Affiliation:
1“Giulio A. Maccacaro” Laboratory of Medical Statistics and Biometry, Department of Clinical Sciences and Community Health, University of Milan, 20122 Milan, Italy
Email: danila.coradini@gmail.com
ORCID: https://orcid.org/0000-0003-3771-4341
Affiliation:
1“Giulio A. Maccacaro” Laboratory of Medical Statistics and Biometry, Department of Clinical Sciences and Community Health, University of Milan, 20122 Milan, Italy
2Scientific Directorate, IRCCS Policlinico San Donato, 20097 San Donato Milanese, Italy
ORCID: https://orcid.org/0000-0001-9358-011X
Explor Med. 2023;4:1079–1093 DOI: https://doi.org/10.37349/emed.2023.00196
Received: July 31, 2023 Accepted: October 26, 2023 Published: December 29, 2023
Academic Editor: Andrea Nicolini, University of Pisa, Italy
The article belongs to the special issue Breast Cancer: Basic and Clinical Advances
Aim: Cholesterol is an essential component of cell membranes and serves as a precursor for several bioactive molecules, including steroid hormones and isoprenoids. Generally supplied by the bloodstream, the de novo cholesterol biosynthesis is activated in response to an increased cell requirement due to normal tissue remodeling or tumor proliferation. In estrogen receptor (ER)-positive breast cancers, cholesterol biosynthesis may promote and sustain tumor growth and concur with the failure of the treatment with aromatase inhibitors.
Methods: In this study, the comparison of gene compared the expression involved in cholesterol biosynthesis was conducted in ER-positive tumors that were responsive and nonresponsive to letrozole; besides, an exploration of their association with genes implicated in estrogen production, the Hippo pathway, and cell cycle control was performed.
Results: In responsive tumors, letrozole significantly decreased the expression of five genes [acetyl-coenzyme A (CoA) acetyltransferase 2 (ACAT2), 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), farnesyl diphosphate synthase (FDPS), and squalene epoxidase (SQLE)] crucial for the biosynthetic process. Conversely, in nonresponsive tumors, these genes were unaffected by letrozole but associated with several genes involved in estrogens production [cytochrome P450 family 19 subfamily A member 1 (CYP19A1), hydroxysteroid 17-beta dehydrogenase 2 (HSD17B2), and sulfotransferase family 1A member 1 (SULT1A1)], cell cycle [control cyclin dependent kinase 4 (CDK4) and CDK6], and Hippo pathway [Yes1 associated transcriptional regulator (YAP1) and baculoviral inhibitor of apoptosis (IAP) repeat containing 5 (BIRC5)].
Conclusions: The findings corroborated the notion that the dysregulation of the mevalonate pathway may contribute to the resistance to letrozole and supported the use of statins to contrast this metabolic dysfunction.
Approximately 80% of postmenopausal breast cancer are estrogen receptor (ER)-positive and, for this reason, usually treated with endocrine therapy. Tamoxifen has been the conventional long-term treatment for several years. Then, to overcome the emergence of tumor resistance, it was first flanked and then replaced by aromatase inhibitors (AIs) both in operable and advanced ER-positive breast cancer achieving excellent results as progression-free and survival time [1–3].
Despite the recognized superiority of AIs over tamoxifen, the response rate in the neoadjuvant setting is only 50% to 70% because of an acquired or de novo resistance to estrogen deprivation therapy. Several putative mechanisms of resistance, mainly based on in vitro experiments, have been proposed. They include intrinsic resistance of tumors to estrogen, aromatase-independent estrogenic hormones, signal transduction by non-endocrine pathways, and selection of hormone-insensitive clones during AI therapy [4, 5], none of which are entirely convincing.
Cholesterol is an essential component of cell membranes because required for membrane composition and fluidity. In addition to this activity at the membrane level, cholesterol is the precursor in several biochemical pathways, including the synthesis of steroid hormones. Generally supplied by the bloodstream, the de novo cholesterol biosynthesis can be activated in response to the increased cell requirement during the physiological tissue remodeling and the proliferation of cancer cells, especially if steroid hormone-dependent. In these latter, it concurs in promoting and sustaining cell growth as the precursor of steroid hormones and may contribute to the failure of the treatment with AIs.
Recent findings [6] indicated that the mevalonate pathway, the core of the biosynthetic process leading to cholesterol, may interplay with the Hippo signaling pathway through the geranylgeranyl pyrophosphate (GG-PP) that plays an essential role in protein isoprenylation, a process which allows some proteins, including small guanosine triphosphatases (GTPases), such as cell division cycle 42 (Cdc42), ras-related C3 botulinum toxin substrate 1 (Rac1), and ras homolog family member A (RhoA), to be anchored at the plasma membrane and enzymatically activated.
Cumulating evidence indicated that the augmented production of isoprenoids, due to the overexpression of the gene coding for the 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase and the following increase in mevalonic acid level, results in the isoprenylation and activation of the small GTPases that interplay with the Hippo signaling pathway by controlling the phosphorylation of Yes-associated protein (YAP)/transcriptional co-activator with PDZ-binding motif (TAZ), the main downstream effectors [7]. Once anchored to the plasma membrane by isoprenylation and activated, GTPases inhibit YAP/TAZ phosphorylation allowing the translocation into the nucleus, the binding to transcriptional enhancer factor (TEA) domain family member 4 (TEAD) transcription factors, and the expression of a wide variety of genes involved in essential physiological processes, such as stem cell functions, cell proliferation and differentiation, organ growth, tissue regeneration, and embryonic development [8–11].
Therefore, the experimental findings indicating that the abnormal accumulation of YAP/TAZ into the nucleus may promote the neoplastic transformation by stimulating cell proliferation, inhibiting apoptosis, inducing the acquisition of additional stem cell properties, and supporting cancer cell dissemination by reducing cell contact inhibition, and triggering epithelial to mesenchymal transition, are expected [12–15]. Likewise, it is unsurprising that several solid tumors, including breast cancer (especially those poorly differentiated), show a nuclear accumulation of YAP/TAZ [16, 17].
The studies also showed that, in tumor cells, the inhibition of the HMG-CoA reductase activity by statins, causing isoprenoid depletion and the unsuccessful activation of small GTPases, promoted the phosphorylation of YAP/TAZ, which is sequestered in cytoplasm and degraded, thus blocking gene transcription [18, 19].
The aim of this in silico study was to compare the expression of the genes involved in the biosynthesis of cholesterol in responsive and nonresponsive tumors from patients treated with a neoadjuvant third-generation AI (letrozole) and explore their association with the genes involved in estrogens production, Hippo signaling pathway, and cell cycle control to verify the potentiality of statins as an additional treatment to overcome the resistance to AIs.
The study was carried out using a publicly accessible dataset retrieved from the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), identified by the GEO accession number GSE5462, and that responded to the following selection criteria: an adequate number of sequential biopsies from the same tumor before and after a short treatment with letrozole; availability of information about the clinical response; gene expression profile obtained using a commercial and therefore reproducible platform.
As described in the original articles [20, 21], postmenopausal patients with large operable or locally advanced breast cancer were recruited prospectively into a neoadjuvant study with the AI letrozole (2.5 mg/day, oral). Women with ER-positive breast cancer had tumor biopsies taken before and after 10 to 14 days of treatment and were assessed for clinical response (based on changes in tumor volume) at three months. Fifty patients had available paired tumor samples: 36 responsive and 14 nonresponsive tumors. All patients consented to provide excess tissues for research purposes, and the study was approved by the ethics committee of the University of Edinburgh, Edinburgh, UK (2001/W/BU/09 and 2001/W/BU/10).
Gene expression data, obtained using Affymetrix Human Genome U133A Array (GEO accession GPL96), were provided as filtered and log2 transformed expression estimates.
According to the aim of the study, a panel of 37 genes was chosen that included: 13 genes coding for the enzymes involved in the biosynthesis of cholesterol (Figure 1), 9 involved in the synthesis of estrogens from cholesterol (Figure S1), 10 coding the essential elements of the Hippo signaling pathway and some representative YAP/TAZ target genes [namely, baculoviral inhibitor of apoptosis (IAP) repeat containing 5 (BIRC5), cellular communication network factor 1 (CCN1), and CCN2] [22, 23] (Figure S2), and 5 [cyclin D1 (CCND1), cyclin dependent kinase 4 (CDK4), CDK6, CDK inhibitor 1A (CDKN1A), and CDKN1B] involved in cell cycle control. The latter genes were chosen on the notion that the estrogen-driven activation of the cyclin D-CDK4/CDK6 complex is essential for the physiologic mammary development and proliferation of ER-positive breast cancers [24, 25], and CDKN1A (P21, Cip1) and CDKN1B (P27, Kip1) are pivotal players in the control of the G1-phase of the cell cycle (details on the selected genes in Table S1).
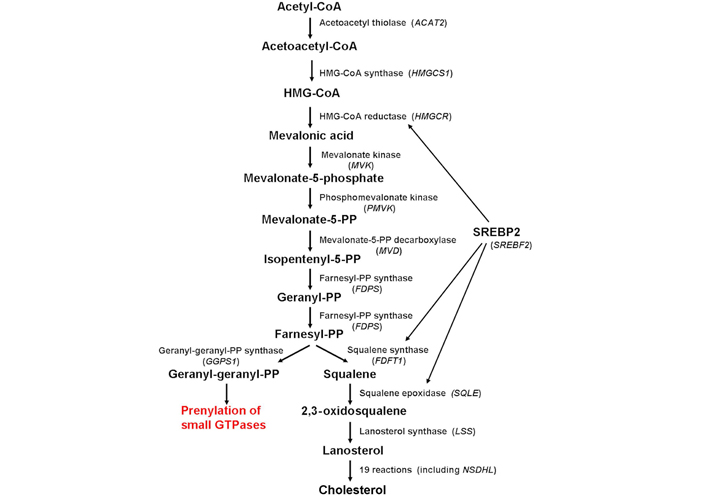
Schematic diagram depicting the enzymes involved in cholesterol biosynthesis (in parenthesis, the corresponding gene). Cholesterol biosynthesis is regulated mainly by the SREBP2, which controls the expression of several genes involved in the biosynthetic pathway, including HMGCR, FDFT1, and SQLE. ACAT2: acetyl-coenzyme A (CoA) acetyltransferase 2; HMGCS1: HMG-CoA synthase 1; HMGCR: HMG-CoA reductase; MVK: mevalonate kinase; PMVK: phosphomevalonate kinase; MVD: mevalonate PP decarboxylase; FDPS: farnesyl diphosphate synthase; FDFT1: farnesyl-diphosphate farnesyltransferase 1; SQLE: squalene epoxidase; LSS: lanosterol synthase; GGPS1: GG diphosphate synthase 1; SREBP2: sterol regulatory element-binding protein-2; SREBF2: sterol regulatory element-binding factor-2; NSDHL: nicotinamide adenine dinucleotide (phosphate) dependent steroid dehydrogenase-like
As each gene may be recognized by several probes, only the best probes were considered for the analysis. Best probes were selected based on the sensitivity and specificity score, where the sensitivity score is the fraction of probes in a probe set that match the gene, and the specificity score indicates to what extent probes of a probe set bind to genes (GeneAnnot version 2.2). When a gene was recognized by two or more best probes, the mean was calculated to obtain a single value of the expression. The nonparametric Wilcoxon signed-rank test for paired samples was used to assess the differential expression of a given gene before and after treatment. The principal component analysis (PCA) was applied to summarize and visualize, as a biplot, the relationship among the different sets of genes (namely, cholesterol biosynthesis, estrogens production, hippo pathway, and proliferation) [26]. In each biplot, any vector represents the two-dimensional projection of the position of the corresponding variable in the multidimensional space. Results interpretation is based on the length, direction, and position of each vector. Specifically, the length of the vector approximates the quality of the representation of the variable: the longer the vector better the variable representation. The direction of the vector reflects the component to which the variable is most strongly related. The position of vectors represents their pairwise correlation: Vectors close to one another are positively correlated, whereas negatively correlated variables are positioned on the opposite side. If vectors are orthogonal, the variables have a small or null correlation. Spearman’s correlation coefficient was used to describe the correlation between genes. All analyses were performed using the open-source software R, version 4.1.2 (http://www.r-project.org), and, in particular, FactoMineR (for the analysis), factoextra (for data interpretation), and ggpubr (for data visualization) packages were used to perform PCA. P < 0.05 was considered statistically significant.
Wilcoxon paired test showed a significant decrease in the expression level of several genes involved in cholesterol biosynthesis in responsive tumors. In particular, a decreased expression level was observed in ACAT2, which encodes the first enzyme of the biosynthetic chain, HMGCS1 and HMGCR, which encode the two enzymes that trigger the mevalonate pathway, FDPS, which codes for the enzyme that catalyzes the synthesis of farnesyl PP (FPP), the precursor for the synthesis of isoprenoids, and SQLE, which encodes the key enzyme for cholesterol commitment (Figure 2). Conversely, no decrease was observed in nonresponsive tumors.
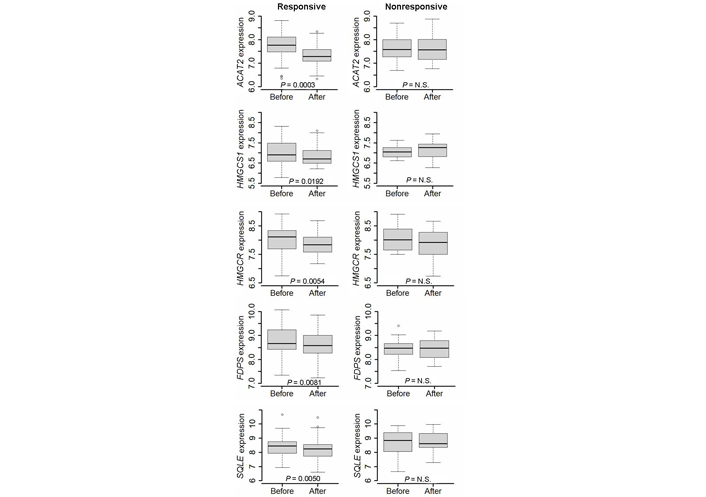
Differential expression of ACAT2, HMGCS1, HMGCR, FDPS, and SQLE gene before and after a 2-week treatment with letrozole in responsive and nonresponsive tumors. Wilcoxon signed-rank test for paired samples was applied. N.S.: non-significant
PCA indicated that the basal expression of the five genes modulated by letrozole showed a different pattern of expression in responsive and nonresponsive tumors (Figure 3).
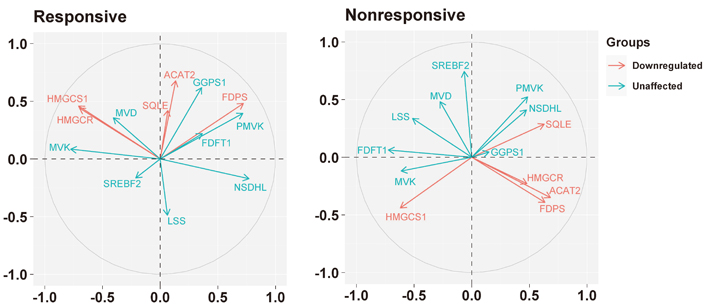
Biplots visualizing the relationship among the genes downregulated by the letrozole treatment (“downregulated”) and the other genes involved in the biosynthesis of the cholesterol (“unaffected”) in responsive and nonresponsive tumors. Correlation between variables is represented by arrows (vectors) within the correlation circle of radius 1. The length of each vector approximates the quality of the representation of the variable: the longer the vector better the variable representation. The position of vectors represents their pairwise correlation: Vectors close to one another are positively correlated, whereas negatively correlated variables are positioned on the opposite side. If vectors are orthogonal, the variables have a small or null correlation
Indeed, in responsive tumors, HMGCR was associated positively with HMGCS1 (r = 0.56, P = 0.0004), and both with MVK (r = 0.54, P = 0.0007, and r = 0.59, P = 0.0002, respectively) and MVD (r = 0.35, P = 0.0386 and r = 0.49, P = 0.0023, respectively); besides, HMGCR and HMGCS1 were associated negatively with NSDHL (r = –0.58, P = 0.0002 and r = –0.56, P = 0.0003, respectively). ACAT2 was associated positively with SQLE (r = 0.41, P = 0.0123), while FDPS was positively associated with PMVK (r = 0.74, P < 0.0001), GGPS1 (r = 0.39, P = 0.0186) and NSDHL (r = 0.35, P = 0.0329).
Conversely, in nonresponsive tumors, ACAT2, HMGCR, and FDPS were associated positively with each other (specifically, ACAT2 vs. HMGCR: r = 0.53, P = 0.0567; ACAT2 vs. FDPS: r = 0.46, P = 0.0972; HMGCR vs. FDPS: r = 0.52, P = 0.0591), and with SQLE (r = 0.43, P = 0.1239; r = 0.34, P = 0.2334; r = 0.44, P = 0.1198, respectively). Besides, ACAT2 and HMGCR were associated negatively with FDFT1 (r = –0.50, P = 0.0704 and r = –0.44, P = 0.1154, respectively). Regarding HMGCS1, it was associated positively with FDFT1 (r = 0.59, P = 0.0271) and MVK (r = 0.39, P = 0.1625) and negatively with PMVK (r = –0.36, P = 0.2129), while FDPS, in addition to the association with ACAT2 and HMGCR, was associated positively with PMVK (r = 0.44, P = 0.1195). SQLE, in addition to the association with ACAT2, HMGCR, and FDPS, was associated positively with NSDHL (r = 0.60, P = 0.0249) and negatively with MVK (r = –0.46, P = 0.1017) and FDFT1 (r = –0.40, P = 0.1510).
The basal relationship between the genes downregulated in responsive tumors and those involved in the production of estrogens differed in nonresponsive tumors compared to responsive ones (Figure 4). In responsive tumors, HMGCS1 and HMGCR were associated positively with hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 (HSD3B2, r = 0.58, P = 0.0002 and r = 0.47, P = 0.0036, respectively) and steroid sulfatase (STS, r = 0.55, P = 0.0006 and r = 0.46; P = 0.0045, respectively); HMGCS1 was also associated positively with cytochrome P450 family 11 subfamily A member 1 (CYP11A1, r = 0.49, P = 0.0023) and CYP17A1 (r = 0.38, P = 0.0218); HMGCR was associated negatively, respectively, with sulfotransferase family 1A member 1 (SULT1A1, r = –0.61, P < 0.0001) and FDPS was associated negatively, with STS (r = –0.58, P = 0.0002).
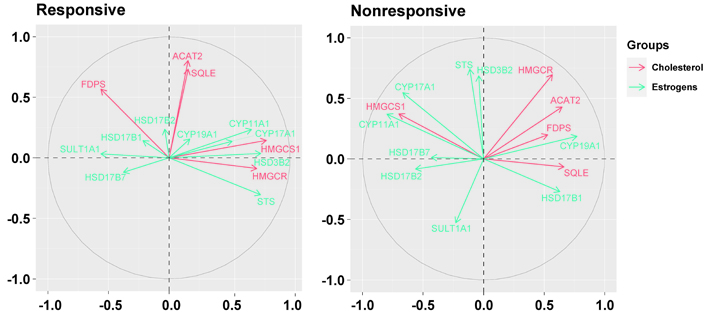
Biplots visualize the relationship among the genes downregulated by the letrozole treatment (“cholesterol”) and the genes involved in the synthesis of estrogens (“estrogens”) in responsive and nonresponsive tumors. Correlation between variables is represented by arrows (vectors) within the correlation circle of radius 1. The length of each vector approximates the quality of the representation of the variable: the longer the vector better the variable representation. The position of vectors represents their pairwise correlation: Vectors close to one another are positively correlated, whereas negatively correlated variables are positioned on the opposite side. If vectors are orthogonal, the variables have a small or null correlation. HSD17B2: hydroxysteroid 17-beta dehydrogenase 2
Conversely, in nonresponsive tumors, ACAT2 and HMGCR were associated positively with CYP19A1 (r = 0.53, P = 0.0522 and r = 0.49, P = 0.0809, respectively) which, conversely, was associated negatively with HMGCS1 (r = –0.59, P = 0.0249). Besides, while HMGCS1 was associated positively with CYP11A1 (r = 0.69, P = 0.0064) and CYP17A1 (r = 0.61, P = 0.0199) similarly to responsive tumors, FDPS and SQLE were associated negatively with CYP17A1 (r = –0.43, P = 0.1239 and r = –0.50, P = 0.0694, respectively) and SQLE was associated negatively also with CYP11A1 (r = –0.48, P = 0.0840). Furthermore, HMGCR and FDPS were associated negatively with HSD17B2 (r = –0.39, P = 0.1656, and r = –0.55, P = 0.0423, respectively), and FDPS was associated negatively also with STS (r = –0.43, P = 0.1281).
PCA showed that, in the nonresponsive tumors, the expression of the genes downregulated in responsive tumors had a different association profile with the genes involved in the Hippo pathway compared to responsive tumors (Figure 5).
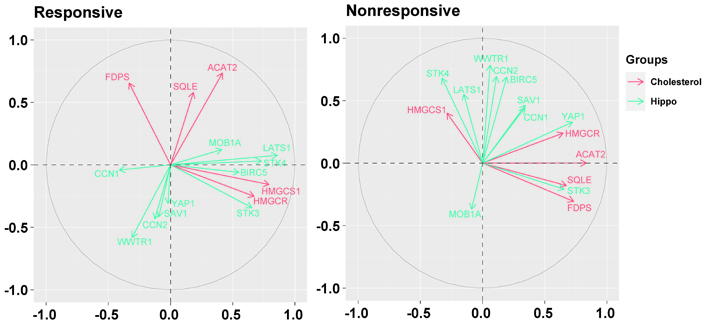
Biplots visualizing the relationship among the genes downregulated by the letrozole treatment (“cholesterol”) and the genes involved in the Hippo pathway (“hippo”) in responsive and nonresponsive tumors. Correlation between variables is represented by arrows (vectors) within the correlation circle of radius 1. The length of each vector approximates the quality of the representation of the variable: the longer the vector better the variable representation. The position of vectors represents their pairwise correlation: Vectors close to one another are positively correlated, whereas negatively correlated variables are positioned on the opposite side. If vectors are orthogonal, the variables have a small or null correlation. LATS1: large tumor suppressor kinase 1; MOB1A: Mps one binder (MOB) kinase activator 1A; SAV1: salvador family WW domain containing protein 1; STK3: serine/threonine kinase 3; WWTR1: WW domain containing transcription regulator 1; YAP1: Yes1 associated transcriptional regulator
In particular, in nonresponsive tumors, HMGCR and FDPS were associated positively with YAP1 (r = 0.38, P = 0.1767 and r = 0.43, P = 0.1237, respectively), and HMGCR showed a positive association also with BIRC5 (r = 0.49, P = 0.0764), one of most representative genes regulated by the YAP/TAZ complex. Besides, ACAT2 and FDPS were associated positively with STK3 (r = 0.70, P = 0.0058 and r = 0.43, P = 0.1216, respectively), and FDPS and SQLE were associated negatively with STK4 (r = –0.46, P = 0.0945 and r = –0.39, P = 0.1635, respectively).
Conversely, in responsive tumors, HMGCS1 and HMGCR were associated positively with STK3 (r = 0.62, P < 0.0001 and r = 0.38, P = 0.0210, respectively), and LATS1 (r = 0.61, P < 0.0001 and r = 0.38, P = 0.0211, respectively).
As shown in Figure 6, in nonresponsive tumors, all but the HMGCS1 gene were associated negatively with CDK6 (specifically, ACAT2: r = –0.56, P = 0.0377; HMGCR: r = –0.52, P = 0.0546; FDPS: r = –0.54, P = 0.0477; SQLE: r = –0.62, P = 0.0184). Besides, in addition to the positive association with CDK6 (r = 0.47, P = 0.0918), HMGCS1 was associated negatively with CCND1 (r = –0.49, P = 0.0756) and CDK4 (r = –0.44, P = 0.1129). In responsive tumors, FDPS was associated positively with CDK4 (r = 0.40, P = 0.0145).
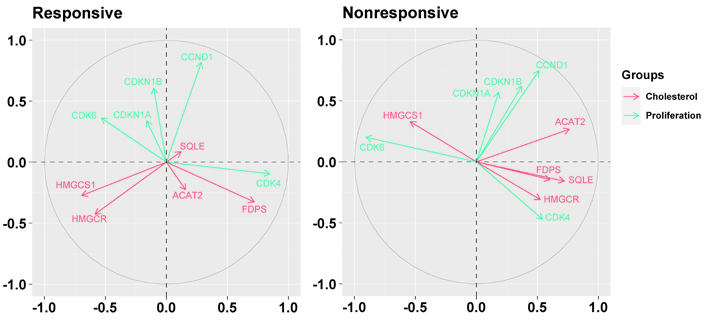
Biplots visualizing the relationship among the genes downregulated by the letrozole treatment (“cholesterol”) and the genes involved in cell cycle control (“proliferation”) in responsive and nonresponsive tumors. Correlation between variables is represented by arrows (vectors) within the correlation circle of radius 1. The length of each vector approximates the quality of the representation of the variable: the longer the vector better the variable representation. The position of vectors represents their pairwise correlation: Vectors close to one another are positively correlated, whereas negatively correlated variables are positioned on the opposite side. If vectors are orthogonal, the variables have a small or null correlation
Present results indicated that in responsive tumors, the treatment with letrozole was associated with a statistically significant decrease in the expression level of five genes that play a crucial role in the biosynthesis of cholesterol: ACAT2, which encodes the first enzyme of the biosynthetic chain; HMGCS1 and HMGCR, which encode the two enzymes that initiate the mevalonate pathway; FDPS, which codes for the enzyme that catalyzes the two-steps condensation of dimethylallyl PP with two molecules of isopentenyl PP (IPP) to form FPP, the precursor of GG-PP; SQLE, which encodes the enzyme that commits cholesterol biosynthesis irreversibly.
This finding could appear unexpected given the specific mechanism of action of letrozole, which inhibits the aromatase by competitively binding to the heme of the cytochrome P450 of the enzyme. However, because some of these genes (especially HMGCR and FDPS) [27, 28] are regulated by estrogen through a functional estrogen response element (ERE) in the promoter region, the decrease in the expression level observed in responsive tumors can be ascribed to the inhibition of the aromatase activity that reduced the estradiol availability. This hypothesis agrees with the finding that, in nonresponsive tumors, the five genes had no evidence of modulation but, taken as a whole, they were associated with several genes involved in estrogen biosynthesis. Indeed, they associated positively with CYP19A1, the gene coding for the aromatase, and negatively with HSD17B2 and SULT1A, which code, respectively, the enzyme that converts the estradiol to the less active estrone and the enzyme that neutralizes the estradiol by converting it into estradiol-sulfate. That is in agreement with the functional cooperation of cholesterol biosynthesis and estradiol production in the promotion and support of cancer cell proliferation: Estradiol acts as a potent growth factor for ER-positive cancer cells, and cholesterol plays the double role of an essential component of cell membranes and precursor for estrogen biosynthesis. Therefore, it is not surprising that, in nonresponsive tumors, the expression of the five genes was associated with that of the genes involved in cell cycle control, and in particular, with CDK4 and CDK6, the genes coding the two CDKs that drive the G1-phase of the cell cycle. Indeed, all but HMGCS1 genes were associated positively with CDK4 and negatively with CDK6, recently recognized as a novel downregulated estrogen-responsive gene that prevents cell proliferation and regulates cell cycle exit during cell differentiation [29].
In addition to the association with the genes involved in cell proliferation, HMGCR, and FDPS were associated positively with YAP1, which codes for the main transcriptional coactivator regulated by the Hippo signaling pathway (Figure 7), and BIRC5, one of the most representative YAP/TAZ-regulated gene that codes for survivin, an anti-apoptosis protein, overexpressed in most tumors including breast cancer [30].
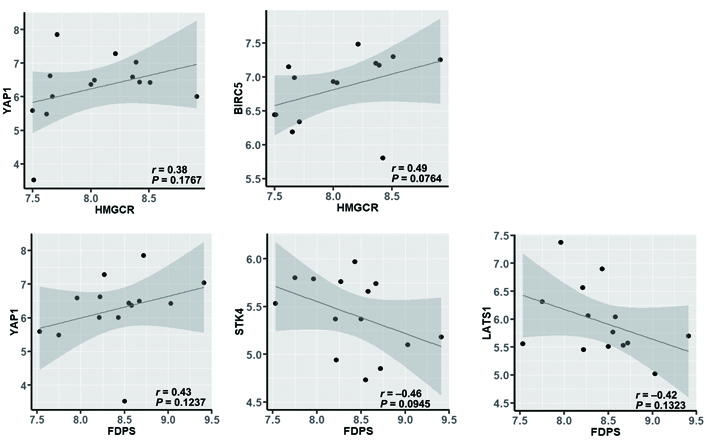
Correlation between HMGCR or FDPS and YAP1, BIRC5, STK4, or LATS1 in nonresponsive tumors (evaluated by using the nonparametric Spearman’s correlation coefficient test)
The positive association between HMGCR or FDPS and YAP1 is of particular interest because, on one side, confirms the functional role of the mevalonate pathway on the activity of the YAP coactivator, through the production of isoprenoids that, anchoring to the membrane and activating the small GTPases, inhibits the phosphorylation of YAP and the following sequestration in the cytoplasm; on the other side, they suggest a relationship between mevalonate and Hippo pathways also at the genomic level. Indeed, the expression of FDPS, which is the regulatory node in the production of isoprenoids, other than associated positively with that of YAP1, was associated negatively with that of STK4 and LATS1, which code for the two STKs that concur in the inactivation of YAP by phosphorylation and can be inhibited by prenylated GTPases (Figure 7). This result is in agreement with the evidence that the expression levels of the kinases involved in the Hippo signaling pathway are downregulated in human breast cancer tissues compared with normal breast tissues [31], and high expression levels of STKs are associated with longer disease-free and overall survival [32]. Besides, some recent studies [33, 34] showed that LATS1 kinase controls human breast cell fate by promoting the ER ubiquitination and ubiquitin-mediated proteasomal degradation and that the YAP/TEAD complex can activate the expression of estrogen-regulated genes by binding to the enhancer region of ERα, thus provide solid pieces of evidence of a close interrelationship among cholesterol biosynthesis, estrogens production, and the Hippo signaling pathway.
Remarkable is also the finding that, in nonresponsive tumors, the HMGCS1 gene, which codes the enzyme turning acetoacetyl-CoA into HMG-CoA (the direct precursor of the mevalonic acid), lost the positive association with HMGCR while retaining the positive association with CDK6 and MOB1A and the negative association with CCND1 and CDK4. That corroborates the notion that the dysregulation of the gene coding for the rate-limiting enzyme HMG-CoA reductase causes, on one side, the abnormal production of bioactive estrogens and, on the other side, the synthesis of the isoprenoids necessary for the isoprenylation of the small GTPases and the subsequent inhibition of YAP inactivation, provides a convincing explanation for the tumor resistance to AIs and supports the use of statins to contrast this metabolic dysfunction. In agreement with this idea, a recent population-based cohort study [35] showed improved patient survival and reduced risk for contralateral breast cancer in postmenopausal women assuming statins for more than five years with conventional AI therapy.
Several treatments have been investigated to overcome tumor resistance to AIs, and the CDK4/CDK6 inhibitors provided the most encouraging results [36–39]. Accordingly, specific CDK4/CDK6 inhibitors such as palbociclib and abemaciclib have become the standard of care in either the adjuvant or second-line setting for patients with ER-positive breast cancer in combination with AI therapy [40–42]. However, despite the substantial improvement in disease-free and overall survival, most tumors fail to respond, and patients ultimately experience disease progression because of acquired resistance [43]. Currently, excluding chemotherapy, no alternative strategies are available to overcome the resistance to CDK4/CDK6 inhibitors.
The cumulating evidence shows that the inhibition of the mevalonate pathway by statins caused the accumulation of the YAP coactivator in the cytoplasm hampering its nuclear translocation and the subsequent transcription of the YAP/TAZ-dependent genes [6, 44, 45] and that statins were effective in preventing and treating several tumors, including breast cancer [46–49], increased the interest for this class of molecules, initially developed as cholesterol-lowering drugs, for the treatment of ER-positive breast cancer with the double advantage of reducing the production of bioactive estrogens in an AI-independent manner and inhibiting the YAP/TAZ-dependent transcriptional activity.
In conclusion, although obtained on a limited number of patients, present results indicated that in nonresponsive tumors, the expression of ACAT2, HMGCS1, HMGCR, FDPS, and SQLE genes, which play a crucial role in the biosynthesis of cholesterol and isoprenoids production was unaffected by the treatment with letrozole and associated with that of some genes involved in estrogens production (CYP19A1, HSD17B2, and SULT1A), cell cycle control (CDK4 and CDK6), and Hippo signaling pathway (YAP1 and BIRC5), corroborating the notion that the dysregulation of the mevalonate pathway contributes to the resistance to letrozole and supporting the clinical use of statins to contrast this metabolic dysfunction.
ACAT2: acetyl-coenzyme A acetyltransferase 2
AI: aromatase inhibitor
BIRC5: baculoviral inhibitor of apoptosis repeat containing 5
CCND1: cyclin D1
CDK4: cyclin dependent kinase 4
CDKN: cyclin dependent kinase inhibitor
CYP11A1: cytochrome P450 family 11 subfamily A member 1
ER: estrogen receptor
FDFT1: farnesyl-diphosphate farnesyltransferase 1
FDPS: farnesyl diphosphate synthase
GG: geranylgeranyl
GGPS1: geranylgeranyl diphosphate synthase 1
GTPases: guanosine triphosphatases
HMG-CoA: 3-hydroxy-3-methylglutaryl-coenzyme A
HMGCR: 3-hydroxy-3-methylglutaryl-coenzyme A reductase
HMGCS1: 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1
HSD17B2: hydroxysteroid 17-beta dehydrogenase 2
HSD3B2: hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2
LATS1: large tumor suppressor kinase 1
MOB1A: Mps one binder kinase activator 1A
MVD: mevalonate pyrophosphate decarboxylase
MVK: mevalonate kinase
NSDHL: nicotinamide adenine dinucleotide (phosphate) dependent steroid dehydrogenase-like
PCA: principal component analysis
PMVK: phosphomevalonate kinase
PP: pyrophosphate
SQLE: squalene epoxidase
STK: serine/threonine kinase
STS: steroid sulfatase
SULT1A1: sulfotransferase family 1A member 1
TAZ: transcriptional co-activator with PDZ-binding motif
YAP: Yes-associated protein
YAP1: Yes1 associated transcriptional regulator
The supplementary material for this article is available at: https://www.explorationpub.com/uploads/Article/file/1001196_sup_1.pdf.
DC: Conceptualization, Investigation, Writing—original draft, Writing—review & editing. FA: Investigation, Writing—review & editing. Both authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
The original study was approved by the local ethics committee of the University of Edinburgh, Edinburgh, UK (2001/W/BU/09 and 2001/W/BU/10).
Informed consent to participate in the study was obtained from all participants.
Not applicable.
Original data are available through the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Supplementary material associated with this article can be found, in the online version.
Not applicable.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Ali Abdul Hussein S. AL-Janabi ... Abdul Razzak Kalaf Hassan
Banashree Bondhopadhyay ... Vishakha Kasherwal
Nadia Islam, Suneela Vegunta
Spoorthi Marada ... Yi Lu
Danila Coradini
Remo Poto ... Gilda Varricchi
Kaoutar Anouar Tadlaoui ... Moulay Mustapha Ennaji
