified cellular viral load in distinct locations of anatomical lesions during mpox disease. This was noted by sample type as follows: saliva (92.3%), oropharyngeal swabs (86.2%), plasma (51.7%), stool (46.1%), urine (9.5%), and semen (5.7%). Viral particles from MPXV DNA were found in non-lesion sites present (up to 67 days), but also in oropharyngeal swabs, with DNA detectable for up to 19 days [64]. Although MPXV diagnostic tests yield satisfactory sensitivity and specificity confirming positive OPXV infections, serology tests in the past could detect other OPXV epitope similarities through cross-reactive antibodies. Recent PCR DNA tests detect specific MPXV genes according to WHO guidelines that include B6R, B7R, E9L, C3L, RPO18, and F3L [65–71]. Currently, there are 3 types of MPXV diagnostic tests available including DNA (87), antigen (18), as well as antibody (14) diagnostics totaling at least 119 to current knowledge (see Table S1).
Unlike other viruses, MPXV does not have a unique receptor for cell entry. Many genes encoding proteins and receptors are described and contribute to MPXV virion particle fusion alongside the intracellular formation of virion particles. Synthesis of intracellular mature virions (IMVs) and enveloped extracellular virions (EEVs) occurs. Specific host proteins are affected by MPXV utilising host-synthesized proteins and affecting immune system responses. For example, cytokines [interleukin 1 (IL-1)] and type I/II interferons (IFNs) are known to be affected by OPXVs due to protein homology and comparative knowledge of IFN signaling [72–74]. The role of type III IFN in OPXVs remains unclear and are therapeutic targets under investigation in other inflammatory pathologies [75, 76].
Poxvirus cellular protein entry occurs via the phospholipid plasma membrane (PM) and endocytosis dependent on low intracellular pH conditions utilising more than eleven conserved fusion proteins to form an entry fusion complex (EFC) [77–79]. Replication in Guarnieri bodies in proximity to cell organelles occurs between the Golgi apparatus and endoplasmic reticulum (ER) [80]. Subsequently, MPXV synthesis of other viral proteins described is encoded by early or late gene products (E/L), that transcribe viral DNA-producing RNA. This is then translated to produce proteins making the IMV/EEV particle utilising host cell proteins that lyse with variable thickness poxvirus cell membrane layers [80]. mpox virion particle entry requires extracellular expressed protein receptors and intracellular proteins as below. Virion particle formation to a stable IMV/EEV requires actin tails synthesised within host cells [81]. It is believed that MPXV and VARV are slightly comparable with estimates of 96.3% genetic homology [82, 83].
Historically, VARV and MPXV gene proteins are denoted by restriction endonucleases described by a capital letter (HindIII restriction endonuclease fragment), number (position within fragment), but also R/L (denoting direction of transcription). Other reviews dictate that it is unknown how poxvirus gene loss and variation occur [84]. Some reviews refer to VACV complement control protein (CCP; VCP) frequently denoting research on CCPs on earlier vaccinia (VACV) strains [77]. Therefore, convention describes researched protein homology in lesser virulent strains of OPXV, initially denoted by VCP (but later as CCP), but also CPXV (BR derived from CPXV British Green proteins), in which these were first characterized [77].
History indicates that differences before clade I and clade II MPXV gene transcripts were that these gene transcripts were initially characterised (A52R, A55R, B17L, C2L, and E5R) [85]. Subsequently, during the 21st century, research on VACV indicated that another gene transcript, H3, is notably homologous between Chordopoxvirinae and Entomopoxvirinae [85, 86]. Other gene transcripts (A27, A33, and L1) had not been confirmed by X-ray crystallography until the 21st century as technological improvements in genome sequencing occurred.
Evidence was found during VACV research therefore that H3 (p35) may not be involved in cell fusion but is involved in OPXV adherence via binding to heparan sulfate and is a plausible immunodominant antigen target [86]. Subsequently, A46R and A52R proteins were shown to be expressed by the lesser virulent VACV potentially homologous that may antagonise host IL-1 and Toll-like receptor (TLR) responses although not to the extent seen prior [72]. Prior research from in vitro studies indicated that H3/A27 binds to heparan sulphate with D8 binding to chondroitin/laminin with both A27/D8 binding to glycosaminoglycans (GAGs) [87].
Shortly around the time of earlier outbreaks of MPXV, reports appeared further clarifying poxvirus inhibitors of complement enzymes (PICEs) generic to previous OPXVs, with other proteins indicated as key to viral replication, cell structure, transcription, as well as immune responses [6–8]. These include morphogenesis factors, profilin-like factors, but also DNA ligases, and actin tail nucleation proteins.
Based on genomic analysis, it was postulated that 5 MPXV genes could affect the immune response: D10L, D14L, B10R, and B14R, alongside B19R which may explain the differences between virulence of previous MPXV clades [88]. As research evolved it became clearer that three proteins, E3L, C3L, and C10L were truncated or fragmented in MPXV, present in the virulent VARV, of which one is considered an IFN resistance protein (E3L) [85]. The E3 homologue gene transcript, F3L, of VARV contains an N-terminal binding domain and a C-terminal double-stranded RNA (dsRNA) binding domain that is also 88% to 92% similar to VACV [89, 90]. The protein encoded is implicated in affecting pattern recognition receptors (PRRs), retinoic acid-inducible gene I (RIG-I), melanoma-differentiation-associated-protein 5 (MDA5), and oligoadenylate synthetase (OAS) enzymes. Each is central to sensing both dsRNA and DNA through TLR signaling largely unknown [89, 90]. In earlier clades of MPXV, therefore it is known that through inhibiting protein kinase R (PKR) and OAS intracellularly, type I/II IFN pathways can be inhibited. Although it is notable that there are four types of OAS enzymes, of which three have typical OAS catalytic activity. The fourth OAS protein is an OAS ligand (OASL) and does not have enzyme catalytic function but is involved in intracellular signaling regulating the human antiviral response.
When MPXV fuses with the host cell PM, a viral particle enters the cytoplasm facilitating cellular replication/synthesis of viral proteins within the host cell by proteins encoded by MPXV genes that may also include A16L, A21L, A28L, F9L, G3L, G9R, H2R, J5L, and L5R [16, 20, 91].
As far as timing and location of OPXV DNA replication occurs, generic DNA synthesis is detectable within 2 h of virion particle cytoplasmic entry [92]. Differences between MPXV clade I/II occurred with virulence proteins still under validation checks. However, most gene transcripts were initially characterised by OPXV homology with VACV/VARV (A49R, A52R, A55R, B17L, C2L, E5R) [85].
During the 21st century CPXV gene transcripts observed were upregulated during cellular infection (BR158, BR203, and BR209). Of these BR203 protein (221 amino-acids) is considered to effect lymphocyte apoptosis while BR209 (126–210 amino-acids) is shared between two MPXV clades affecting IL-1 function as an IL-1β binding protein [85, 91]. Below is an example of the MPXV genes and proteins outlined herein (see Figure 2 and Tables S2–4).
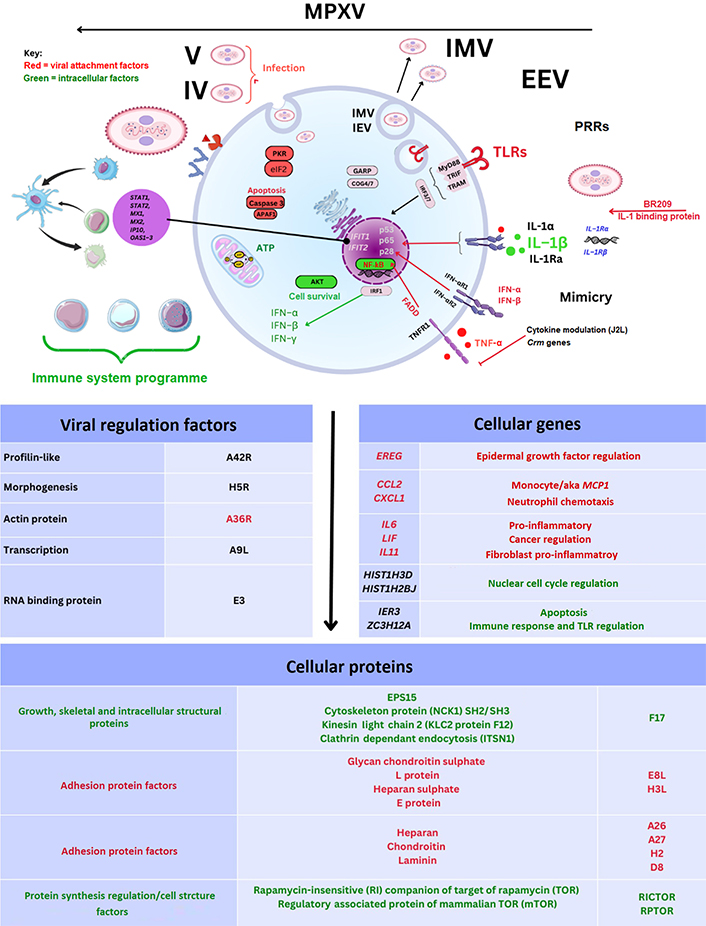
MPXV cell genes and proteins. TNF-α: tumour necrosis factor α; IL-1Ra: IL-1 receptor (IL-1R) antagonist; TNFR1: TNF receptor 1; FADD: Fas-associated protein with death domain-containing adaptor-inducing IFN-β; NF-κB: nuclear factor-kappa B; MyD88: myeloid differentiation factor 88; TRIF: Toll/IL-1R domain; TRAM: translocation chain associating membrane protein; IRF3: IFN regulatory factor 3; GARP: Golgi-associated retrograde protein; COG4: conserved oligomeric Golgi 4; eIF2: eukaryotic initiation factor 2; APAF1: apoptotic protease activating factor 1; STAT1: signal transducer and activator of transcription 1; MX1: myxoma resistance 1; IP10: IFN-γ inducible protein 10; EREG: epiregulin; CCL2: C-C motif chemokine ligand type 2; CXCL1: C-X-C motif chemokine ligand type 1; LIF: leukaemia inhibitory factor; HIST1H3D: histone H3.1; EPS15: epidermal growth factor receptor substrate 15; IER3: immediate early response 3; ZC3H12A: zinc finger CCCH-type containing 12A; IFIT1: IFN-induced with tetratricopeptide repeat 1; IFN-αR1: IFN-α receptor 1; Crm: cytokine release modulator; aka: also known as; MCP-1: monocyte chemoattractant protein 1
A notable article in 2008 examined MPXV gene transcript expression. This was examined in monocytes with MPXV gene transcripts observed including ribonucleotide reductase (C10L) and others [93]. Also described were an array of encoded RNA polymerases (G6R, E7R, and F4L), and DNA ligase enzymes (A50R) necessary for host cell propagation [93]. Prior reviews suggest that D14L is missing in MPXV clade II as well as B10R and B14R which plausibly may affect lymphocyte apoptosis and MPXV virulence [94]. Considerable divergence in OPXV protein naming became more specific with the term mpox inhibitor of complement enzyme (MOPICE) being used to describe proteins encoded by the above gene transcripts. While identification of the D14L gene transcript in gene knockout in vivo experiments confirmed that increases in viral load were observed, the MPXV MOPICE protein is considered to affect the complement pathway C3 and C5 convertases through binding to C3b and C4 complement proteins [88]. The findings above were relevant in explaining changes between MPXV clades with suggestions that the loss of D14L may potentially generate a successful adaptive immune response [88].
Activation by cytoplasmic DNA of the cyclic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) synthase (cGAS) and resulting intracellular synthesis of 2’3’-cyclic GMP-AMP (2’,3’-cGAMP) and stimulator of IFN genes (STING) remains of interest. During OPXV research these may act as intracellular regulators of other IFN-stimulating gene (ISG) factors (IFIT and ISG) including many affecting cellular IFN synthesis.
In 2017, it was additionally confirmed that GARP complexes were the retrograde transporters that the endosomal cellular transport system utilises encoded by four vacuolar protein sorting (VPS) genes (VPS51–54) [95]. This was followed up shortly after by the discovery of a 2’,3’-cGAMP-specific nuclease, described within a family of poxin proteins in 2018. These were inactivated in VARV infection and may affect innate immune response signaling [96, 97]. Recently, it has been further clarified that after the virion particle enters a host cell, GARP and eight COG body proteins may facilitate cell entry, fusion, and virion particle formation (COG3/4/7/8) around ER [77, 78, 95]. Specifically, VPS52 and VPS54 knockout MPXV infected cells in vitro incurred significant loss of function in actin tail formation; thereby resultant effect was that these were not required for IMV formation but for maturation towards EEV virion particles [95, 98]. The same group then further identified this COG complex as being composed of eight heterodimeric proteins of two subtypes (lobe A COG1–COG4 and lobe B COG5–COG8) with COG4 and COG7 considered to be more important for viral fusion and egress [95, 98].
In October 2022, the first article detailing a specific MPXV protein characterised by crystallography was published to our knowledge [70]. Gene A42R (gp153), homologous to cellular profilin proteins, was found to be conserved across OPXVs with 98% homology. It has been described that the A42R protein may have some affinity to actin whilst simultaneously being a cytotoxic T (TC) cell epitope, and TC cells are denoted phenotypically by CD molecule, CD8+ [70, 71].
Although not involved in MPXV replication, reports in November 2022 documented a novel MPXV protein that may affect intracellular host proteins and be of therapeutic value [99]. Researchers discovered the synthesis of the conserved OPXV enzyme H1 phosphatase during MPXV infection [99]. It is expected to dephosphorylate STAT1 and downregulate IFN signaling [99].
While bioinformatics research from earlier MPXV genomic analysis recently implicated, in yet-to-be-peer-reviewed reports, that eight MPXV genes were upregulated [IER3, IFIT2, IL11, ZC3H12A, EREG, NF-κB light polypeptide gene enhancer (NFKBIE), IFIT1, and amphiregulin (AREG)]. Significance was associated with the suppression of two antiviral gene transcripts (IFIT1/2) regulating IFN synthesis [100, 101]. Interestingly it was noted that a chemokine ligand gene transcript (CXCL1) was predicted to be significantly activated with CCL2, but slightly less so in comparison to CPXV and VACV [101]. Furthermore, the authors predicted upregulation of an early response IER3 gene transcript [101, 102]; but also, IL11 upregulation of a potential cytokine protein that could be considered proinflammatory in fibroblasts [101–104]. EREG is encoded by the EREG gene and is a member of the epidermal growth family (EGF) signaling through EGF receptors present in epithelial cells [105–108]. Furthermore, NFKBIE noted above follows nuclear transcription and NF-κB activation occurring during myelopoiesis [109]. AREG gene transcript expression is indicative of influence on cell proliferation of keratinocytes and fibroblasts in skin epithelial cell layers [110].
More recently, immunoinformatics prediction on potential peptide vaccine candidates to HLA-II DRB*0101 that is highly expressed (99.74%) in global populations suggests that a peptide could be designed that would bind to major histocompatibility complex (MHC) class I/II molecules and stimulate both TLR3 and TLR4 receptors [111]. These were interesting observations as HLA alleles can underpin differential immune responses and would be very interesting to see experimental research on.
However, other bioinformatics studies imply that the top three cellular pathways affected during MPXV infection are cytokines (TNF and IL-17), immune cells [e.g., T helper 17 (TH17)], and nuclear transcription (NF-κB) pathways [101, 112]. These are hypothetical, however, given that CXCL1 has a short-half-life, it is biologically plausible that infection by other factors may upregulate cytoplasmic CXCL1 synthesis leading to neutrophil and monocyte migration during MPXV infection, like TNF-α, expressed in epithelial cell layers [112].
MPXV protein antigens were evidenced around 2001 as replicating in epithelial cells, macrophages (Mϕ), dendritic cells (DCs), and fibroblasts utilising anti-vaccinia polyclonal antibodies and anti-MPXV polyclonal antibodies [113]. MPXV proteins outlined above are encoded by at least twenty-four gene transcripts that cause cellular changes upregulated 1 day after infection in two other key cell subtypes infected. The immune cell type that matures into Mϕ and presents pathogenic antigens, namely monocytes, is better characterised in the context of other recent viral infections [114]. OPXVs are known through VACV/MPXV/CPXV research in their ability to modulate antiviral immunity that led to smallpox eradication utilising the first. The role of monocyte markers and Mϕs in antigen presentation can be considered further as type I/II IFN remains crucial in viral clearance of infected cells [115–117]. The underlying mechanisms of the success of earlier VARV eradication remain unclear to this day but likely involve a host cell TC cell response unknown to date.
It is therefore necessary to consider the role of TLRs in MPXV which are both intracellular/extracellular transmembrane proteins. There are ten known TLR types in humans, but specifically six may be more relevant to immune cell recognition of OPXV infection, like TLR2/3/4/5/6/7, which act as cell membrane and vesicular sensors during viral and/or bacterial infections [118–121]. Below is an example of the TLR perspective in OPXVs (see Figure 3).
During 2009 reports emerged indicating that one TLR receptor, TLR2 researched in vitro, may affect CD8+ TC cell proliferation utilising the phosphatidylinositol-3-kinase (PI3K) activation of PKB (AKT) for cell proliferation [118]. During VACV research it was suggested that T cells, denoted by γδ, can proliferate and express MHC class I and act similarly to other antigen-presenting cells (APCs) dominant in peripheral blood that produce type II IFN-γ [122, 123]. Shortly after, in 2011, observations were made in vivo (n = 8) that B cell responses potentially could be partially abrogated in MPXV D14L deficient infection in comparison to a biphasic T cell response [88]. This occurred up to 2 weeks with a gap and then peaking at over 3 weeks, which would correspond to the induction of other characterised T cell types since 2000 that include regulatory T (TREG) and TH17 cell phenotypes. However, this T cell response is indicated within the CD4+ effector memory T (TEM) cells at 1 week and then encompassing the CD8+ TC phenotypes following at 2 weeks, with secretion of type II IFN (IFN-γ) and TNF-α expression. It is indicated the TH cell response is ongoing up to 48 days after infection [88].
Notable artificial intelligence immunoinformatic mapping indicates a second TLR of consideration. Other authors suggested that seven potential MPXV-specific epitopes exist, recognised by both TH cells and TC cells as well as B cells, which are antigenic, non-allergic, activate IFN-γ, and are non-toxic [124]. Towards this end suggestions were also of TLR5 within TC cells that are normally flagellin-activated, however further research would be required to substantiate this claim [124]. There are 10 types of TLR that are differentially pathogenic activated and according to current protein sequencing, it is unclear what role A47R, encoding 240 amino-acids, plays in clade IIb MPXV proteins and is indicated as TLR-like or IL-1-like [124]. Cell signaling pathways can be affected by many proteins during virus elimination and immune cell regulation [37, 125].
Now TH17 cells and TREG cells were beginning to be clarified in research between 2006 and 2013 that could explain this. Uniquely, in 2009, Van Vliet et al. [126] extrapolated VACV genes and MPXV genes to confirm cell fusion genes (10), and pH conditions, alongside two groups of two VACV genes that inhibited cell fusion. The report identifies more proteins (n = 164) within OPXV virion particles associated with both VACV and MPXV during the maturation of the virion particle [126]. Also identified were putative roles for other structural proteins (e.g., actin, tubulin, transgelin, laminin, vimentin, and cofilin) [126].
Therefore, as OPXVs also express IL-18 binding protein homologues, this represents another potential route of OPXV immune response modulation affecting delayed type II IFN-γ release. In addition, all OPXVs contain serpin genes with serpin 2 (B13R) in earlier MPXV clades [127, 128]. There are 180 serine proteases regulated by 37 serine protease inhibitors (SERPINs) in humans regulating haemostasis, inflammation, tissue remodeling, or angiogenesis. Furthermore, other roles for TNF modulation by homologous virus-encoded receptors [TNFR superfamily member 1B (TNFSFR1B; TNFR2)/p75] are plausible as Crms are known in other OPXV infections [128, 129].
Immunological responses to OPXVs occur across natural environments in hosts, but MPXV specifically, which are dependent on at least four key factors that include B cells, T cells alongside APCs (monocytes, Mϕs, and DCs), and natural killer (NK) cells. In recent serological studies investigating residual VARV immune responses 23 years after eradication, further research clarification came. It was observed (n = 204) that residual immune cell memory to VACV remained during the 2003 MPXV USA outbreak. This was measured by B cell antibodies present in a total of 68.5% of those receiving one dose and 79.5% of those receiving two doses aged over age 35 [130]. Estimates of smallpox immunity longevity are largely unknown [131, 132].
Shortly after Hammarlund et al. [133] in 2008 examined MHC expression, and during MPXV infection it was shown, in comparison to VACV, that MHC expression was comparatively not affected by either synthesis or maturation indicative of a role of T cell receptor signaling with antigenic peptide fragments presented.
During in vivo research (2013), reports indicated increases in total NK cells during MPXV infection in combination with a significant reduction in the overall percentage of a specific NK cell phenotype (CD56dimCD57+) [134]. Below are shown overall immune responses (see Figure 4).
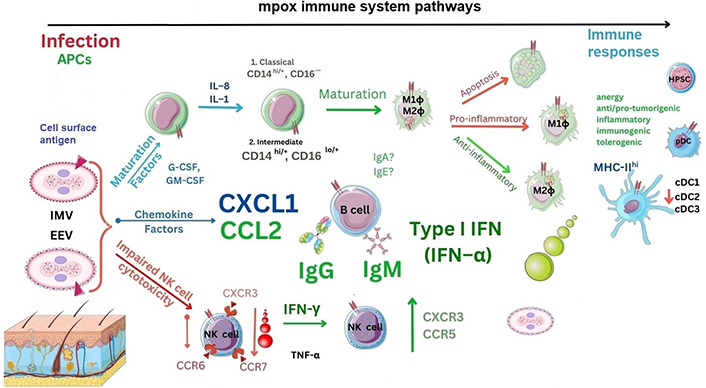
Immune responses to MPXV. CCR6: C-C motif chemokine receptor type 6; CXCR3: C-X-C motif chemokine receptor type 3; G-CSF: granulocyte colony stimulating factor; GM-CSF: granulocyte monocyte colony stimulating factor; IgA: immunoglobulin A; cDC1: conventional DC type 1; HPSC: haematopoietic stem cell. ?: no data
In a recent MPXV analysis in 2022 (n = 17), it was confirmed that up to 3 days after MPXV infection there is a temporal reduction of CD4+ T cells with an increase in CD8+ T cells representing TH and TC cells, respectively [135]. Within the adaptive T cell compartment, naive T (TN) cells expressing the lymphocyte common antigen, CD45 (CD45RA+CD27+) increase alongside increases in effector TEM cells without CD45RA– or CD27– occurred [135]. During mpox disease, T cells observed had concurrent increases in CD38 receptors within both T cell (CD4/CD8) phenotypes normalising at around three weeks [135]. Summary reports were indicative of an OPXV TH1 cell-specific profile, but notably, there was no difference in immune cells during HIV infection or in non-HIV-affected individuals, the significance of which remains unknown [135]. Notably, these authors indicated that between 2–4 days after infection, key chemokine changes in NK cells occurred that were CXCR3, CCR7, and CCR6 which were temporarily reduced [133–136]. Furthermore, between days 5–8, NK cell frequencies were expanded significantly before reducing [134, 136]. In contrast, NK cell chemokines expressed were upregulated including CXCR3 and CCR5 at days 7–8. The role of NK cells during MPXV infection is unknown currently. Recent transcriptome NK cell transfection research indicated gene transcript upregulation of granzyme B/K alongside both TNF-α and TNF-related apoptosis-inducing ligand (TRAIL) genes in comparable other DNA viruses and could be indicative of release from other TC cells or γδ T cells [137–139]. Other authors concur that CD160 in the context of viral infection affecting NK cells could be a worthy target of investigation [138–140].
Notably, the longevity of protection provided by initial vaccinia vaccines was unknown with CD nomenclature not designated until the 1980s [141]. In 2005 during in vivo research, investigators examined antibody responses and T cell responses utilising VACV [141]. It was then found that memory B cells expressing CD20 were essential to OPXV B cell plasmablast generation, but also that the T cell response could be abrogated with host survival [114, 141]. Therefore, it is notable, at least with SARS-CoV-2 infection, where there was the appearance of MHC downregulation on certain immune cell subtypes, that this may not be the case with MPXV. Interestingly, here it was seen that infected CD14+ monocytes could still produce type II IFN (IFN-γ) and TNF-α but indeed be non-responsive to host and VACV infected cells with NK cell phenotypes clarified by CD proteins (CD56+, CD16+, CD16–CD56–, CD16+, CD56+) [136].
Up to 2007, other studies (n = 76) examined the overall OPXV response to indicate that anti-OPXV IgM and IgG antibodies produced by B cells were key to immunological responses with IgM only associated with MPXV or rather OPXV infection [142]. As above, LN swelling can occur after infection indicative of GC leukocyte production; however, the exact mechanisms remain unclear. It was indicated with another OPXV (VARV) that upregulation occurs of gene transcripts PKR, STAT1, STAT2, MX1, MX2, IP10, OAS1, OAS2, and OAS3 as well as both type I IFN and type II IFN gene transcripts [11]. It is notable that, during mpox disease, in vivo research showed immune cells secrete or express IL-1Ra, IL-2, IL-6, IL-8, IFN-γ, CCL2, CCL5, G-CSF, and GM-CSF with soluble CD40 (sCD40) upregulated, of which two (G-CSF and GM-CSF) are known Mϕ differentiation factors [143]. Moreover, in recent in vitro cell culture experiments, it was seen that there was statistically significant expression of other gene transcripts encoding chemokines, cytokines, and growth factors including CXCL1, IL11, colony stimulating factor 2 (CSF2) but also pentraxin 3 (PTX3) [144]. CXCL1 protein was confirmed in vitro to be upregulated in CPXV and MPXV monocytes in other studies alongside IL-1 and IL-8, which would therefore make it possible that these are classical or intermediate monocytes [145].
There have been suggestions that MPXV clades selectively downregulate host cellular responses through reports comparing West African and Congo Basin clades (I/II). So far, it is indicated that MPXV reduces fibroblast growth factor (FGF) signaling, B cell receptor (BCR), growth hormones, and the apoptotic signal Fas (CD95) pathway with defective phosphorylation of both c-mesenchymal-epithelial transition factor (c-met) and stem cell growth factor receptor (SCFR, CD117 or c-kit) TFs with caspase 3 [114, 146–148]. Recently of relevance it has been acknowledged in transcriptome reports that the protein signaling pathways now emerging include G protein-coupled receptors (GPCRs), heat shock protein (HSP60/70), histamine, as well as plasmin alongside multiple HIST markers [144, 149]. Therefore, further clarifying some of the unknown into how atypical mpox vesicular rashes may occur.
As we discussed in our last article, chemokine receptors and ligands can be considered directional system markers that influence cytotoxic cell immune responses, some of which are considered therapeutic targets [114, 150]. In 2013, it was seen that there was potentially a cross OPXV CD8+ TC specific epitope in one of two peptides from E9L (amino-acids 562–570), with a possible third CD8 TC epitope (amino-acids 107–115) that could be an immunodominant T cell epitope [136, 151].
More recently in December 2022, it was further clarified that there were potentially a further 318 CD4+ and 659 CD8+ T cell epitopes specific to OPXVs [152]. In early 2023, further reports started to clarify that 124 amino-acids within the MPXV A35R protein generate a comparable frequency of B cell (CD19+) IgG from plasmablasts. Researchers compared MPXV infection to VACV-immunised individuals to conclude that A35R/H3L could be a potential additional serological B cell marker [153].
As VARV was eradicated, adapting usage of modified vaccinia as a vector occurred, and cellular mechanisms underlying this immunogenicity is required. Originally, during VARV outbreaks, VACV was utilised in immunisation until the late 1970s during which ongoing research showed that serial passage of a vaccinia strain (denoted by a strain from Ankara), attenuates VARV infection. This attenuation could have potential beyond that originally envisaged. Subsequent research and the discovery of DCs by Steinman and Cohn in 1973 at Rockefeller University was a key milestone discussed in 2012 by Rowley and Fitch [154]. Shortly after, in August 1975, Köhler and Milstein discovered how to produce specific monoclonal antibodies [154]. DCs were further characterised recently using single-cell RNA sequencing [147]. Furthermore, DCs are unique in being able to express elevated levels of type I IFN early in infection, but also elevated levels of MHC class II molecules. These affect both innate and adaptive immune system phenotypes in viral pathologies and cancer [114, 155–158]. Approximately 50 years after the original discovery, the complexities of DCs are still being discovered.
It is known that DC maturation and cellular differentiation may have potential anti-tumourigenic effects and antiviral tolerance but also stimulatory effects. It is commonly believed that the original TH cell response is required to be immunologically beneficial, but that this can be affected by two other APCs (monocytes and Mϕs).
In 2011, the role of VACV, through expression in recombinant MVA vector, was researched utilising CPXV in vitro to explore the differential response within leukocytes [159]. Uniquely, MVA has a modulatory effect on DC maturation that may direct other APCs and induce a TH and TC response [160]. It was shown that DCs express a chemokine, CCR7, expressed by most immune cell phenotypes; but in addition, CXCL10, TNF-α, IL-6, and importantly IL-12 were found to be cytokines that can affect immune cell phenotypes and are expressed during VACV cellular infection [158, 159, 161].
Therefore, the usage of CPXV in research which shares many homologies with other OPXVs has further clarified non-productive infection of DC cell phenotypes. Notably, DCs can be broadly classified into plasmacytoid DC (pDC), myeloid-derived DC (mDC), and into three further conventional sub-types, cDC1, cDC2, but also cDC3 that remain crucial [114, 162]. DCs can develop into immunogenic or inflammatory monocytic cells. The tolerogenic profile can be anergic, as well as pro-tumourigenic or anti-tumourigenic in characteristic during recognition of pathogenic antigens and/or tumour-associated antigens (TAAs).
Unique properties of OPXVs indicate that DCs have been shown to be permissively infected by certain types within this family of viruses, where mDCs and monocyte-derived DCs (moDCs) show vacuolar formation with loss of characteristic dendrites and syncytial cell formation. On the other hand, mDCs excessively vacuolate while immature pDCs show less vacuolation and syncytial cell formation. CPXV infection was shown to differentially inhibit DCs during maturation with suppression TLR-stimulated cytokine responses from early CPXV viral proteins. Alternatively in 2018, a cowpox protein (CPXV012) inhibited proteins linked to the ER lumen which are associated with transporting antigen presentation (TAP) proteins [163, 164]. It is known that DCs can present antigens either dependent or independent of TAP localised around the ER lumen. Therefore, resultant effects on β2 microglobulin, and MHC class I modulation could interfere with consequent viral peptide presentation by immune cells [163, 164].
More recently in 2020, Leite Pereira et al. [165] used mass cytometry to investigate the expression of seventeen cell surface receptors in leukocytes after ex vivo infection of human whole-blood samples with MVA to show downregulation for most of the characteristic cell surface markers in specific leukocytes. This MVA infection resulted in significant downregulation of CCR5 by CD4+ T cells, CD8+ T cells, B cells, and three different DC phenotypes with upregulation of MHC class II (HLA-DR) expression on DCs [165]. Furthermore, Leite Pereira et al. [165] indicated that MVA-infected APCs can directly transfer endogenous viral proteins into the MHC class II pathway to efficiently activate CD4+ T cells. To this end, through in vivo research and chemical inhibitors, it was elucidated that subcellular pathways including proteasomes and autophagy processes have a further role in endogenous MHC class II peptide presentation. Surprisingly, the involvement of both transporters associated with antigen presentation (TAP), and lysosomal-associated membrane protein 2 (LAMP-2) did not occur [166]. Therefore, MHC class I/II antigen presentation during intracellular OPXV infection is crucial to understanding how permissive infection affects cellular apoptosis.
It was further explored that MVA could produce a reduction in B-cell lymphoma 2 (BCL-2) expression as a key regulatory protein inducing cellular apoptosis [167]. Other cellular markers CD80, CD86, and CD83 are known as B and T cell signaling molecules expressed during DC maturation [168–171]. However, smallpox genes, through MVA research, have clarified that early gene expression during DC infection occurs during maturation on either immature or mature mDCs [168, 170, 172].
In addition, DC maturation is known to occur and produce type I IFN-α, within 18 h of infection, with apoptosis occurring simultaneously through virus antigen-specific MHC class I peptide-dependent CD8+ TC responses [173]. Type I IFNs have been found to be differentially elicited in cDCs, and not pDCs, during MVA infection via TFs (IRF3/IRF7) mediated by the IFNR (IFNAR1) sensitive to both type I IFNs, IFN-α and IFN-β. This occurs through the cGAS/STING pathway and is dependent on TLR3 and Tank-binding kinase 1 (TBK1). Laboratory studies of MVA in DC infection clarify that endosomal or lysosomal enzymes, like cathepsin B, can attenuate this IFN response through VACV E gene transcripts [174]. This may occur with the production of virulence factors affecting IRF3 and IFNB [174]. Furthermore, cDC synthesis of IFN and secretion could be independent of MDA-5, mitochondrial antiviral signaling (MAVS) protein, TLR3, or Toll/IL-1R domain-containing adaptor-inducing IFN-β (TRIF) [90, 174].
Cell cycle virulence genes, like p28, have also been implicated in playing a role in OPXV infection. For example, during ECTV and CPXV infection of Mϕ [117]. It has been indicated that OPXVs have modulatory functions, and other authors suggest that there are unknown ubiquitinating ligands intracellularly that regulate a cell cycle protein, p28 [117, 175]. Deficiency of p28 was investigated in vivo to show abrogation of OPXV replication in Mϕ cells. Moreover, p28 is active in DCs and NK cells, and forms a subunit of the cytokine IL-27 that is produced by DCs. Uniquely, p28 appears to perform a multi-functional role in not only DC/NK cells but is also key in the proteasomal degradation of p53 affecting both tumour cell regulation and bacterial infection [125, 175]. The exact nature and effects of PRRs, damage-activated molecular patterns (DAMPs), and how these relate to IFN stimulation and release from multiple immune cell subtypes remains unknown [176, 177]. Therefore, both IL-12 and IL-2 represent both cellular maturation and inhibitory cytokines that can be secreted by DCs and regulate type I and type II IFN secretion as well as the maturation of other immune cells.
Current data available indicates that smallpox vaccination prior to MPXV infection may have a protective effect against MPXV and prevent symptoms. Younger generations have not received these since 1980 [54, 55]. At present, there are three smallpox vaccines utilised, including ACAM2000® (IMVAMUNE), and Aventis Pasteur smallpox vaccines (APSV) in the USA [178–180]. For example, ACAM2000® (IMVAMUNE) is utilised in active immunization against smallpox disease in high-risk populations and comprised of live VACV during MPXV outbreaks [180]. Additionally, the third vaccine from MVA-Bavarian Nordic (BN; MVA-BN) is distributed under two brand names that are MVA and BN virus. Currently, these are approved as JYNNEOSTM (MVA-BN strain) in the USA; but also, IMVANEX (European Union; EU), and IMVAMUNE in Canada. With the exception of JYNNEOSTM the other vaccines were originally authorized for use against smallpox and are under intensive monitoring by the WHO, Center for Disease Control and Prevention (CDC), Food and Drug Administration (FDA), European Medicines Agency (EMA) and others (see Supplementary materials) [181, 182]. Other recent articles indicate that vaccine effectiveness of 1st generation to 3rd generation smallpox vaccines adapted to target mpox disease appears in the range 58–89% [183]. Below are some of the historical perspectives (see Figure 5).
For specific antiviral therapy in adults and children weighing over 13 kg, early indications that an antiviral tecovirimat approved in the EU may be beneficial (see Supplementary materials). Other antivirals of consideration for MPXV infections include vaccinia Igs intravenous (VIGIV) and brincidofovir [184, 185]. These indications potentially could include the treatment of other OPXV infections like VACV, MPXV, and CPXV, although this would need to be confirmed. Tecovirimat is considered an OPXV inhibitor, with brincidofovir a broader range DNA polymerase inhibitor. Since 2005, tecovirimat has been considered a relevant therapeutic to target OPXV infection. The original name was ST246, considered to inhibit extracellular virion production. This occurred through targeting the conserved F13L viral host protein and was active against not only VACV, but also MPXV, CPXV, as well as ECTV (mousepox) and camel poxvirus (see Supplementary materials) [186]. Tecovirimat was initially discovered at the National Institute of Allergy and Infectious Diseases (NIAID), in Bethesda, with other departments under the National Institute of Health (NIH). It was developed by a cooperation between Viropharma with scientists at the United States Army Medical Research Institute of Infectious Diseases (USAMRIID). Since then, it has been manufactured by Siga Technologies, a pharmaceutical company conducting research on bioweapons defence developing the drug under a governmental contract following a USA request by tender. Due to its significance for bioterrorism, the FDA granted tecovirimat fast-track status. On July 13, 2018, the FDA announced the approval of tecovirimat under the brand name TPOXX® as a prior OPXV antiviral indicated in vitro to have no kidney cytotoxic effects. An EU marketing authorization under exceptional circumstances was granted in January 2022 [187]. Other antiviral drugs considered for the treatment of MPXV infections include brincidofovir (CMX001) developed by Chimerix and marketed under the brand name Tembexa. Brincidofovir is an experimental antiviral agent against several viruses and is a competitive substrate inhibitor of viral dsDNA polymerases [188]. Brincidofovir has been conjugated to a lipid for slower plasma release, which is then cleaved and metabolised to the active ingredient cidofovir diphosphate intracellularly for plasma release to prevent extracellular virion release [185, 188]. However, it is also effective against Ebola virus (EBOV; see Supplementary materials) [189–192]. Brincidofovir has been shown to be effective against a variety of viruses including herpes simplex virus 1 (HSV1), adenovirus (AdV), human polyomavirus 1, as well as other poxviruses [189–192]. No specific recommendations concerning the management of HIV patients having a risk of exposure to MPXV have been made at the date of this report, although further information may become available. Recently a phase 2/3 clinical trial evaluation national clinical trial (NCT) was completed evaluating MVA-BN (NCT02038881/NCT02977715) [193, 194]. As of 21st February, there are currently 20 registered clinical trials listed as awaiting, or in progress (see Table S5).
The first question in addressing the topic of pathogenic infections like mpox is whether prior outbreaks caused by any pathogen will evolve. Currently, there is a low likelihood or risk with suggestions that the indicative R0 is 1.39; however, both VARV and MPXV remain infectious diseases with the former extinct in nature. Although OPXVs may share some homologous protein structure, the virulence, pathogenicity, and transmission seen with VARV before 1980 require analysis. It was seen before and after 2017 that MPXV in endemic countries had increased in localised countries. Transmission requires close contact, and unlike SARS-CoV-2, MPXV infection has demonstrated comparatively less mortality, although some cases present with severe forms of disease (particularly immunocompromised individuals). Contagion, when symptomatic is usually noticeable, and is dependent on social, healthcare, and other factors. Therefore, the chances of unnoticed transmission are limited. Given that MPXV has adjusted between climates or is now more visible due to technological advances and geographical surveillance, MPXV appears a potential health threat regardless of low fatality and transmission rates [195]. Risk factors remain unclear. This was emphasized in a routine recent survey in the UK screening blood donations for transfusion (n = 10,896), at a time of high prevalence of circulating MPXV, by PCR to the TNFR gene and a non-variola OPXV gene [196]. Whilst this study aimed to establish potential contamination of blood donations, it was therefore confirmed following the MPXV outbreak, the absence in the general population of MPXV infection measured by antibodies. Therefore, this serves as a useful reminder of the surveillance tools available, but also more importantly the impact viruses may have undetected [114, 196]. In addition, the risk of animal pet transmission can be considered since the 2022 increase in clade IIb MPXV cases. Recent reports clarify no pet transmission in cats or dogs (n = 154) currently, at least in the UK, in households with confirmed MPXV infections [197]. With regards to the contribution of T cells during co-infection with immune-deficiency disorders, this remains unknown. Emerging reports examining whether quantifying high or low T cells below or above 350 cells per mm3 as a guideline to adaptive immune responses could represent potential use in clinical settings awaiting further clarification (see Supplementary materials) [198]. At the time of drafting this report, R0 describing the transmissibility of the original infectious pathogen has previously been estimated with smallpox in the range 5.5–6.8; however, MPXV R0 data requires further clarification, and although estimates have been described, transmission rate remains unclear currently. Much research draws similarities between OPXV proteins of VACV, HPXV, CPXV, and MPXV due to homology, as we describe above; however, both require further clarification within the research protocols available. The MPXV infection 2022 outbreak currently has not yet classified MPXV as a sexually transmitted infection (STI) by the WHO [199]. Even though it is logical to consider that underlying immunosuppression may affect the outcome of MPXV disease, this assumption is yet to be determined. Nonetheless, this recent outbreak strongly suggests that HIV-infected patients, particularly those with low T cell (CD4+) cell counts could manifest signs and symptoms differently [55]. Studies conducted in Africa during earlier outbreaks reported cases of co-infection which are variable globally [200–206].
Homologous proteins described and current serological assays remain in development largely unknown in 2000 with OPXV prevalent across host animal populations. Data outlined above has been checked on genomic sequencing databases, like UniProt, whilst some undergo further validation. Since 2019, other studies have documented polymorphisms within the APOBEC3 genes, however much remains unknown in this regard [207]. Other known proteins and assays to date of this report are detailed (see Supplementary materials).
In conclusion, there are currently 90,656 confirmed cases of MPXV globally from January 1, 2022 up until October 11, 2023. Even though the MPXV causal agent of mpox disease could be considered milder than other OPXVs with lower fatality rates, mpox disease is still a major issue as a public health threat. The surveillance, research, and prophylaxis will require further prioritisation. Medical scientists and physicians necessitate further knowledge of the complexities of proteins, immunological mechanisms, and pathogenesis. This is pertinent to MPXV infection to expand on our findings, further clarifying missing literature, whilst improving guidance on potential therapeutic targets and individual health outcomes worldwide. With SARS-CoV-2 research extensive, in comparison, OPXVs remain of scientific interest. Surveillance of other existing OPXVs, like CPXV, appears comparatively more diverse in understanding zoonotic transmission across many of the species historically known. With VARV eradicated in nature some years ago, similarly to prior HPXV, further investigations into OPXVs will require further research. Smallpox experimentation is outside the remit of biosafety level 4 (BSL-4) laboratories and is currently banned and subject to WHO guidelines due to the incidence and severity of prior eradicated VARV as the only human virus to have ever been eradicated.
APCs: antigen-presenting cells
CCL2: C-C motif chemokine ligand type 2
CCP: complement control protein
CCR6: C-C motif chemokine receptor type 6
CD4+: cluster of differentiation 4
cDC1: conventional dendritic cell type 1
COG4: conserved oligomeric Golgi 4
CPXV: cowpox virus
CXCL1: C-X-C motif chemokine ligand type 1
CXCR3: C-X-C motif chemokine receptor type 3
DCs: dendritic cells
dsDNA: double-stranded deoxyribonucleic acid
ECTV: ectromelia virus
EEVs: enveloped extracellular virions
ER: endoplasmic reticulum
EREG: epiregulin
EU: European Union
FDA: Food and Drug Administration
GARP: Golgi-associated retrograde protein
G-CSF: granulocyte colony stimulating factor
GM-CSF: granulocyte monocyte colony stimulating factor
HIST1H3D: histone H3.1
HIV: human immunodeficiency virus
HPXV: horsepox virus
IER3: immediate early response 3
IFIT1: interferon-induced with tetratricopeptide repeat 1
IFNs: interferons
IFN-αR1: interferon-α receptor 1
IgA: immunoglobulin A
IL-1: interleukin 1
IL-1R: interleukin 1 receptor
IMVs: intracellular mature virions
IRF3: interferon regulatory factor 3
mDC: myeloid-derived dendritic cell
MHC: major histocompatibility complex
mpox: monkeypox
MPXV: monkeypox virus
MVA: modified vaccinia Ankara
MVA-BN: modified vaccinia Ankara-Bavarian Nordic
MX1: myxoma resistance 1
Mϕ: macrophages
NF-κB: nuclear factor-kappa B
NK: natural killer
NR1: non-repeat 1
OAS: oligoadenylate synthetase
OPXVs: orthopoxviruses
ORFs: open reading frames
PCR: polymerase chain reaction
pDC: plasmacytoid dendritic cell
PKR: protein kinase R
PRRs: pattern recognition receptors
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2
STAT1: signal transducer and activator of transcription 1
TAP: transporting antigen presentation
TC: cytotoxic T
TEM: memory T
TFs: transcription factors
TH17: T helper 17
TLR: Toll-like receptor
TNFR1: tumour necrosis factor receptor 1
TNF-α: tumour necrosis factor α
UK: United Kingdom
USA: United States of America
VACV: vaccinia virus
VARV: variola virus
VPS: vacuolar protein sorting
VZV: Varicella zoster virus
WHO: World Health Organisation
The supplementary material for this article is available at: https://www.explorationpub.com/uploads/Article/file/1003119_sup_1.pdf.
BB: Conceptualization, Methodology, Formal analysis, Writing—review & editing, Data curation, Writing—original draft, Visualization, Software, Supervision. IF: Formal analysis, Data curation, Writing—review & editing, Software, Visualization. CI: Formal analysis, Data curation, Writing—review & editing. AAPG and PM: Formal analysis, Writing—review & editing. ECC and OAB: Writing—review & editing.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Requests for accessing the datasets could be direct to the corresponding author (abrownbscmsc@gmail.com) or to Supplementary materials.
Not applicable.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Marc H.V. Van Regenmortel
Christine Jacomet
Vladimir N. Uversky
Brent Brown ... Ingo Fricke
Chittaranjan Baruah ... Bhabesh Deka
Om Saswat Sahoo ... Subhradip Karmakar
Mikolaj Raszek ... Alberto Rubio-Casillas
Ankit Kumar ... Vijay Mishra
Yulia Desheva ... Irina Isakova-Sivak
