Affiliation:
1Department of Pediatrics, Medical University Sofia, 1431 Sofia, Bulgaria
2Pediatric clinic, University Hospital Alexandrovska, 1431 Sofia, Bulgaria
Email: poli.mu@gmail.com
ORCID: https://orcid.org/0000-0002-3012-1861
Affiliation:
1Department of Pediatrics, Medical University Sofia, 1431 Sofia, Bulgaria
2Pediatric clinic, University Hospital Alexandrovska, 1431 Sofia, Bulgaria
Affiliation:
3Department of Dermatology and Venereology, Medical University Sofia, 1431 Sofia, Bulgaria
ORCID: https://orcid.org/0000-0002-7669-1368
Affiliation:
1Department of Pediatrics, Medical University Sofia, 1431 Sofia, Bulgaria
2Pediatric clinic, University Hospital Alexandrovska, 1431 Sofia, Bulgaria
ORCID: https://orcid.org/0000-0001-8168-742X
Explor Asthma Allergy. 2024;2:65–75 DOI: https://doi.org/10.37349/eaa.2024.00029
Received: July 15, 2023 Accepted: December 01, 2023 Published: February 29, 2024
Academic Editor: Ingrid Terreehorst, University of Amsterdam, The Netherlands
The article belongs to the special issue The Different Faces of Food Allergy
Food allergy is characterized by an abnormal immune reaction that occurs reproducibly upon exposure to a specific food. This immune response can lead to a variety of symptoms, the prevalence of food allergies has increased in recent decades, most likely due to environmental factors that likely play a role in the expression of genetic susceptibility. Recent understanding of the immunopathogenesis of allergic diseases has suggested that these atopic diseases may be due to monogenic mutations associated with inborn errors of immunity (IEI). Aspects to be assessed in suspected IEI involve the onset of atopic disease within the initial months of life, the progression of the condition, and the response to conventional therapy. A prospective study was conducted on 385 patients admitted to the clinic with suspected immunodeficiency. Most children were referred for recurrent respiratory infections, but almost half had concurrent atopy (44%), atopy and autoimmunity (3%), autoimmunity (6%) and malignancy (1%). The results of the study underline the importance of the allergic phenotype and suggest that children with more severe allergic diseases should be screened for possible underlying inborn defects of immunity. If a congenital disorder of immunity is suspected, comprehensive immunologic testing is required, and genetic testing is essential to identify the specific genetic abnormalities. Molecular diagnosis provides a comprehensive understanding of congenital immune disorders, allowing tailored interventions and personalized surveillance strategies.
Food allergy (FA) is characterized by an abnormal immune reaction that occurs reproducibly upon exposure to a specific food. This immune response can lead to a variety of symptoms, including skin reactions (urticaria, angioedema), anaphylaxis [including food-dependent exercise-induced anaphylaxis (FDEIAn)], and oral allergy syndrome due to immunoglobulin E (IgE)-mediated reactions, as well as food protein-induced enterocolitis syndrome (FPIES), allergic proctocolitis and enteropathy, food-induced pulmonary hemosiderosis due to non-IgE-mediated reactions. Both IgE- and non-IgE-mediated mechanisms may be involved in chronic inflammatory diseases such as eosinophilic esophagitis, eosinophilic gastroenteritis, and atopic dermatitis. FA often lasts a lifetime and results in a significant healthcare burden worldwide. FA prevalence has increased in recent decades. Most FAs are acquired in the first or second year of life. The peak prevalence of FAs is around 6 percent to 8 percent at the age of one year. In Europe, the pooled lifetime and point prevalence of self-reported FA was 19.9% and 13.1%, respectively. The point prevalence of sensitization based on specific IgE (sIgE) was 16.6%, the skin prick test (SPT) was 5.7%, and the positive food sample was 0.8% [1]. The reasons for this increasing prevalence are not known, but environmental factors are likely to play a role in influencing the expression of genetic susceptibility. Factors such as the mother’s diet during pregnancy, the mode of delivery, the use of antibiotics after birth, the integrity of the skin barrier, the timing of the introduction of allergenic foods into the infant’s diet, and others influence the development of FA in the child.
The highest risk factor for FA at 3 years of age is early-onset (1–2 months) and persistent eczema [2]. Environmental factors, including allergens, pollutants (such as car exhaust and industrial fumes), and pathogenic microbes, have the potential to disrupt epithelial barrier functions. When epithelial cells become dysfunctional, they trigger T helper type 2 (Th2)-mediated immune responses, leading to eosinophil infiltration, mast cell degranulation, IgE production, and pathological changes involving fibroblast and endothelial activation. These inflammatory reactions contribute to the development of various allergic diseases, including atopic dermatitis, FA, and asthma [3]. According to Lack [4], the dual allergen exposure hypothesis proposed that the timing and balance of cutaneous and oral exposure play a crucial role in determining whether a child develops allergy or tolerance. Rural exposure may provide an allergy-protective effect by influencing microbial translocation and colonization, thereby altering the composition and metabolic functions of the human microbiome [3]. The immaturity of physiological and immunological barriers in infants diminishes the efficacy of their mucosal defenses, likely contributing significantly to the elevated prevalence of gastrointestinal infections and the onset of FAs during the initial years of life. In a recent study investigating fecal metabolomics in early childhood FAs, various modules of functionally related intestinal metabolites were linked to the presence of FA [5]. Notably, in children with confirmed FAs, reduced levels of bile acids, amino acids, steroid hormones, and sphingolipids were observed. The authors even proposed a hypothesis suggesting that diminished bile acids might be associated with caffeine from breast milk [5].
A recent review highlights the importance of dietary fiber in the diet and emphasizes its impact on the microbiome, immune regulation, the importance of timing of intervention to take advantage of potential opportunities, and potential mechanisms for dietary fiber in the prevention and treatment of allergic diseases [6]. One mechanism for modulating the immune system influenced by dietary fiber involves microbial fermentation and secretion of bioactive metabolites. Dietary fibers such as galacto-oligosaccharides (GOS), inulins, pectin, and β-galactomannan have been shown to support a functional intestinal epithelial barrier [6]. In vitro studies suggest that dietary fibers can influence the secretion of cytokines and chemokines from epithelial cells, macrophages, and dendritic cells (DCs) through the activation of C-type lectin receptors (CLRs) and Toll-like receptors (TLRs, TLR-2 and TLR-4) [7]. Conversely, substances such as pectin can inhibit TLR-2-induced cytokine secretion. It is unlikely that a single type of dietary fiber can provide all immune-related signals, and regular consumption of different dietary fibers may offer advantages over single fiber supplementation.
Recent research indicates that micronutrient deficiencies are prevalent among atopic children experiencing conditions like atopic dermatitis, FAs, rhinitis, and asthma [8]. Specifically, iron deficiency can compromise immune activation, with prolonged deficiencies in iron, zinc, vitamin A, and vitamin D associated with a Th2 signature, the maturation of macrophages and DCs, and the formation of IgE antibodies. As micronutrient deficiency can mimic infection, the body’s innate response is to restrict access to these nutrients, preventing their dietary absorption [9]. Iron deficiency significantly affects immune cells, promoting the survival of Th2 cells, altering Ig classes, and priming mast cells for degranulation. During pregnancy, iron deficiency increases the risk of atopic diseases in children, and individuals with allergies, both children and adults, are more likely to experience anemia. Conversely, an improved iron status seems to provide protection against the development of allergies. Many allergens, such as lipocalins and pathogenesis-related proteins, have the ability to bind iron and either remove or supply iron to immune cells. Consequently, locally induced iron deficiency triggers immune activation and allergic sensitization [10].
Additional research suggests that food itself possesses an immunomodulatory effect [11]. Examining the correlation between diet and immune function, overall health, epithelial barrier function, and the gut microbiome in the context of allergies, the authors proposed a sustainable immune-supportive diet [11]. This diet comprises a diverse range of fresh and minimally processed plant and fermented foods, supplemented with moderate amounts of nuts, omega-3-rich foods, and animal products in proportions similar to the EAT—Lancet diet. This includes (fatty) fish, (fermented) full-fat dairy products, eggs, lean meat, or poultry that may be free-range or organically grown, serving as a complement to other therapies for allergic diseases [11]. Another study observed signs of dysbiosis and a less pronounced production pathway for short-chain fatty acids (SCFAs) in the stool of children with FPIES within the first 6 months of life, suggesting a potential role in its pathogenesis [12].
Recent advancements in understanding the immunopathogenesis of allergic diseases reveal that these atopic conditions may stem from monogenic mutations within the context of congenital defects of immunity [13]. Inborn errors of immunity (IEI) or primary immunodeficiencies (PIDs) represent a growing spectrum of diseases characterized by various combinations of severe and/or recurrent infections, autoimmunity, lymphoproliferation, inflammation, atopy, and malignancy. This encompassing term not only includes immunodeficiencies (IDs) but also encompasses diseases characterized by immune system dysregulation and autoinflammatory disorders. It’s important to note that infections only scratch the surface of the clinical manifestations of IEI [13].
If a monogenic mutation is the cause of the allergy, it falls into the category of primary atopic disorders [14]. IDs associated with atopy include selective IgA deficiency (SIgAD) and common variable ID (CVID). Individuals with SIgAD are more prone to asthma, upper respiratory allergies, and FAs [15, 16].
The presence of atopic diseases (asthma, eczema, allergic rhinitis, and urticaria) may be indicative of CVID in children compared to CVID in adults. In both age groups, the absence of IgE is common, with 97% having no detectable allergen-sIgE [17]. Nevertheless, in one pediatric cohort, 38% of patients had signs of allergic disease, including asthma, FA, eczema, urticaria, and rhinitis [18], and in another cohort, 83% had asthma [19]. In a series of 45 patients, five had atopic dermatitis and six had eosinophilic esophagitis [20].
IDs characterized by elevated IgE levels include hyper-IgE syndrome (HIES) due to mutations in signal transducer and activator of transcription 3 (STAT3), interleukin (IL)-6 signal transducer (IL-6ST), zinc finger protein 341 (ZNF341) deficiency, dominant-negative defect of caspase recruitment domain family member 11 (CARD11), phosphoglucomutase 3 (PGM3) deficiency, IL-21 receptor (IL-21R) deficiency, ERBB2 interacting protein (ERBIN) deficiency, actin related protein 2/3 complex subunit 1B (ARPC1B) deficiency, Wiskott-Aldrich syndrome (WAS), Omenn syndrome, immunodysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome, dedicator of cytokinesis 8 (DOCK8) deficiency, and lipopolysaccharide-responsive beige-like-anchor-protein (LRBA) deficiency. Hypersensitivity to food antigens may be a feature of IPEX or a competing diagnosis. IPEX patients often suffer from IgE-mediated FAs (see Table 1) [21, 22]. In cases where concurrent dermatitis is present, serum testing for allergen-sIgE is usually preferable to SPT to determine the presence of food-sIgE. Testing for non-IgE-mediated FAs, such as FPIES and celiac disease, should also be considered.
Conditions associated with FA
| Condition | Key features |
|---|---|
| Dominant-negative mutations in STAT3 (Job’s syndrome) | Elevated IgE levels, recurrent skin and lung infections |
| WAS | X-linked disorder can lead to eczema, low platelet counts, and recurrent infections |
| Omenn syndrome | Severe combined ID (SCID) is characterized by skin rash, failure to thrive, recurrent and severe infections, chronic diarrhea, lymphadenopathy, and hepatosplenomegaly |
| Hyper IgM syndrome | Weakened ability to fight infections and an increased susceptibility to recurrent bacterial, viral, and fungal infections |
| STAT3 gain-of-function (GOF) | Various immune-related issues, including autoimmune diseases and allergies |
| Netherton syndrome | Severe skin abnormalities, including chronic skin inflammation, eczema, and hair shaft abnormalities |
| SIgAD | Significant decrease or absence of IgA antibodies in the bloodstream. May be asymptomatic in some individuals, but it can lead to recurrent respiratory and gastrointestinal infections |
| CVID | Impaired immune function, particularly in antibody production. It results in recurrent and severe bacterial, viral, and fungal infections, along with an increased risk of autoimmune conditions and lymphomas |
| IPEX | X-linked disorder that leads to severe immune system dysregulation. Characterized by autoimmune reactions that affect multiple organs, leading to conditions such as enteropathy, polyendocrinopathy, and skin disorders |
| IL-6ST | Elevated IgE levels, bacterial sinopulmonary infections, cold skin abscesses, severe eczema, FA, pneumatoceles, and skeletal abnormalities |
| ERBIN deficiency | Elevated IgE levels, eosinophilic esophagitis, FA, allergic rhinitis, hyperextensibility, and vascular anomalies |
| Loeys-Dietz syndrome | Connective tissue abnormalities, FA, allergic rhinitis, asthma, eczema, skeletal and vascular anomalies, and developmental disability |
| DOCK8 deficiency | Elevated IgE levels, eczema, recurrent cutaneous viral infections and candidiasis, and FA; increased susceptibility to malignancy and autoimmunity |
In general, the rates of FA and anaphylaxis in patients with HIES are significantly lower than in patients with atopic dermatitis and similarly elevated IgE levels (FA 38 percent versus 58 percent and food-induced anaphylaxis 8 percent versus 33 percent) (Figure 1) [23].
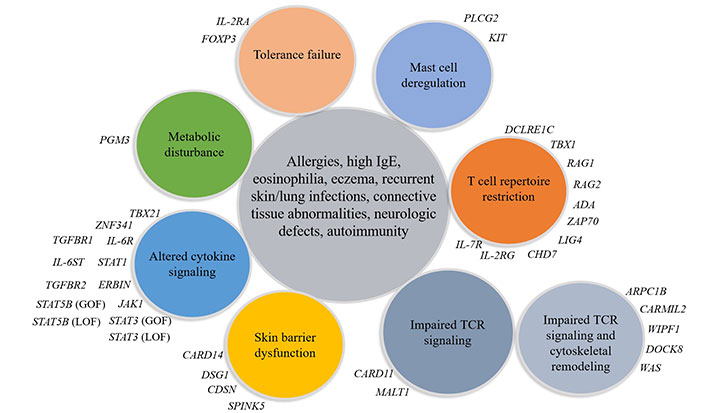
Currently defined monogenic primary atopic disorders. PLCG2: phospholipase C-gamma 2 gene; TCR: T cell receptor; FOXP3: forkhead box protein P3; DSG1: desmoglein-1; CDSN: corneodesmosin; WIPF1: WAS protein interacting protein family member 1; IL-2RG: IL-2R gamma; CHD7: DNA-binding protein 7; ZAP70: zeta-chain associated protein kinase 70; ADA: adenosine deaminase; RAG1: recombination-activating gene 1; IL-2RA: IL-2R antagonists; TBX21: T-box transcription factor 21; TGFBR1: transforming growth factor beta receptor 1; SPINK5: serine protease inhibitor Kazal-type 5; MALT1: mucosa-associated lymphoid tissue 1; CARMIL2: capping protein regulator and myosin 1 linker 2; LIG4: DNA ligase IV; DCLRE1C: DNA cross-link repair 1C; KIT: proto-oncogene, receptor tyrosine kinase; LOF: loss-of-function; JAK1: Janus kinase 1
IEI associated with atopic syndrome, including autosomal dominant HIES, autosomal recessive HIES, Netherton syndrome, hypogammaglobulinemia, SIgAD, IgM deficiency, IPEX, chronic granulomatous disease (CGD) and PLCG2 gene-associated antibody deficiency and immune dysregulation, and many other genetic PID disorders [14, 24, 25].
It is now clear that the genes involved in IEI and the genes involved in the mechanism of atopy overlap [24]. Different mutations in the same gene can cause many different phenotypes and, on the other hand, mutations in many different genes can reproduce a common phenotype. Some common features are important to recognize in suspected IEI with atopic phenotype (Figure 2) [14]. These patients usually present with specific clinical symptoms and laboratory findings that need to be carefully analyzed to determine the underlying disease. It is important to consider the presence or absence of a positive family history for PID and/or consanguinity. Factors to consider include the onset of atopic disease at birth or in the first months of life, the severity of the atopic disease, and its response to conventional therapy. Atopic features associated with autoimmunity, malignancy, recurrent infections (especially opportunistic pathogens and herpesviridae) or failure to thrive, elevated serum IgE levels (total IgE > 2,000 kU/L), high eosinophil levels (> 1,500 cells/mm3) and immunological abnormalities should be considered [26].
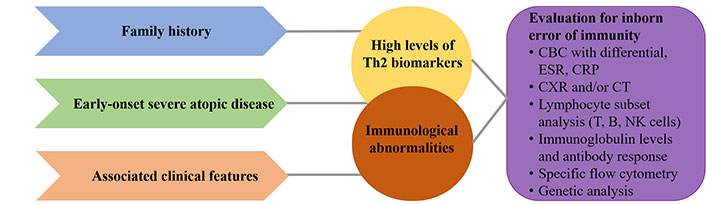
Clinical signs and diagnostic approach in inborn IDs. This diagnostic process must be completed in order to make the correct diagnosis and initiate optimal treatment. CBC: complete blood count; CRP: C-reactive protein; CXR: chest X-ray examination; CT: computed tomography; ESR: erythrocyte sedimentation rate; NK: natural killer
In patients with suspected IgE-mediated FA, a detailed clinical history focusing on the allergy is recommended as the first step in the diagnostic work-up, while genetic testing should be the final step. Genomics in FA investigates genetic and epigenetic changes associated with the disease. This is evidenced by the finding that the concordance in peanut allergies between identical twins is 64.3%, while the prevalence in the general population is around 2.9% [27]. This suggests that genetics plays a key role in the development of FAs [27]. Studies in this area would make it possible to identify heritable risk factors for FAs. Proteomics of skin strips identifies 45 proteins as principal component 1 with the highest expression in children with FA and atopic dermatitis [28]. These proteins are keratin intermediate filaments, proteins associated with inflammatory responses (S100 calcium binding proteins, alarmins, protease inhibitors), glycolysis, and antioxidant defense enzymes. They are associated with abnormalities in the integrity of the skin barrier and may increase the risk of epicutaneous sensitization to food allergens.
Various mechanisms underlie primary atopic disorders—mast cell signaling and granule dysfunction, altered cytokine signaling/JAK1, STAT3/STAT6, transforming growth factor (TGF)-β signaling, STAT3 pathway, TCR signaling/coalbed methane (CBM) complex, and actinopathies [29, 30]. Novel mechanism causing severe allergic dysregulation due to GOF mutations in the transcription factor STAT6 [31–34]. These discoveries open new perspectives in terms of treatment options for patients with IEI with atopic phenotypes. Monoclonal antibodies such as dupilumab are likely to play a role in the therapy of some of these diseases and need to be further investigated. Personalized proactive therapy for eczema and early introduction of oral allergenic foods in minute amounts are essential for the prevention of FA. Genetic counseling should also be part of the management of IEI.
A prospective study was conducted on 385 patients referred to the clinic with suspected ID [35]. The study covered a period of 9 years—from January 2014 to December 2022. Most children were referred for recurrent respiratory infections, but almost half had concurrent atopy (44%), atopy and autoimmunity (3%), autoimmunity (6%), and malignancy (1%) (Figure 3). No abnormal immunological parameters were found in 20 children (5.19%), while 45 children (11.68%) were diagnosed with ID (Figure 4).
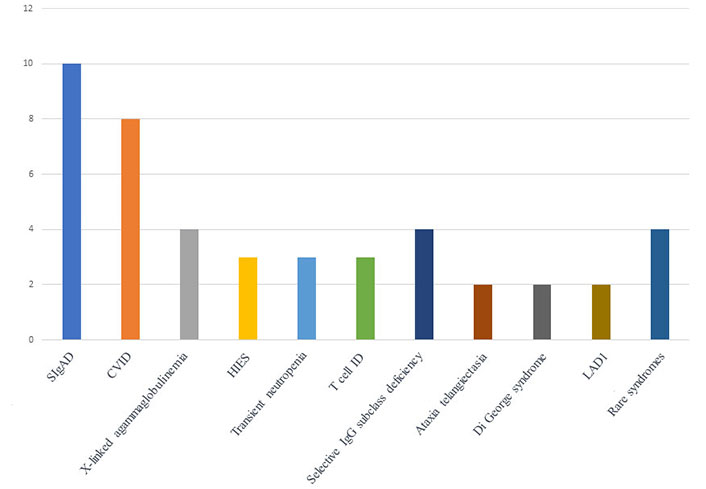
Distribution of the confirmed IEI in 45 patients. LAD1: leukocyte adhesion deficiency type 1
Most of them were children with SIgAD—10 children in total, CVID in 8 children, X-linked agammaglobulinemia in 4 children, HIES in 3 children, transient neutropenia in 3 children, T cell IDs in 3 children, 4 children had selective IgG subclass deficiency, 2 children had ataxia telangiectasia, 2 children had Di George syndrome, 2 children with LAD1, 1 child had Netherton syndrome, 1 child with LIG4 deficiency, 1 child with Castelman disease, and 1 child had STING-associated vasculopathy with onset in infancy (SAVI) (Figure 4).
A comparison of some basic data between the children with confirmed immune deficiency and the rest of the group is presented in Table 2.
Basic characteristics and demographics of the children in the study
| Characteristics | Patients with confirmed immune deficiency (n = 45) | Patients without confirmed immune deficiency (n = 340) | P value |
|---|---|---|---|
| Gender (male:female) | 25:20 | 189:151 | P = 0.84* |
| Age (years) ± SD | 5.10 ± 3.88 | 5.73 ± 3.75 | P = 0.16# |
| Comorbidity (none):atopy:autoimmunity:autoimmunity and atopy | 19:16:6:4 | 182:129:18:11 | P = 0.023* |
| Infection confirmed yes:no | 21:24 | 143:197 | P = 0.34* |
| IgE (kU/L) ± SD | 786.79 ± 3,035.86 | 248.90 ± 1,400.38 | P = 0.01# |
| Elevated IgE (n, %) | 16, 35.55% | 76, 20.54% | P = 0.05* |
* Chi-square; # analysis of variance (ANOVA). SD: standard deviation
In a study involving 313 patients with IEI, atopic manifestations were observed in 16.3% of patients [36]. This encompassed FAs in 10.2%, atopic dermatitis in 6.7%, and allergic asthma and allergic rhinitis in 1.3% of patients. Notably, allergic diseases served as the initial manifestation in 40% of atopic patients. The majority of atopic patients fell into the combined ID (CID) category (74.5%), followed by SCID (9.8%), generalized variable ID (5.9%), CGD (5.9%), SIgAD (2.0%), and leukocyte adhesion defect (2.0%). CID patients had significantly higher rates of atopic dermatitis (P = 0.001) and FAs (P < 0.001) compared to other IEI groups, with FAs and atopic dermatitis being the most common comorbidities in atopic patients with CID and SCID [36]. The results of this study again highlight the importance of not underestimating the allergic phenotype. Children with allergic diseases should be thoroughly investigated for an underlying IEI. In a multinational study of 30 centers in 23 countries, caring for a total of 8,450 patients with IEI, allergic diseases were identified in 16.3% of IEI patients [37]. These allergic diseases included bronchial asthma (3.6%), atopic dermatitis (3.6%), allergic rhinitis (3.0%), and FAs (1.3%). Allergies were newly diagnosed in 8% of IEI patients and 7.5% reported a delay in diagnosis. The highest incidence of allergic disease occurred in antibody deficiency, followed by CID. This study emphasizes the importance of raising awareness and educating healthcare providers about the link between allergic diseases and IEI [37].
In cases where clinical features of atopy predominate, IEI could be misdiagnosed. An underlying IEI should be considered, especially in severe cases of atopic disease with concomitant signs of autoimmunity and recurrent infections, an unusual clinical course, and non-response to classical treatment strategies. It is recommended to re-examine children with FAs at regular intervals, depending on age, food, and medical history, to detect possible development of spontaneous tolerance. Once IEI is suspected, comprehensive immunologic testing is required and genetic testing is essential to identify the specific genetic abnormality. Molecular diagnosis allows a comprehensive understanding of congenital immune disorders and thus tailored interventions and personalized surveillance strategies. In the future, advances in research into FAs and inborn defects of immunity through precision medicine and genetic testing could lead to personalized treatments that allow for tailored care for these individuals. More precise diagnostic tools will support early detection of IEI and enable better patient care. Targeted therapies, including gene therapy and immunomodulators, could provide solutions for correcting the gene mutations responsible for IEI. New biological therapies and strategies to induce immune tolerance may offer new options for the treatment of FAs. Improved education and advocacy networks will empower patients and caregivers and emphasize patient-centered care. Research into preventative measures, early interventions, and the influence of environmental factors will help to reduce the overall burden of these conditions. Collaboration between healthcare professionals, data sharing, and global research initiatives will enable progress in understanding the complex interplay between FAs and IEI. Promoting improved health policies and facilitating access to specialized care will ensure that people with these conditions receive the support they need. Ultimately, these advances will improve the quality of life of those affected by FAs and IEI.
CARD11: caspase recruitment domain family member 11
CID: combined immunodeficiency
CVID: common variable immunodeficiency
DOCK8: dedicator of cytokinesis 8
ERBIN: ERBB2 interacting protein
FA: food allergy
FPIES: food protein-induced enterocolitis syndrome
GOF: gain-of-function
HIES: hyper-immunoglobulin E syndrome
ID: immunodeficiencies
IEI: inborn errors of immunity
IgE: immunoglobulin E
IL: interleukin
IL-21R: interleukin-21 receptor
IL-6ST: interleukin-6 signal transducer
IPEX: immunodysregulation, polyendocrinopathy, enteropathy, X-linked
PIDs: primary immunodeficiencies
SCID: severe combined immunodeficiency
SIgAD: selective immunoglobulin A deficiency
SIgE: specific immunoglobulin E
STAT3: signal transducer and activator of transcription 3
TCR: T cell receptor
Th2: T helper type 2
TLRs: Toll-like receptors
WAS: Wiskott-Aldrich syndrome
PK: Conceptualization, Validation, Investigation, Resources, Data curation, Writing—original draft, Visualization. VP: Validation, Investigation. MS: Validation, Resources, Visualization. GP: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing—review & editing, Visualization, Supervision. All authors have read and agreed to the published version of the manuscript.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
The datasets for this manuscript are not publicly available but can be requested from the corresponding author (Polina Kostova, poli.mu@gmail.com).
Not applicable.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Angel Mazon ... Antonio Nieto
Valentina Faihs ... Knut Brockow
Ana Muñoz-Urribarri
Leonel Pereira, Ana Valado
Purificación González-Delgado ... Anna Nowak-Wegrzyn
