Affiliation:
1School of Pharmacy, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong 999077, China
2State Key Laboratory of Chemical Biology and Drug Discovery and Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hong Kong 999077, China
ORCID: https://orcid.org/0000-0001-9817-3898
Affiliation:
3Department of Food Science and Nutrition, The Hong Kong Polytechnic University, Hong Kong 999077, China
ORCID: https://orcid.org/0000-0003-4005-978X
Affiliation:
2State Key Laboratory of Chemical Biology and Drug Discovery and Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hong Kong 999077, China
ORCID: https://orcid.org/0000-0003-4984-7109
Affiliation:
3Department of Food Science and Nutrition, The Hong Kong Polytechnic University, Hong Kong 999077, China
ORCID: https://orcid.org/0000-0003-3526-7808
Affiliation:
2State Key Laboratory of Chemical Biology and Drug Discovery and Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hong Kong 999077, China
Email: kf.chan@polyu.edu.hk
ORCID: https://orcid.org/0000-0003-1475-288X
Affiliation:
1School of Pharmacy, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong 999077, China
Email: sharon.leung@cuhk.edu.hk
ORCID: https://orcid.org/0000-0002-0194-7058
Affiliation:
4College of Food Science, Southwest University, Chongqing 400715, China
Email: yilanhuaswu@163.com
ORCID: https://orcid.org/0000-0001-5263-3406
Explor Drug Sci. 2023;1:210–220 DOI: https://doi.org/10.37349/eds.2023.00014
Received: February 22, 2023 Accepted: April 23, 2023 Published: June 30, 2023
Academic Editor: Marcello Iriti, Milan State University, Italy
The article belongs to the special issue Bioactive Peptides discovery and development
Aim: This study aims to report an engineered peptide zp39 with favorable bioactivity against enterohemorrhagic Escherichia coli (E. coli, EHEC). Its antibacterial mechanisms and application in a real food system are assessed.
Methods: Spatial conformation of synthetic peptide zp39 (GIIAGIIiKIKk-NH2, lowercase letters indicate dextrorotatory amino acids) was predicted by PEPstrMOD and its secondary structure was further determined by circular dichroism (CD) spectroscopy. Then, standard E. coli O157:H7 strain ATCC 43888 was used to evaluate the bioactivity of zp39. A double dilution method was applied to investigate its efficacy in normal broth medium, serum, and highly saline conditions. Its effects on cell membrane permeability and potential were measured by fluorescent assays. Thereafter, morphological changes of E. coli O157:H7 cells were monitored by electron microscopy technologies. Finally, the potential application of zp39 in controlling EHEC in food was tested with spinach juice and the Galleria mellonella larvae model was employed to assess the in vivo efficacy.
Results: Peptide zp39 presented an amphiphilic helical structure. It effectively inhibited the growth of E. coli O157:H7 at a concentration of 4 μmol/L in a bactericidal mode. Mechanistic studies revealed that it affected membrane permeability and potential in a dose-dependent manner. Moreover, zp39 maintained satisfactory bioactivity against E. coli O157:H7 even in the presence of 70% serum or 1,000 μmol/L chloride salts. In spinach juice application, > 90% E. coli O157:H7 cells were killed within 2 h after exposure to 64 μmol/L zp39. In vivo study proved that treatment with 64 μmol/L zp39 could effectively boost the survival ratio of infected larvae by 50%.
Conclusions: This study depicts a synthetic dodecapeptide that shows the potential application in controlling EHEC. This molecule may be developed into a highly effective antimicrobial agent applied to prevent food contamination and associated infections.
Enterohemorrhagic Escherichia coli (E. coli, EHEC) has long been recognized as an important foodborne pathogen [1]. Thereinto, E. coli O157:H7 is the most typical serotype, associated with strong toxicity and severe clinical symptoms [2]. EHEC O157:H7 is transmitted to humans primarily through consuming contaminated meat, dairy products, or vegetables because food is one of the most major carriers of this bacteria [3]. A multistate outbreak of E. coli O157:H7 in the USA, which led to 116 hospitalized and 5 deaths in 2006, was attributed to bagged spinach [4]. Eleven years later in 2017, another outbreak was found to be associated with E. coli O157:H7 contamination in soy nut butter, resulting in 32 cases (> 80% occurred in children) [5]. These incidents suggest the current food safety control strategy may not be sufficient to combat this foodborne pathogen. In this context, developing novel alternatives to control the rapid spread of E. coli O157:H7 is highly demanded.
Peptides and their derivatives are a class of antibacterial molecules demonstrating great potential [6]. To date, multiple peptide-based compounds have been used in the fields of medicine and food industry [7]. For instance, glycopeptide vancomycin was applied to treat bacterial infections, especially against Gram-positive strains [8]. Nisin, a naturally isolated cyclic peptide, is the only bacteriocin that was approved to be used as a food additive [9]. Pexiganan, a linear peptide with 22 residues, has achieved commoditization and is employed as a high-efficient template to design novel analogs [10]. Furthermore, the latest advances have shown that antimicrobial peptides could be promising weapons against E. coli O157:H7 [11, 12].
Previously, we have reported a linear engineered peptide GIIAGIIIKIKK-NH2 (denoted as zp3) showing potent bioactivity against E. coli ATCC 25922 with a minimal inhibitory concentration (MIC) at 8 μmol/L [13]. By substituting isoleucine at position 8 and lysine at position 12 to their corresponding dextrorotatory enantiomers, the new peptide GIIAGIIiKIKk-NH2 (denoted as zp39) has shown better protease stability. In the present study, we investigated its potential application in controlling EHEC O157:H7 which has been proven to cause more damage to the human body than E. coli ATCC 25922 [14], offering better clinical insights into its potential application.
Peptide zp39 was customized by Synpeptide Co., Ltd (Nanjing, China) with > 90% purity. The structural model of zp39 was predicted by an online website PEPstrMOD (https://webs.iiitd.edu.in/raghava/pepstrmod/) [15]. The obtained predictive file was visualized by Molsoft interactive PDB fast viewer (https://molsoft.com/pdbv.html). Helical wheel projection was generated by NetWheels with a few modifications to highlight the unnatural residues (http://lbqp.unb.br/NetWheels/).
For circular dichroism (CD) spectroscopy, zp39 powder was dissolved in water (1 mM). Then, the peptide solution was diluted to 0.05 mM by 10%, 30%, 50%, 70%, and 90% trifluoroethanol (TFE) in water respectively. Spectra were recorded by a J-810 spectrometer (Jasco, Japan). Detailed parameters: room temperature; start wavelength 260 nm; end wavelength 190 nm; data interval 0.1 nm; bandwidth 1 nm; scanning speed 100 nm/min. The original [θ]obs was then transferred to [θ] by the formula: [θ] = [θ]obs/10rlc, where [θ]obs is the observed CD values; [θ] = residue ellipticity; r = amino acid residues (12); l = optical path (1 cm); c = sample concentration (0.05 mM). The helical ratio was roughly calculated by the formula: helical (%) = (–[θ]222 + 3,000)/39,000 [16], where [θ]222 is the calculated [θ] at the wavelength of 222 nm.
The standard E. coli O157:H7 strain, whose identifier is ATCC 43888, was obtained from our in-house bacterial bank [17]. The strain of ATCC 43888 does not produce either Shiga-like toxin I or II and does not possess the genes for these toxins. The MIC value of zp39 was determined following the broth double-dilution method [18]. Briefly, single-colony bacteria grown on Mueller-Hinton agar (MHA, Hopebio, China) was resuspended in Mueller-Hinton broth (MHB, Hopebio, China) to 600 nm optical density (OD600) at 0.1. Next, a total of 5 μL of the bacterial solution was mixed with 150 μL of MHB medium containing various amount of zp39 (0, 2, 4, 8, 16, and 32 μmol/L). The OD600 values were recorded by a microplate reader (CLARIOstar® Plus Microplate Reader, BMG labtech, Germany) after 18 h culture at 37℃. The MIC value was determined as the lowest peptide concentration that resulted in no observed bacterial growth.
E. coli O157:H7 was inoculated into MHB medium, and then mixed with various amount of zp39 (0, 2, 4, 8, 16, and 32 μmol/L). The bacterial cells were incubated at 37℃ and OD600 values were recorded every 2 h for 12 h.
The initial cell density of E. coli O157:H7 was adjusted to around 107 colony forming units (CFU) per milliliter (CFU/mL). Then, bacterial cells were treated with various amount of zp39 (0, 4, 16, and 32 μmol/L). At specific intervals (1, 2, 4, and 8 h), a sample of 10 μL aliquot was withdrawn for serial10-fold dilution. Prepared solutions (10 μL) were dropped on the surface of MHA plates and incubated overnight at 37℃. Subsequently, bacterial colonies (3–30 valid) were counted and the primary CFUs were calculated.
This experiment was performed according to the manual of the LIVE/DEADTM BacLightTM Bacterial Viability Kit (Invitrogen, USA). In brief, logarithmic phase E. coli O157:H7 cells were collected by 4,000 rpm centrifugation (Eppendorf 5418, Germany) and resuspended in sterile 0.9% NaCl solution (Dieckmann, Hong Kong). Then, bacterial cells were treated with various amount of zp39 for 4 h. After centrifugation, collection, and resuspension, cells were stained by the dye mixture (prepared as per the specification) for 20 minutes. E. coli O157:H7 cells were finally rinsed thrice and pipetted onto a clear microscope coverslip for imaging (Nikon Eclipse Ti2E Live-cell Fluorescence Imaging System, Japan).
These experiments were performed following the MIC broth double-dilution method [18], except fetal bovine serum (HyClone, USA) or salts (NaCl, CaCl2, MgCl2, Dieckmann, Hong Kong) were added to the medium. For serum test, MHB mediums were replaced with equivalent volumes of serum (10–70%) for individual groups during incubation. For salt test, extra salts (50–1,000 μmol/L) were added to MHB mediums during incubation. After 18 h culture at 37℃, the MIC value was determined as abovementioned protocol.
Cytotoxicity was determined by thiazolyl blue tetrazolium bromide (MTT, BioFroxx, Germany) assay [13]. Briefly, human embryonic kidney cells (HEK293) were treated with zp39 at various concentrations for 24 h. Next, a total of 10 μL MTT solution (5 mg/mL) was used to stain for another 4 h. The precipitates were dissolved in 150 μL dimethylsulfoxide (Dieckmann, Hong Kong). Absorbance values at a wavelength of 570 nm were recorded by a microplate reader (Clariostar, BMG LABTECH, Germany). The group treated with phosphate buffered saline (PBS, Sigma-Aldrich, USA) was set as a control (100% survival).
E. coli O157:H7 cells (OD600 = 0.6) were treated with zp39 (0, 4, 8, 16, and 32 μmol/L) for 4 h. Next, SYTOX Green (1 μmol/L, ThermoFisher, USA) was applied to stain for 10 minutes at room temperature. Under the excitation wavelength at 488 nm, emission spectrums of each group from 500 nm to 600 nm were recorded (Clariostar, BMG LABTECH, Germany). The maximum fluorescence units can be observed at 523 nm.
The potentiometric probe 3,3’-dipropylthiadicarbocyanine iodide [DiSC3(5)], ThermoFisher, USA) was applied to evaluate membrane potential dissipation [19]. Briefly, E. coli O157:H7 cells at the logarithmic phase were adjusted to OD600 value of 0.2 in PBS. Next, cells were stained by 0.5 μmol/L DiSC3(5) for 1 h. Then, cells were subjected to the treatment of PBS, valinomycin (1 μmol/L), and zp39 (4 μmol/L and 16 μmol/L) respectively for 5 minutes. Finally, the fluorescence units were measured using a microplate reader (Clariostar, BMG LABTECH, Germany) with an excitation wavelength of 610 nm and an emission wavelength of 660 nm.
E. coli O157:H7 cells (OD600 = 0.6) were divided into two groups. One was treated with PBS and another was treated with 32 μmol/L zp39 for 2 h. Afterward, cells were collected and fixed with 2.5% glutaraldehyde (Tokyo Chemical Industry, Japan) at 4℃ overnight. Then, cells were subjected to a series of gradient dehydration by ethanol (Dieckmann, Hong Kong) solutions (50, 70, 90, and 100%, 10 minutes for each concentration). Finally, the specimens were air-dried, coated with gold, and visualized under the scanning electron microscopy (SEM) apparatus (Tescan VEGA3, Czech).
The preparation of E. coli O157:H7 cells was the same with SEM till fixed in 2.5% glutaraldehyde. Subsequently, cells were washed with PBS and stained with 2% osmium tetroxide (Sigma-Aldrich, USA). Next, cells were dehydrated with 100% ethanol (Dieckmann, Hong Kong) for 10 minutes. A total of 2 μL solution was added to a copper grid for scanning transmission electron microscopy (STEM) imaging (JEOL JEM-2100F, Japan).
Fresh spinach leaves purchased from a local market were squeezed to get the semisolid mash. The solid precipitate was then removed with filter paper. The remaining spinach juice was sterilized through a 0.22 μm filter. Next, E. coli O157:H7 cells were added to tested juice samples with the final concentration of ~ 106 CFU/mL. Then, spinach juice was divided into four groups and treated with 0, 4, 16, and 64 μmol/L zp39, respectively, for 2 h at 4℃. Finally, a total of 10 µL solution was pipetted on the MHB plate from each group. On the second day, the colony for each plate was counted and the original live bacteria was calculated accordingly. All testing concentrations were performed in quintuplicate.
Spinach juice was prepared as above. E. coli O157:H7 cells were added into the juice with the final concentration of ~106 CFU/mL. Then, spinach juice was divided into six groups and five of them were treated with PBS, 4, 16, 64 μmol/L zp39, and 64 μmol/L meropenem respectively, for 2 h at 4℃. Subsequently, a total of 10 μL juice was injected into each larva (10 larvae per group). The survival ratio was monitored for 36 h. The larvae group injected with sterile spinach was set as a negative control and the larvae group injected with PBS-treated E. coli O157:H7-contaminated juice was set as a positive control.
All experiments were performed in triplicate unless specified. The results were expressed as mean ± standard error. The normality of data was tested by Shapiro-Wilk test. Levene test was used to check for homogeneity of variances from three or more samples. Difference was analyzed by one-way analysis of variance (ANOVA) and Duncan test. The F-test was used to check for homogeneity of two variances and the Student’s t-test was used to check for differences. * represents P < 0.05, ** represents P < 0.01, *** represents P < 0.001, and **** represents P < 0.0001.
The chemical structure of zp39 is depicted in Figure 1A. Note that two unnatural amino acids, dextrorotatory isoleucine, and dextrorotatory lysine are highlighted in blue. The predicted spatial conformation is shown in Figure 1B. As illustrated in Figure 1C, basic polar residues (lysine) are presented in the pink diamond shape while nonpolar residues (glycine, isoleucine, and alanine) are presented in the green hexagon shape. Two amino acids enclosed by hollow purple squares are dextrorotatory residues. They are linked by gray lines to generate a helical wheel projection.
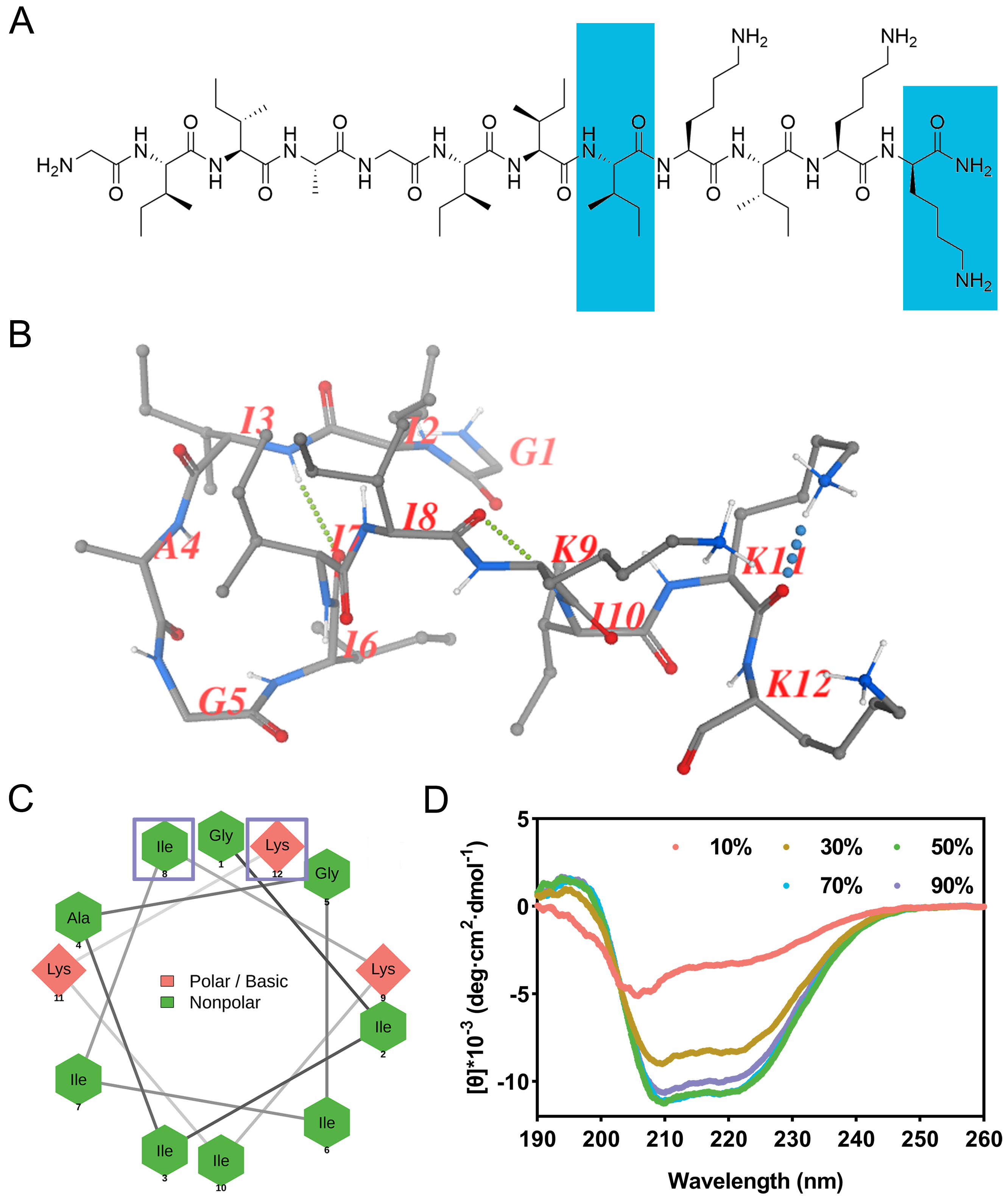
Structural characterization. (A) Chemical structure of zp39 and dextrorotatory residues are highlighted in blue; (B) predicted spatial conformation of zp39; (C) helical wheel projection of zp39 and dextrorotatory residues are enclosed by hollow purple squares; (D) CD spectra of zp39 in different ratio of TFE solutions
In CD spectra (Figure 1D), negative peaks at 208 nm and 222 nm can be detected for all testing conditions. As the ratio of TFE increased from 10% to 50%, the tendency of zp39 to form a helical structure was more obvious. According to the algorithm of beta-structure selection (BeStSel), the helical conformation ratio increased gradually from 4.0% (in 10% TFE) to 24.6% (in 50% TFE). As the proportion of TFE continued to rise, it had little effect on the helical structure. The summary of the proportion of each structure in different concentrations of TFE was shown in Table 1.
The proportion of each structure in different concentrations of TFE
| TFE (%) | Helix | Antiparallel | Parallel | Turn | Others |
|---|---|---|---|---|---|
| 10 | 4.0% | 28.9% | 2.4% | 16.0% | 48.7% |
| 30 | 19.7% | 12.5% | 7.2% | 16.0% | 44.6% |
| 50 | 24.6% | 13.2% | 7.1% | 15.2% | 39.8% |
| 70 | 23.8% | 13.1% | 7.3% | 14.4% | 41.4% |
| 90 | 23.5% | 12.6% | 7.0% | 14.9% | 41.9% |
Adopting the broth double dilution method, the MIC of zp39 against E. coli O157:H7 ATCC 43888 was determined as 4 μmol/L. Growth kinetics in the presence of zp39 was monitored in Figure 2A. After 12 h of incubation, no obvious increases in OD600 values were detected in testing bacterial samples containing 4, 8, 16, and 32 μmol/L zp39. In contrast, the OD600 value of the control group (in the absence of zp39) increased to approximately 1.8. For the 2 μmol/L treatment group, the growth rate comparatively slowed down to a certain extent, but it still reached more than 1.5 in 12 h.
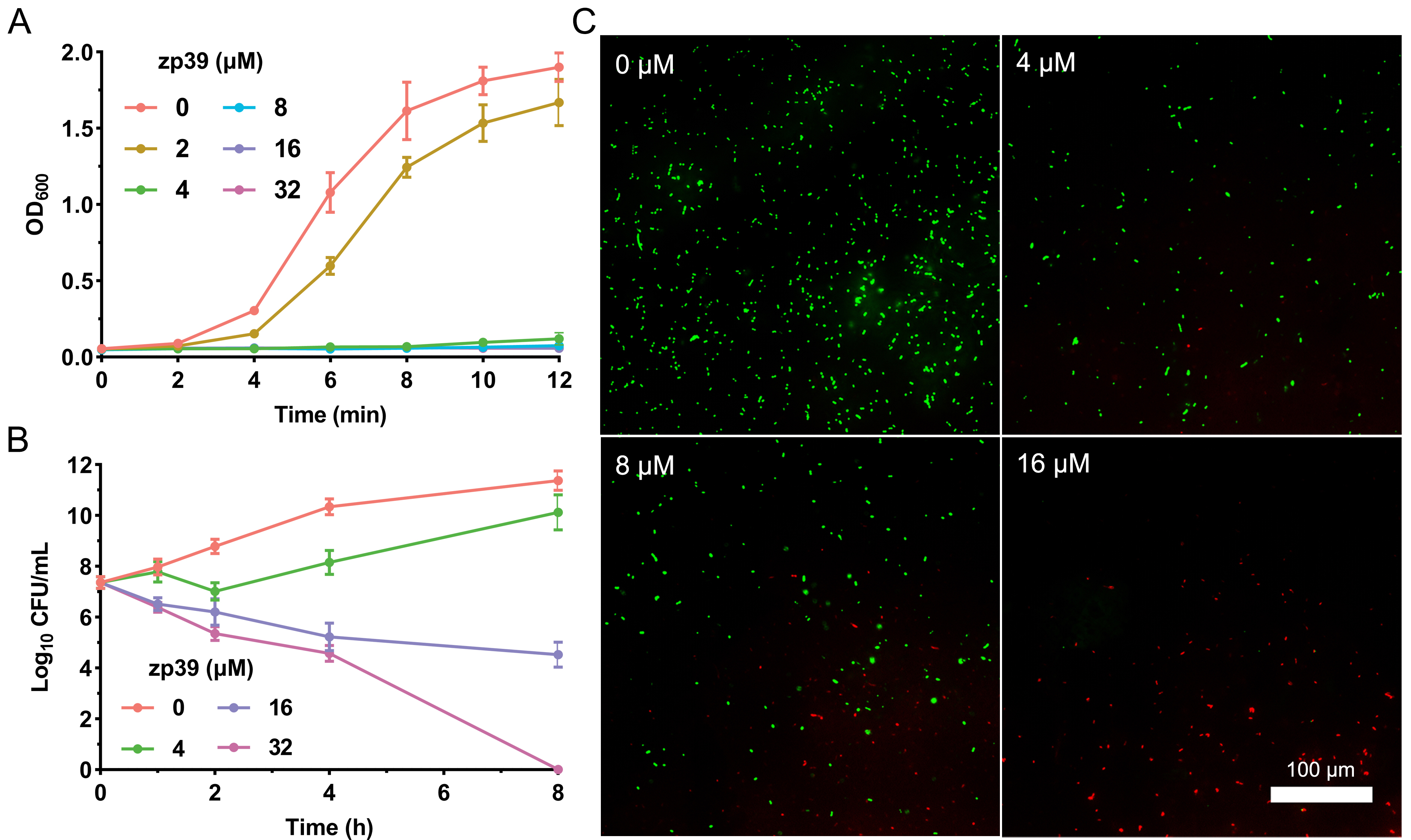
Bioactivity tests of zp39 against E. coli O157:H7. (A) Growth curve; (B) time-kill curve; (C) LIVE/DEAD staining
The time-kill curve illustrated in Figure 2B indicated that within 4 h, live colonies of E. coli O157:H7 cells decreased from 7.4 log10 CFU/mL to 4.6 log10 CFU/mL in the presence of 32 μmol/L zp39. After 8 h of incubation, the CFU in the highest concentration group was below the lowest detection limit. For other testing groups, live colonies were 11.4 (0 μmol/L), 10.1 (4 μmol/L), and 4.5 (16 μmol/L) log10 CFU/mL respectively.
For LIVE/DEAD staining shown in Figure 2C, E. coli O157:H7 cells emitted bright green fluorescence without zp39 treatment. While after incubating with zp39, the green fluorescence gradually decreased and the red fluorescence gradually increased in the visual field in a dose-dependent manner from 0 μmol/L to 4, 8, and 16 μmol/L respectively.
Serum and salt tolerance of zp39 was determined by broth double dilution method. As shown in Figure 3A, in the environment containing serum, the MIC of zp3 and zp39 against E. coli O157:H7 gradually increased. In the presence of 60% serum, fold change in MIC of zp3 reached 64 μmol/L and that of zp39 was 16 μmol/L. For salt tolerance tests (Figure 3B), even adding extra 1,000 μmol/L chlorides, zp39 remained favorable antibacterial activity against E. coli O157:H7 (4, 8, and 16 μmol/L for NaCl, CaCl2, and MgCl2 groups, respectively).
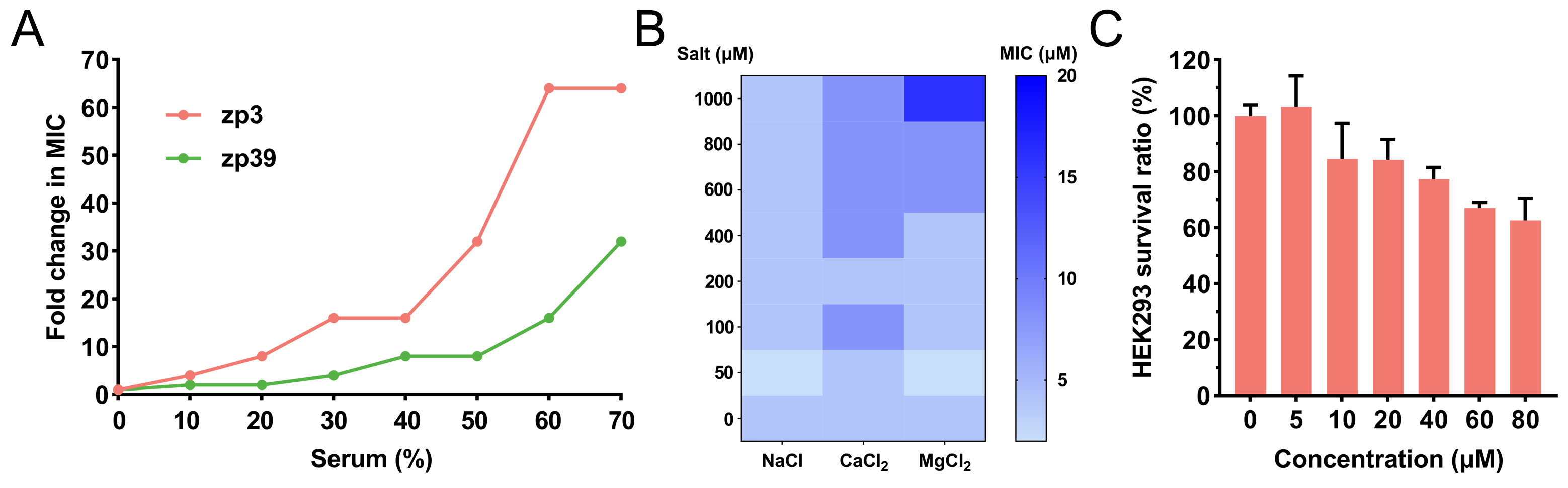
Bioactivities in varied situations and cytotoxicity. (A) Fold change in MIC of zp39 against E. coli O157:H7 in the presence of serum; (B) MIC of zp39 against E. coli O157:H7 in the presence of extra salts; (C) survival ratio of HEK293 cells after zp39 treatment
HEK293 cells are employed to evaluate the cytotoxicity of zp39 to mammalian cells (Figure 3C). Compared with the control group, HEK293 cells exposed to 80 μmol/L zp39 demonstrated a > 60% survival ratio.
SYTOX Green assay was applied to measure the membrane permeability change after the zp39 treatment (Figure 4A). Higher fluorescence units suggest enhanced cell membrane permeability. In this test, positive peaks at 523 nm (maximum emission wavelength) exhibited a significant dose-dependent effect. Compared with the control group (0 μmol/L), membrane permeability of zp39-treated E. coli O157:H7 cells increased 1.2-fold (4 μmol/L), 1.3-fold (8 μmol/L), 1.7-fold (16 μmol/L) and 3.0-fold (32 μmol/L), respectively.
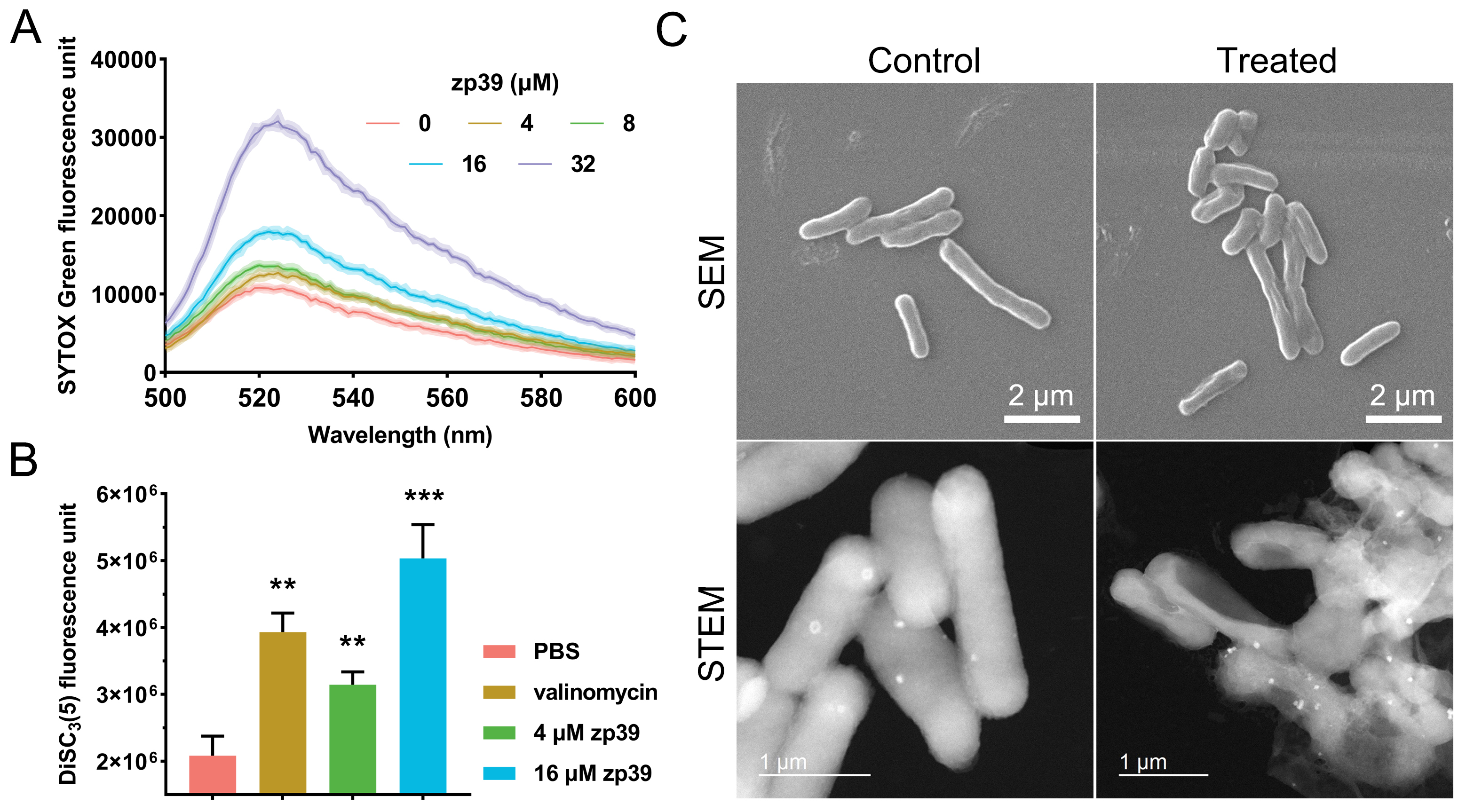
Mechanistic studies. (A) Membrane permeability test; (B) membrane potential test and valinomycin concentration was 1 μmol/L. ** P < 0.01, *** P < 0.001; (C) SEM and STEM images of E. coli O157:H7 before and after zp39 treatment
As indicated by Figure 4B, after merely 10 minutes of treatment by zp39, fluorescence units of E. coli O157:H7 membrane potential indicator DiSC3(5) have increased 1.5-fold (4 μmol/L) and 2.5-fold (32 μmol/L), showing a statistically significant difference with the PBS-treated group. As a reference, a well-known membrane potential modulator valinomycin (1 μmol/L) caused an approximate 2-fold signal increase under the same period.
Cellular morphologies of E. coli O157:H7 before and after zp39 treatment were captured by SEM and STEM (Figure 4C). Normally, bacterial cells appeared as plump rod shapes. However, zp39 treatment (32 μmol/L, 2 h) could lead to marked cellular shrinkage and deformation. In the STEM image of zp39 treated E. coli O157:H7, hollow cells, and even broken cellular fragments can be identified.
Next, the antibacterial activity of zp39 in spinach juice was investigated. The results shown in Figure 5A demonstrated that the number of living bacteria had dropped significantly after 16 and 64 μmol/L zp39 treatment in 2 h. On average, the values of log10 CFU/mL were 6.0 (control), 5.7 (4 μmol/L), 5.1 (16 μmol/L), and 4.4 (64 μmol/L), respectively. In other words, > 90% of E. coli O157:H7 cells were killed within 2 h in the presence of 64 μmol/L zp39.
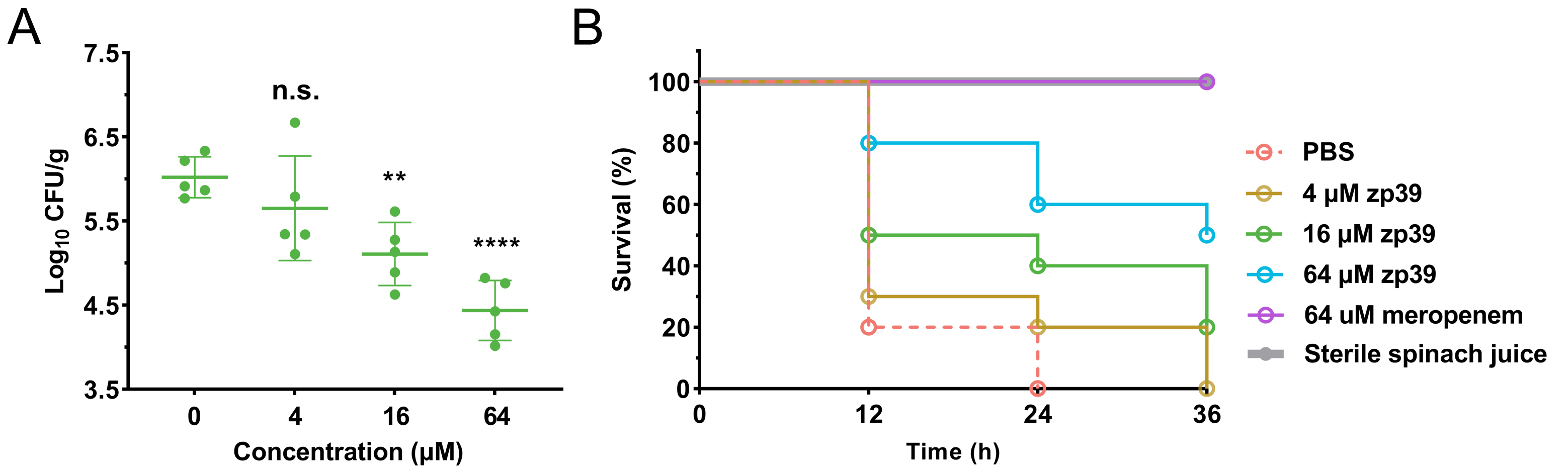
Antibacterial potency of zp39 in spinach juice. (A) Living bacterial colonies in the different treatment groups. ** P < 0.01, **** P < 0.0001, n.s. means no significant difference; (B) larvae survival ratio in the different treatment groups (n =10 in each group)
Feeding larvae with this contaminated spinach juice, survival curves for each testing group can be summarized (Figure 5B). For negative control which sterile spinach juice was injected, 100% of larvae survived after 36 h of exposure. For positive control, PBS-treated E. coli O157:H7-contaminated spinach juice was injected, all larvae died after 24 h of exposure. In the low concentration group (4 μmol/L), the final death time of 20% of larvae extended from 24 h to 36 h after E. coli O157:H7 infection. In medium concentration (16 μmol/L) and high concentration (64 μmol/L) groups, 20% and 50% of larvae survived, respectively, after 36 h of exposure.
In this work, we first synthesized peptide zp39 and investigated its possible conformation. CD data indicated that a modest amphiphilic environment like a TFE solution was conducive to the formation of the helical structure of zp39. Similar to many cationic amphiphiles [20], zp39 may change its spatial structure under the induction of the bacterial cell membrane, which could facilitate its binding to the surface of bacteria via electrostatic attraction and hydrophobic affinity.
The growth curve showed that zp39 could significantly reduce the reproduction rate of E. coli O157:H7. Subsequent time-kill assay further proved that zp39 worked in a bactericidal mode. LIVE/DEAD staining images hinted that zp39 probably inflicted damage on the membrane of E. coli O157:H7 cells because the dye propidium iodide can only bind to DNA through an impaired cell envelope, thus emitting red E. coli fluorescence [21].
Due to the incorporation of two dextrorotatory amino acids, zp39 is relatively robust against hydrolysis by proteases comparing with its analog zp3 containing no unnatural residues. Furthermore, the presence of a high concentration of salts had no significant impacts on the antibacterial activity of zp39 against EHEC, particularly attractive for its future application in foods with heavy salts [22]. Cytotoxicity test revealed that zp39 had a great selectivity index of > 20, determined as the half maximal inhibitory concentration to HEK293 cells (> 80 μmol/L) to the MIC against E. coli O157:H7 (4 μmol/L).
Membrane permeability is an important indicator of whether antibacterial molecules can enter the interior of bacteria [23]. Our study illustrated that peptide zp39 was able to augment the permeability of E. coli O157:H7, which may seriously hinder the normal physiological function of the cell membrane. Meanwhile, cellular membrane potential was dissipated after zp39 treatment, indicating that the interaction between zp39 and bacterial cell membrane could lead to prompt cationic electrical movement across the phospholipid bilayers. The influence on membrane permeability and potential was adverse to the existence of bacteria [24]. The attack of zp39 also distorted E. coli O157:H7 cells. Damaged peripheral structures, dissipation of membrane potential, and possible cytoplasmic leakage collectively contributed to the death of this pathogen.
In many cases, humans get infected by EHEC through the consumption of contaminated food [25]. Fresh-squeezed fruit/vegetable juices and ready-to-eat foods are especially risky because of the lack of high-temperature cooking and other sterilization procedures [26]. Worse still, they often contain a lot of ingredients, including salts and enzymes, which may reduce the bioactivity of antibacterial peptides. Here, we showed that the studied peptide zp39 could resist the influences of such factors and maintained a highly effective bactericidal effect in spinach juice. Its satisfactory stability may be partly due to two unnatural amino acids, dextrorotatory isoleucine, and lysine. Galleria mellonella assay also confirmed that zp39-treated E. coli O157:H7-contaminated spinach juice showed reduced toxicity to larvae. The improved survival ratio (at medium and high concentration groups) proved that zp39 may be a potential antibacterial agent in precaution of E. coli O157:H7-associated infection.
CD: circular dichroism
CFU: colony forming unit
DiSC3(5): 3,3’-dipropylthiadicarbocyanine iodide
E. coli: Escherichia coli
EHEC: enterohemorrhagic Escherichia coli
MHB: Mueller-Hinton broth
MIC: minimal inhibitory concentration
OD600: 600 nm optical density
PBS: phosphate buffered saline
SEM: scanning electron microscopy
STEM: scanning transmission electron microscopy
TFE: trifluoroethanol
We thank the University Research Facilities in Life Sciences (ULS) of The Hong Kong Polytechnic University for providing facilities.
PZ: Conceptualization, Methodology, Investigation, Writing—original draft. XY: Methodology. KYW: Resources, Supervision. SC: Resources, Supervision. KFC: Resources, Supervision. SSYL: Resources, Supervision, Project administration, Funding acquisition. LY: Writing—review & editing, Project administration, Funding acquisition. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
This work was supported by the National Natural Science Foundation of China [32102032] and the Health and Medical Research Fund Hong Kong [21200782].
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Qiushi Chen ... Xuechen Li
Shoko Tanaka ... Kohei Sato
Feliciana Real-Fernandez ... Paolo Rovero
Chloé O. Sebilleau, Steven J. Sucheck
Qian Zhang ... Chunqiu Zhang
Suze Ma ... Qi Zhang
Yanyan Liao, Xuefeng Jiang
Qian Wang ... Shiyu Chen
Jarais Fontaine, Jianfeng Cai
Oscar Noya ... Sandra Losada
