Affiliation:
1Institute of Chemistry of Organometallic Compounds (ICCOM), National Research Council of Italy (CNR), 50019 Sesto Fiorentino, Italy
ORCID: https://orcid.org/0000-0001-5747-2285
Affiliation:
2Interdepartmental Research Unit of Peptide and Protein Chemistry and Biology, Department of NeuroFarBa, University of Florence, 50019 Sesto Fiorentino, Italy
ORCID: https://orcid.org/0000-0001-7790-9886
Affiliation:
2Interdepartmental Research Unit of Peptide and Protein Chemistry and Biology, Department of NeuroFarBa, University of Florence, 50019 Sesto Fiorentino, Italy
ORCID: https://orcid.org/0000-0003-3075-8878
Affiliation:
3Interdepartmental Research Unit of Peptide and Protein Chemistry and Biology, Department of Chemistry “Ugo Schiff”, University of Florence, 50019 Sesto Fiorentino, Italy
ORCID: https://orcid.org/0000-0002-2947-7107
Affiliation:
2Interdepartmental Research Unit of Peptide and Protein Chemistry and Biology, Department of NeuroFarBa, University of Florence, 50019 Sesto Fiorentino, Italy
Email: paolo.rovero@unifi.it
ORCID: https://orcid.org/0000-0001-9577-5228
Explor Drug Sci. 2023;1:377–387 DOI: https://doi.org/10.37349/eds.2023.00025
Received: May 09, 2023 Accepted: June 29, 2023 Published: October 26, 2023
Academic Editor: Xuechen Li, The University of Hong Kong, China
The article belongs to the special issue Bioactive Peptides discovery and development
Protein therapeutics are extensively used in the treatment of autoimmune diseases, but a subset of patients appears to be refractory to these treatments, mainly due to the development of an immune response to the drug. A better understanding of the mechanism underlying the therapeutic drug’s failure becomes fundamental for the development of new and more effective treatments. Unfortunately, there are few cases where the exact mechanisms through which drugs bypass immunological tolerance and provoke immunogenicity have been studied. In this context, peptide epitope identification gained increasing importance in investigating the molecular mechanism of therapeutic drug’s immune responses. Despite peptide identification and use to monitor anti-drug antibody (ADA) profiles is a promising research field, their use is far away from a wide application both at the research and at the commercial level. Herein it is reported a compilation of studies in which peptides are directly involved in anti-drug immune responses, becoming the molecular key step for a better understanding of refractory reactions in therapeutic drugs. An overview on T-cell and B-cell peptide recognition is given, showing the growing potential and advantages of peptides when used in the field of refractoriness to drugs. This review includes studies describing antigenic peptides that enable enhanced ADA detection directly in patients’ sera, as well as the proof of concept that asses the use of peptides instead of proteins, to facilitate the identification of neutralizing ADA.
The human immune system uses a complex collection of protective mechanisms to monitor, usually eliminating, exogenous organisms and toxins. This active reaction to the external environment is defined as immune response and it can be divided into innate and adaptive immune responses. The former is a non-specific and rapid response to any sort of pathogens, and it involves physical barriers such as skin and mucosa, soluble proteins, and bioactive small molecules that are either constitutively present in biological fluids (such as the complement proteins) or that are released from cells as they are activated (including cytokines). The adaptive immune response, indeed, is triggered against specific antigens and it is based primarily on the antigen-specific receptors expressed on the surfaces of T- and B-lymphocytes and generates a strong and long-lasting response. When an organism interacts for the first time with a new pathogen, it starts producing T- and B-cell effectors. The production of these effector cells can be divided into two different responses: the first, called “primary immune response”, is the result of the first-time exposure to an antigen, and the second, the so-called “secondary immune response”, involves memory T- and B-cells [1]. Even if innate and adaptive immune responses are described separately, they must be considered as opposite sides of the same coin and closely related to each other, where the innate response represents the first line in host defense, and the adaptive response becomes prominent after several days, as T- and B- cells undergo clonal replication. B- and T-lymphocytes, responsible for humoral and cell mediated immunity, do not interact with the whole pathogen, but rather with molecular components named antigens. These antigens are recognized by specific receptors exposed on the cell surface of both B- and T-cells, and the recognition strictly depends on the receptor-exposing cells. In fact, B-cells recognize solubilized antigens through antigen receptors, named B-cell receptors (BCRs), consisting of membrane-bound immunoglobulins (Igs), also known as antibodies, that can mediate humoral adaptive immunity [2]. T-cell receptors (TCRs), located on the T-cell surface, allow the recognition of antigens presented on the surface of antigen-presenting cells (APCs) bound to major histocompatibility complex (MHC) molecules. The epitopes are presented by MHC I and MHC II molecules that are recognized by two distinct subsets of T-cells: cluster of differentiation 4 positive (CD4+) and CD8+ T-cells, respectively. In this situation, the immune response must be carefully controlled to prevent these harmful mechanisms from being released against the mammalian host’s own tissues, and indeed failure of self-tolerance underlies the broad class of autoimmune diseases [3].
In this scenario, it is important to consider that therapeutic proteins, highly interesting from a pharmaceutical point of view as diagnostics and therapeutics, may be processed as foreign antigens by the immune system. As opposed to small molecule immunomodulators, they have a defined mode of action, more precise binding, and low toxicity, and, among them, therapeutic antibodies (tAbs) play a pivotal role. Since the first approach described in the literature by Köhler and Milstein [4] in 1975 to produce monoclonal antibody (mAb), thanks to the continuous technological progress of molecular biology, more than one hundred of them have become approved drugs, establishing the class of tAb [5–7]. However, the intrinsic immunogenicity of murine antibodies has initially hampered their therapeutic application, subsequently solved, at least in part, by the development of chimeric, humanized, and finally fully human mAbs [8, 9]. Nevertheless, their use is not completely devoid of problems, and the initial euphoria of the first applications of antibodies as therapeutics was quickly overtaken by concerns when it became apparent that these compounds had significant issues when utilized as drugs [10]. In fact, anti-drug antibodies (ADAs) can be formed in response to any tAb, and this happens, e.g., up to 16% for patients receiving infliximab (INF) and to 44% for those receiving adalimumab (ADL), mAbs against the tumor necrosis factor-α (TNF-α), widely used in the treatment of autoimmune diseases [11]. The exponential interest brought by ADA is explicitly illustrated in the scientific literature: as reported in Web of Science, in the period between 2004 and 2013, 128 research articles and 43 reviews including the keywords “anti-drug antibodies” were published, in comparison to the 980 research articles and 237 reviews published in the timeframe 2014–2023 (consulted in March 2023). ADAs bind to the tAb and may cause allergic reactions as well as a reduction in the drug’s availability, and monitoring drug and anti-drug levels is a crucial tool for tailoring therapy to each patient’s unique needs. Moreover, regulatory authorities now require assessing the immunogenicity of new therapeutic protein drug candidates before granting market authorization [12]. To manage these immunogenicity problems, it has become extremely important to develop a screening methodology capable of detecting the presence of ADA in patients treated with tAb. In this scenario, a deep understanding of the molecular insights into the mechanism of immune response to drugs is becoming a cutting-edge research focus.
The individual response to drug antigens is triggered by one or more immunodominant regions on the antigen surface, which can involve adjacent amino acids (continuous epitope) or non-adjacent amino acids that form a specific conformational epitope. In light of this, it has been almost 40 years that peptides, especially the ones presented by the MHC, are considered likely candidates as main antigens [13–16] toward which the immune response is directed. In particular, crystallography and conformational studies, boosted by increasingly advanced technologies such as the cryo-electron microscopy, revealed the possible antigenic significance of short peptide sequences by analyzing the structure of the antigen-antibody complex. As an example, it may be considered the recent case of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein interaction with a human mAb, studied by crystallography, which led to the development of vaccines against coronavirus disease 2019 (COVID-19) [17]. In fact, designed peptide antigens are nowadays taking hold both for diagnostic and vaccinal purposes [18–22], either alone or in combination with carriers, intended to increase peptide bioavailability [23]. Moreover, the quest for clinically useful peptides, both in the pharmaceutical industry and in academia, has been inspired by the recent significant advancements obtained by peptide synthetic technologies.
While it is now clear that the use of therapeutic proteins can induce immunogenicity and alter the nature and grade of the adaptive immune response, less information is available for the exact mechanisms through which drugs bypass immunological tolerance and provoke immunogenicity (Figure 1). Indeed, the identification of peptides both in T-cell dependent or independent cell mediated pathways become in any case of great interest in understanding the insights of immune response mechanisms.
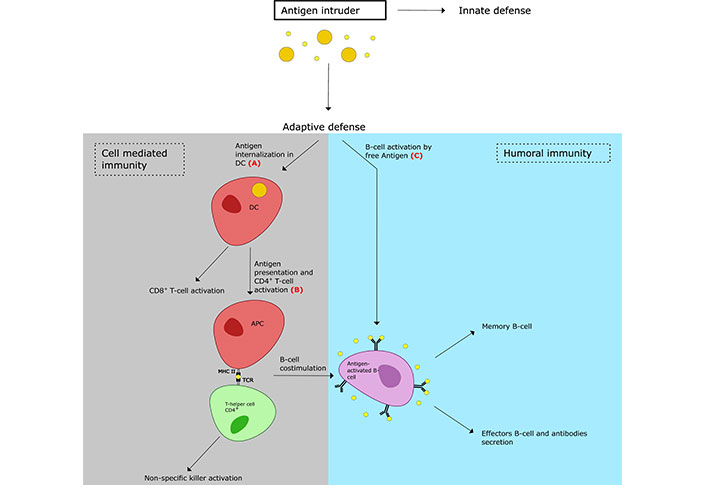
Schematic representation of primary immune response. The studies reported in this review are directed at the following steps: (A) internalization of the antigens by dendric cells [25, 28, 32, 33]; (B) Ag presentation by APC [36, 37]; (C) B-cell activation by free antigens [26, 42, 43, 45, 48–50]. DC: dendritic cell
Despite not being directly responsible for antibody formation itself, a crucial step in the occurrence of ADA is the activation of CD4+ cells. Following the internalization of the therapeutic protein by the APCs, which process it and present antigenic peptides as epitopes on MHC II, naive lymphocytes are activated to help T-cells in the presence of an additional secondary signal. The engagement of activated T-cells and B-cells induces the formation of plasma cells sustaining IgG production after antigen exposure and the isotype switching from IgM to IgG. IgM may form when a strong CD4+ T-cell response is lacking, but the low affinity and their short lives make them less likely to interfere with drugs compared to IgG [24].
The correlation between T-cell responses and ADA development was studied by Kalluri et al. [25] in the interferon-β (IFN-β) therapy, used for the treatment of multiple sclerosis. Overlapping 20-mer peptides covering the whole IFN-β sequence were synthesized and investigated, along with the whole protein, using peripheral blood mononuclear cells (PBMCs) from patients treated with IFN-β and healthy donors. The study showed that IFN-β treated patients showing ADAs occurrence presented higher T-cell responses than those not developing ADAs. Moreover, despite this type of response was directed against different IFN epitopes, only IFN-β125–159 was proportional to ADA responses while IFN-β1–40 was independent from the ADA status [25]. Moreover, both these peptide epitopes overlapped with B-cell epitopes associated with neutralizing ADA [26].
Therefore, a better investigation of MHC-presented peptides derived from biotherapeutics is a fundamental part of the study of immunogenicity risk assessment of the drug. The peptides presented by APCs varies depending on both the enzymes that the protein encounter on their pathways and on the receptivity of MHC II, however, in recent years, different factors that may affect epitope presentation have been studied [27].
Casasola-LaMacchia et al. [28] attempted to study the effect on peptide presentation in human primary monocyte-derived DCs caused by biotherapeutic antibody-target immune complexes. The case of the study was the TNF and its tAb blockers: INF, ADL, and a single-armed antigen-binding fragment (Fab’). The immunopeptidome derived by DCs of fourteen different donors, loaded with the antibodies or their immune complexes with TNF, was analyzed by trapped ion mobility time-of-flight mass spectrometry. The results showed that among the approximately 12,000 unique MHC II-associated peptides per preparation identified 0.3% were anti-TNF sequences. Among various factors that are likely to play a role in epitope presentation (e.g., the abundance of MHC II proteins, donor MHC II haplotype, inherent characteristics of the antigenic sequences), they found out an enhanced CD4+ T-cell epitope presentation for drugs complexed with their target and a higher presentation for INF, followed by ADL, and Fab’, in accordance with the size scale of the complex, suggesting that high-molecular weight drug-target structures could reinforce the immunogenic response [28].
Another anti-TNF biotherapeutic is certolizumab pegol (CZP), consisting of a single Fab derived from a humanized anti-TNF antibody functionalized with a branched polyethylene glycol (PEG) through a maleimide linker. The PEG moiety improves the water solubility of the molecule and its blood half-life, reducing its glomerular filtration [29, 30]. PEG-ylation induces additional immunogenic variables that may alter the response to the therapeutic protein [31], so de Bourayne et al. [32] investigated the role of PEG of CZP in peptide presentation to T-cells. They generated T-cell lines raised against CZP and CZP not-pegylated (CZNP) and showed that CZP-primed fewer T-cell compared with CZNP. The whole sequence of the heavy chain variable domain (VH) and of the light chain variable domain (VL) of CZP was mapped using overlapping 20-mer peptides that were used to evaluate the response of the primed lines to the epitopes: CZP-primed lines responded to 5 epitopes, CZNP-primed lines responded to 7 epitopes, and 4 epitopes were recognized by both. Moreover, the flow cytometric assay showed a minor internalization of CZP by DCs, and the low impact of PEG in CZP proteolysis was demonstrated using in vitro digestion assay with cathepsin B. The mechanism is still unclear, but a hypothesis is that considering that no receptors are involved, the PEG-ylation might interfere with the uptake, reducing the local protein concentration, and limiting the interactions with cell surface components [32].
Another factor impacting the uptake and the subsequent peptide presentation could be the microenvironment during the uptake, as studied by Lubich et al. [33] on the functional factor VIII (FVIII). FVIII is administrated to patients who suffer from hemophilia A, a congenital bleeding disorder caused by the lack of this factor in circulation, resulting in ineffective coagulation upon injury. FVIII administration, however, leads to ADA development in about 30% of the patients and thus various studies focused on this protein [34]. Various endogenous proteins bind to FVIII during its internalization and these different complexes impact the epitope presentation, thus the study investigated the recognition of FVIII peptides by FVIII-specific CD4+ T-cell hybridoma clones, modifying the microenvironment during the FVIII uptake, more in detail, FVIII was compared with FVIII complexed with von Willebrand factor (VWF, the plasma chaperon of FVIII), FVIII activated by thrombin (activation that occurs at the bleeding site), and FVIII accompanied by a concurrent blockade of APCs receptors. Evaluating 99 novel FVIII-specific CD4+ T-cell hybridoma clones (expressing TCR that recognizes a given peptide-MHC complex), generated using a whole human FVIII peptide pool consisting of 15-mer peptides shifted by 3 amino acids, it was possible to investigate the presentation of which epitope was altered. VWF was found to not inhibit the general uptake of FVIII, but rather influence some peptides but not others; thrombin activation, instead, modified the whole peptide repertoire presented by APCs, thus suggesting a different presentation in bleeding and non-bleeding sites; finally, neither blocking the mannose receptor nor the asialoglycoprotein receptors seemed to alter the epitope presentation by APCs, while downregulation appeared blocking low-density lipoprotein receptors and bivalent ion-dependent receptors [33].
As previously said, T-cell response is strictly related to the presented peptides and these peptides are not easy to characterize, making the study of anti-drug immunity difficult. This is greatly facilitated using T-cell hybridoma clones, which however requires the knowledge of the sequence, a peptide pool that covers entirely the protein and, moreover, the repertoire is related to various factors which are difficult to predict, as seen above. So, another strategy could involve leveraging existing tools to investigate the immune response, as shown by Zerra et al. [35], who engineered FVIII to contain a peptide (323–339) derived from the model antigen ovalbumin (OVA), commonly used in the field of antigen-specific immunity, to evaluate the CD4+ T-cell response specific to OVA323–339. They demonstrated that the incorporation of model antigens in therapeutic protein could be used to study antigen-specific responses and they also found out that CD4+ T-cell response to FVIII-OVA is enhanced by pre-existing anti-FVIII antibodies while suffering from a poor response when these antibodies lack [35].
Peptides are useful not only to overcome protein therapeutics immunogenicity but also to induce antigen-specific immune tolerance, preventing the immune response. Immune tolerance occurs with the generation of CD4+ T-cells with a regulatory phenotype which are anergic, produce the anti-inflammatory cytokine interleukin-10 (IL-10), suppress naive T-cell activation, and lack the expression of the transcription factor forkhead box P3 (FoxP3). Pletinckx et al. [36] proposed a peptide-base antigen-specific immunotherapy to induce immune tolerance to FVIII using apitopes (antigen-processing-independent epitopes). They identified and designed two peptides from human FVIII immunodominant T-cell epitopes that mimed the peptides derived from the natural antigen process and a combination of both, termed ATX-F8-117, was administrated to transgenic mice, resulting in the promotion of FVIII-specific T-cell tolerance and suppression of FVIII inhibitor formation, showing how the peptide administration could be a promising strategy to induce immune tolerance [36].
A similar approach using peptides to induce immune tolerance was investigated by Maldonado et al. [37], functionalizing biodegradable poly(lactide-co-glycolide) nanoparticles with both peptides and rapamycin, a tolerogenic immunomodulator. Using the model peptide OVA323–339 it was possible to suppress specific OTII T-cell responses, showing an antigen-specific T-cell tolerance.
The specific humoral immune response against drugs is up to now mainly evaluated by identifying ADA in treated patients. While the molecular mechanisms of ADA generation are still unclear, both patient and drug characteristics are definitely involved [38]. Hence, further discussion about peptides in drug immunogenicity must be conducted through the recognition of B-cell epitopes. In fact, antibody production is turned out after B-cell activation, which terminally differentiates into plasma cells that produce antibody molecules closely modeled after the receptors of the precursor B-cell [39]. B-cell activation takes place during the antigen presentation, in which a direct binding of the BCR to specific foreign antigens triggers the immune response. Antigenic molecules, which become immunogenic after their binding ability to MHC, are known as haptens in the context of immunology and have been deeply studied since their discovery by Karl Landsteiner in the early 20th century [40]. Haptens bound peptides form a hapten-carrier complex (HCC) which can evoke powerful immune reactions [41].
As indicated in the Introduction, the triggering factors in B-cell activation and the subsequent ADA formation were attributed to the murine origin of tAb, and therefore the evolution leading first to chimeric (mouse-human) and later to fully human mAbs was the logical attempt to overcame immunogenicity problems. Despite this strategy sensibly reduced immunogenicity, mAbs derived from human germline sequences were not enough for the development of immunologically inert drugs. In this context, thanks to the use of peptides, Kosmač et al. [42] explored the ADA antigenic binding sites of INF by testing overlapping peptides covering the entire antibody sequence. They demonstrate that ADA was directed to discontinuous segments of variable domains of INF by assessing antibody reactivity. The underlying facts under this induction of the immune response are not clear, and protein unfolding, and aggregation phenomena could be involved. Following this idea, Eyes et al. [43] used a custom microarray to identify peptide epitopes enhanced by aggregation on the single chain fragment variable (scFv) format antibody. Their data indicated that the immune response directed to specific regions of the therapeutic scFv revealed by partial denaturation occurs only on exposure to the aggregate. Despite this study was conducted in mice, these peptides could be used in human clinical samples to evaluate where aggregation-specific ADAs pose an immunogenicity safety issue.
In fact, the exploitation of peptides reproducing specific epitopes recognizing antibodies holds great potential to answer many unsolved questions in chemical immunology at the molecular level [44]. Thanks to the ability of peptides to mimic specific portions of protein antigens, the development of synthetic probes interacting with high affinity specific antibodies can contribute to setting up diagnostic/prognostic peptide-based immunoassays particularly useful to guide therapeutic treatments. Despite this extremely interesting potential of peptides, their use in ADA detection assays has been limited. In the literature, a few interesting examples of peptide-based enzyme-linked immunosorbent assay (ELISA) enhancing ADA detection directly in patients’ sera are available. In 1999, Brickelmaier et al. [45] used IFN-β linear peptide epitopes to identify ADAs specific to the native active form of the drug in treated multiple sclerosis patients. This peptide-based ELISA was able to overcome epitope exposure problems of the whole IFN-β protein in the assay plate, providing information on the nature of antigenic forms involved in patients’ B-cell stimulation. In fact, the use of peptides is a step forward in understanding failures in full protein-based ELISA due to problematic epitope recognition/exposition, reproduction of mutations, unfolded sequences, or lack of post-translational modifications.
Recently, Ruff et al. [46] investigated the immunogenicity of natalizumab, a therapeutic mAb targeting the α4 chain of α4β1 and α4β7 integrin widely used for the treatment of multiple sclerosis, applying a phage-display general method for the development of peptide-based immunoassays to quantify therapeutic mAbs [47]. In practice, they identified the natalizumab binding pocket that was further used to quantify drug arm exchange during treatment [48]. In fact, natalizumab undergoes in vivo Fab arm exchange to form a monovalent bispecific antibody, and the development of this assay that can distinguish and quantify between bivalent (unexchanged) and monovalent (exchanged) forms of natalizumab in clinical samples is useful for optimizing extended interval dosing and reducing the risk of progressive multifocal leukoencephalopathy.
Another promising example of a peptide-based assay with clinical purposes was developed by Rusche et al. [49] which identified ADL derived synthetic peptides with maximum immunogenicity. In this work, the ADL complementary determining regions were used as hypothetical epitopes, identifying two immunogenic peptide sequences: HC3 from the VH and LC3 from the VL of ADL. Despite these two peptides completely differ in their amino acid sequences, they showed similar ADA reactivity. Moreover, peptides were able to detect a set of ADAs, which appear to be linked to disease activity and, probably, to the biologic efficacy and response of the drug in juvenile idiopathic arthritis. In fact, the clinical correlation observed with this peptide-based ELISA is significantly increased when compared to commercially available ELISAs, based on an ADL sandwich format, probably thanks to its ability to identify only neutralizing antibodies.
Other techniques, e.g., liquid-chromatography tandem mass spectrometry, have been applied to characterize antigenic peptides. Gao et al. [50] used this technique to identify a surrogate peptide for the quantification of an anti-sclerostin mAb in cynomolgus monkey serum. Thanks to the identified peptides, authors demonstrated that decreased drug concentrations were due to the clearance refereed by ADAs rather than the interference of ADAs in the detection platform.
The use of peptides reproducing specific epitopes holds great potential to answer many unsolved questions in therapeutic protein immunogenicity. Thanks to the ability of peptides to mimic specific portions of protein antigens, the identification of epitopes that interact with high affinity with their specific immunological targets can help elucidate at the molecular level mechanisms that can be exploited to improve the therapeutic use of proteins. Despite this extremely interesting potential of peptides, their diagnostic or therapeutic use has been limited. Herein it has been reviewed the studies in which peptides proved to be useful not only to overcome protein therapeutics immunogenicity but also to induce antigen-specific immune tolerance, demonstrating that peptides can supply valuable practical solutions to deal with protein therapeutics immunogenicity. In fact, this review has shown that peptides are helpful and efficient resources for a deep understanding of immune responses. Therefore, following the extensive use of biological therapies, these studies should open the doors to the application of peptides in clinical studies targeting the different steps of immune activation.
ADA: anti-drug antibody
ADL: adalimumab
APCs: antigen-presenting cells
CD4+: cluster of differentiation 4 positive
CZNP: certolizumab pegol not-pegylated
CZP: certolizumab pegol
DC: dendritic cell
ELISA: enzyme-linked immunosorbent assay
Fab’: single-armed antigen-binding fragment
FVIII: functional factor VIII
IFN-β: interferon-β
Igs: immunoglobulins
INF: infliximab
mAb: monoclonal antibody
MHC: major histocompatibility complex
OVA: ovalbumin
PEG: polyethylene glycol
tAbs: therapeutic antibodies
TCRs: T-cell receptors
TNF-α: tumor necrosis factor-α
FRF: Conceptualization, Investigation, Writing—review & editing. FE and ADS: Investigation, Writing—review & editing. AMP: Conceptualization, Writing—review & editing. PR: Conceptualization, Investigation, Writing—review & editing, Supervision. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Qiushi Chen ... Xuechen Li
Ping Zeng ... Lanhua Yi
Shoko Tanaka ... Kohei Sato
Chloé O. Sebilleau, Steven J. Sucheck
Qian Zhang ... Chunqiu Zhang
Suze Ma ... Qi Zhang
Yanyan Liao, Xuefeng Jiang
Qian Wang ... Shiyu Chen
Jarais Fontaine, Jianfeng Cai
Oscar Noya ... Sandra Losada
