Affiliation:
Institute of Molecular Pathobiochemistry, Experimental Gene Therapy and Clinical Chemistry (IFMPEGKC), RWTH University Hospital Aachen, D-52074 Aachen, Germany
Email: rweiskirchen@ukaachen.de
ORCID: https://orcid.org/0000-0003-3888-0931
Explor Drug Sci. 2025;3:1008124 DOI: https://doi.org/10.37349/eds.2025.1008124
Received: July 01, 2025 Accepted: July 31, 2025 Published: August 11, 2025
Academic Editor: Juergen Reichardt, James Cook University, Australia
The article belongs to the special issue Advances and Innovations in Gene-Based Medicine
Gene-based medicine is transforming modern healthcare by offering precise, personalized interventions that target the genetic causes of disease. Breakthroughs in gene editing technologies, including clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) nuclease technologies (CRISPR-Cas9), base editing, and prime editing, are enabling promising therapeutic applications for rare inherited disorders and complex conditions like cancer. Furthermore, improvements in both viral and non-viral delivery methods are expanding clinical possibilities and enhancing safety measures. Despite these advancements, challenges such as off-target effects, ethical considerations, production complexities, and high costs continue to hinder widespread adoption. This review explores current innovations in gene-based medicine, addresses remaining obstacles, and outlines future directions, emphasizing the transformative potential of genomic-driven therapies for patients worldwide.
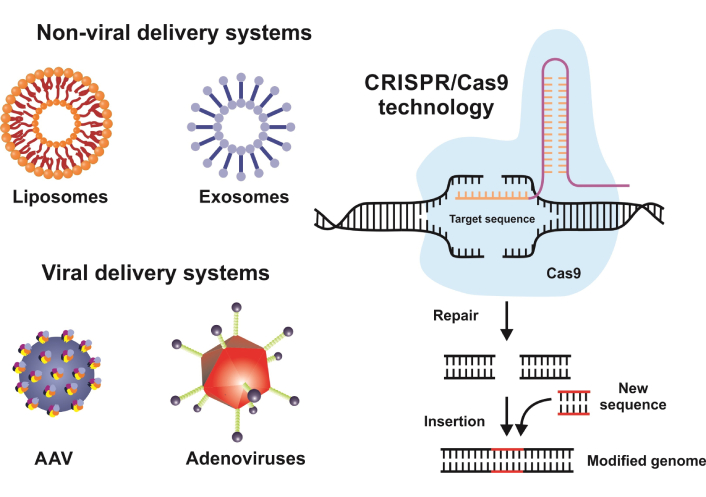
Gene-based medicine traces its origins to landmark experiments in the late 20th century, when researchers began exploring the feasibility of delivering functional genes to correct inherited disorders [1]. Early breakthroughs, such as the first successful gene therapy for adenosine deaminase deficiency in the early 1990s, demonstrated the potential to treat diseases at their genetic roots rather than merely managing symptoms [2, 3]. Since then, the field has rapidly diversified, incorporating RNA-based strategies, refined viral and non-viral vectors, and powerful genome-editing tools to address a broader range of conditions. In essence, gene-based medicine seeks to intervene at the molecular blueprint of life (i.e., the genome), offering a radical shift from symptomatic care to targeted, potentially curative therapies. These foundational advances established the framework for modern gene therapy, setting the stage for the transformative technologies and clinical applications explored in this paper.
Gene-based medicine has quickly emerged as a transformative force in modern healthcare, fundamentally altering our approach to diagnosing, treating, and preventing disease [4]. By directly targeting the genetic underpinnings of various conditions, researchers and clinicians are increasingly able to develop highly precise and personalized treatments. Central to this wave of innovation is the advent of powerful gene-editing technologies, such as clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) nuclease technologies (CRISPR-Cas9), which have democratized the ability to manipulate genetic material with unprecedented speed and accuracy [5]. Equally important are the expanding efforts to translate preclinical findings into clinical settings, leading to successful gene therapy approvals for inherited disorders, certain cancers, and neurological diseases [6]. However, the rapid ascent in this field also brings forth pressing questions about safety, regulatory oversight, ethical boundaries, and cost-effectiveness [7]. This review seeks to chart a path forward by highlighting major breakthroughs, examining ongoing challenges, and envisioning future directions that could propel gene-based medicine from a specialized therapeutic niche to a mainstay of healthcare worldwide.
The recent surge in gene-based medicine owes much to scientific discoveries and methodological advances that have reshaped our grasp of disease etiology. The following subsections detail these crucial developments, ranging from novel gene-editing platforms to innovative delivery systems, while elaborating on emerging trends that promise to influence both research and clinical practice.
CRISPR-Cas9 stands at the forefront of gene editing due to its relative simplicity, efficiency, and versatility [5]. By leveraging the bacterial immune response system, scientists can introduce targeted cuts to the DNA double helix, enabling the repair or replacement of faulty genetic segments. Beyond CRISPR-Cas9, there has been growing interest in base editing and prime editing, which allow for precise single-nucleotide modifications and more refined editing processes (Figure 1). These refined tools expand the scope of what is genetically feasible, opening the door for therapeutic interventions that can tackle disorders at their source.
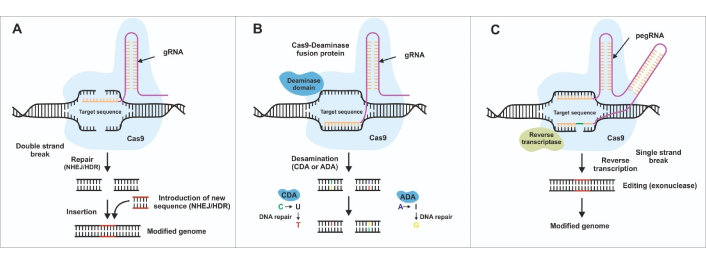
Schematic representation of gene editing mechanisms. This figure illustrates three major gene editing approaches: (A) CRISPR-Cas9; (B) base editing; and (C) prime editing. CRISPR-Cas9 works by introducing double-stranded DNA breaks through a Cas9 nuclease guided by a guide RNA (gRNA). These breaks can be repaired through non-homologous end joining (NHEJ), which may result in insertions/deletions, or through homology-directed repair (HDR) for precise corrections. Base editing utilizes a catalytically impaired or “nickase” Cas9 fused with a deaminase enzyme [either a cytidine deaminase (CDA) or an adenosine deaminase (ADA)] to convert single nucleotides (e.g., C-to-T or A-to-G) without creating double-stranded breaks. Prime editing combines a nickase Cas9 with a reverse transcriptase enzyme and a prime editing gRNA (pegRNA) that directs both binding and template extension to enable more diverse and precise edits. These methods expand the range of possibilities for genetic interventions, each balancing ease of use, efficiency, and off-target considerations. For more details on these editing techniques, please refer to [8]. CRISPR: clustered regularly interspaced short palindromic repeat; Cas: CRISPR-associated
The potential to correct hereditary mutations, eliminate disease-causing viral DNA, combat age-related diseases, or neutralize oncogenes has ignited extensive research, although the challenges of off-target effects and unpredictable genomic outcomes persist [4, 9–11].
There are clear core distinctions between CRISPR-Cas9 (introducing double-stranded breaks), base editing (targeted, single-nucleotide changes), and prime editing [precise insertions, deletions, and substitutions guided by a prime editing guide RNA (pegRNA)]. Key factors of these methodologies, such as mechanism of action, DNA break requirements, off-target effects, tissue specificity, and clinical maturity, are listed in Table 1.
Comparison of CRISPR-Cas9, base editing, and prime editing
| Key features | CRISPR-Cas9 | Base editing | Prime editing |
|---|---|---|---|
| Mechanism of action | Utilizes a Cas9 nuclease and a gRNA to create DSBs at specific DNA loci. | Uses a Cas9 nickase fused to a deaminase enzyme to convert one nucleobase to another without introducing DSBs. | Involves a Cas9 nickase fused to a reverse transcriptase domain and uses pegRNAs to install insertions, deletions, or substitutions. |
| Types of edit | Disrupts or replaces targeted DNA via NHEJ or HDR. | Enables precise single-nucleotide modifications, such as C-to-T or A-to-G at specific locations. | Capable of making more varied edits, such as base changes and small insertions/deletions, without generating full DSBs. |
| DNA breaks required | Yes, it creates DSBs. | Typically, only a nick is made in one strand. | No DBS breaks. Instead, it relies on a nick in a single strand for polymerase extension. |
| Off-target considerations | Off-target cleavage can occur if the gRNA has partial homology to non-target sites, but high-fidelity variants reduce this risk. | Generally, there are fewer off-target edits compared to Cas9-induced DSBs, but some off-target deaminations may still occur. | This method generally has reduced off-target activity compared to standard CRISPR-Cas9, although optimization is ongoing. |
| Efficiency and accuracy | While often highly efficient, results can vary depending on the cell type and repair pathway. | The precision for single-base changes is typically high, with efficiency depending on the accessibility of the target site and the surrounding base context. | It offers moderate to high precision, but efficiency can be lower than base editing, with ongoing improvements to pegRNAs and enzymes. |
| Tissue-specific efficiency | Success depends on vector choice, promoter specificity, and cellular repair machinery. | Similar factors affect the efficiency, which may vary depending on the tissue or cell line due to differences in deaminase expression and DNA accessibility. | It is currently under active investigation and optimal tissue tropism depends on delivery methods and pegRNA design. |
| Toxicity profile | Potential for cytotoxicity resulting from DSBs, which can be improved by using high-fidelity Cas9 variants. | This is typically lower cytotoxicity since no DSBs are created, but the risk of off-target deamination remains a concern. | There is a lower risk of large genomic rearrangements, but low-level off-target events are still possible. |
| Stage of development | Widely studied and is being used in multiple clinical trials, as well as in ex vivo therapies. | There is growing clinical interest with some proof-of-concept studies conducted in genetic and acquired diseases. | The technology is rapidly evolving with preclinical studies showing high promise, but broad therapeutic use is still in progress. |
| Complexity and size | This technology requires minimal resources: only a single Cas9 protein plus gRNA (100 nt). | The combination of Cas9 nickase and deaminase adds approximately 1 kb of extra size, requiring careful design to prevent off-target changes. | The system includes Cas9 nickase, reverse transcriptase, and more complex pegRNAs for design flexibility. |
DSB: double-stranded break; gRNA: guide RNA; HDR: homology-directed repair; NHEJ: non-homologous end joining; pegRNA: prime editing gRNA; CRISPR: clustered regularly interspaced short palindromic repeat; Cas: CRISPR-associated
While lipid nanoparticles (LNPs) have received significant attention for their low immunogenicity and strong encapsulation capabilities, various other non-viral vectors (such as silicon-based nanoparticles, metal nanoparticles, polymers, dendrimers, polysaccharide nanoparticles, and carbon-based nanoparticles), as well as exosomes, show promise for targeted drug and gene delivery [12, 13] (Figure 2).
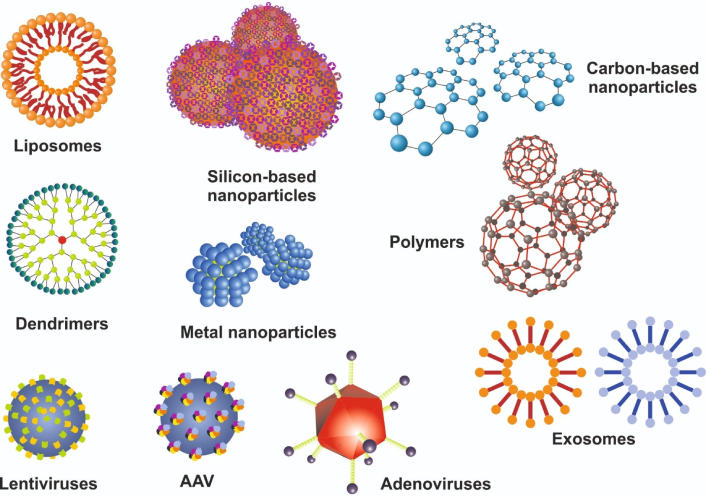
Classification of viral and non-viral delivery systems for gene-based therapies. This figure outlines the principal strategies for delivering gene-editing components or therapeutic genes to target cells. On the non-viral side, lipid nanoparticles (LNPs), silicon-based nanoparticles, metal nanoparticles, polymers, carbon-based nanoparticles, dendrimers, exosomes, and many other synthetic carriers provide an alternative for transient, re-dosable delivery with lower immunogenic risk. On the viral side, adeno-associated virus (AAV), lentivirus, and adenovirus serve as common vectors, each offering distinct advantages in cargo capacity, tropism, and duration of expression but varying in immunogenicity and safety profiles. Figure 2 is modified from [14]. © 2023 by the authors. CC BY
Exosomes demonstrate natural biocompatibility and the potential for targeted cargo release, while liposomes and solid LNPs offer structurally defined systems that can improve stability and protect therapeutic payloads. Polymeric nanocarriers, on the other hand, may allow for modular approaches to loading and controlled release but could raise concerns about biodegradability and manufacturing scalability. To provide a clearer contrast between these technologies, Table 2 summarizes key attributes, such as immunogenic profiles, gene loading capacities, and suitability for various clinical settings, across different carrier types. Overall, these options highlight that there is no universally optimal delivery platform. Instead, selecting the most suitable system depends on disease-specific requirements, the intended therapeutic payload, and practical considerations such as cost, safety, and patient accessibility.
Comparative overview of AAV vectors, LNPs, and exosomes
| Delivery platform | Delivery efficiency | Tissue specificity | Immunogenic risk | Advantages | Limitations |
|---|---|---|---|---|---|
| AAV vectors | Generally, high transduction efficiency | Specific serotypes exhibit tropism for muscle, liver, retina, etc. | Moderate to high; pre-existing neutralizing antibodies are common | Long-term gene expression; well-characterized biology | Limited transgene capacity (~ 4.7 kb); risk of immune response can reduce efficacy |
| Lipid nanoparticles (LNPs) | Moderate to high (depending on formulation) | Variable; targeting ligands can be incorporated for cell-type specificity | Relatively low, though some infusion-related reactions are possible | Non-viral, potentially allowing repeated dosing; scalable manufacturing | Potential instability in circulation; optimizing composition for each target remains complex |
| Exosomes | Variable and still under investigation | Intrinsic targeting properties reflect parent cell origin; some potential for tissue-specific engineering | Generally low; derived from endogenous vesicles, but risk varies by source | Natural biocompatibility; can carry diverse cargos (RNA, proteins, or small molecules) | Large-scale production and purification remain challenging; cargo loading and consistent characterization are still evolving fields |
AAV: adeno-associated virus
Delivering therapeutic genes into patient tissues efficiently and safely remains one of the central obstacles in gene-based therapy. Viral vectors, including adeno-associated viruses (AAVs), have traditionally been favored for their relatively high transduction rates [5]. Yet issues like immunogenicity, limited packaging capacity, and the possibility of insertional mutagenesis continue to spur interest in non-viral alternatives. LNPs, for example, have emerged as a promising approach, offering lower immunogenicity and the flexibility for repeated dosing. Exosomes, which represent the own extracellular vesicles of an individual, are another avenue under investigation due to their biocompatibility and capacity for cargo encapsulation of drugs and potential gene therapeutic active compounds [15–17]. As vector and nanoparticle research converges with tissue-specific promoters and cell-targeting ligands, the quest for optimal delivery solutions is poised to yield next-generation platforms capable of reaching previously inaccessible targets.
Personalized medicine in the context of gene-based approaches capitalizes on genomic, transcriptomic, and proteomic data to tailor interventions. Instead of adopting one-size-fits-all protocols, clinicians can now stratify patients by genetic risk factors, individual molecular signatures obtained by transcriptomics, proteomics, metabolomics, and the usage of artificial intelligence (AI) algorithms combined with machine learning, ultimately offering safer and more effective treatments [18]. In oncology, for instance, identifying specific driver mutations in a tumor of a specific patient can guide the selection of therapies that directly interfere with those genetic aberrations [19, 20]. Pharmacogenomics complements this customization by determining the response of patient to specific drugs, thereby mitigating adverse effects and optimizing treatment outcomes [21]. Gene-based medicine reinforces the central premise of personalized healthcare: no two individuals share an identical genetic landscape, and hence, treatment strategies ought to reflect that uniqueness.
Growing evidence suggests that AI plays an essential role in enhancing genomic research and the development of gene-based therapies. Machine learning algorithms can process complex genomic datasets more efficiently, helping to pinpoint critical genetic markers and therapeutic targets with high precision. This capability is particularly crucial in personalized medicine, where interventions are tailored to individual molecular profiles. AI-driven approaches also show promise for guiding the selection and optimization of delivery vectors, as computational models can predict how efficiently different nanoparticle formulations or viral constructs will reach intended tissues. By integrating these insights with emerging tools like exosomes and lipid platforms, researchers can expedite the translation of novel gene therapies into clinical applications. Recent work highlights how AI-driven methodologies streamline the analysis of both large-scale genomic data and nanoparticle behavior, underscoring the growing convergence of computational intelligence and gene-based medical innovation [22, 23]. Importantly, for the shaping of these initiatives, it will be essential to establish a regulatory network with standardized language, which includes information about terminology and guidance for promoting best practices in automation and digitalization [24].
From rare genetic disorders, such as spinal muscular atrophy, to more prevalent conditions like certain forms of cancer, the clinical success of gene-based therapies has advanced beyond experimental proof-of-concept. The US Food and Drug Administration (FDA) has approved 45 cellular and gene therapy products as of May 15, 2025 [25] (Table S1), highlighting the rapidly expanding range of strategies used to treat various diseases. These therapies come from various manufacturers, including large pharmaceutical companies such as Bristol-Myers Squibb, Novartis, and Pfizer, as well as smaller specialized biotech firms. Many of these products focus on hematologic malignancies, such as multiple myeloma, B-cell lymphomas, and acute lymphoblastic leukemia, illustrating the effectiveness of chimeric antigen receptor (CAR) T-cell and other autologous T-cell immunotherapies (e.g., ABECMA, YESCARTA, KYMRIAH). These therapies involve collecting own T cells from patients, genetically modifying them to target cancer cells, and then reintroducing them to trigger a specific immune response.
Several hematological and hereditary disorders have also seen significant therapeutic progress. Products like HEMGENIX and BEQVEZ, which use AAV vectors to deliver functional copies of the factor IX gene, offer transformative treatments for hemophilia B. Similarly, ZYNTEGLO offers an AAV-independent gene therapy approach for β-thalassemia, potentially leading to curative results. Additionally, two gene editing-based approaches (CASGEVY and LYFGENIA) represent cutting-edge interventions for sickle cell disease. In the field of muscular dystrophies, ELEVIDYS utilizes AAV technology to deliver a shortened yet functional form of the dystrophin gene to patients with Duchenne muscular dystrophy.
Aside from inherited conditions, a subset of therapies leverages viral vectors or engineered cells to address complex disorders such as type 1 diabetes (e.g., LANTIDRA for islet cell transplantation), macular telangiectasia (ENCELTO employing an encapsulated cell-based gene therapy), and various immunodeficiencies. More recently, oncolytic viral therapies (e.g., IMLYGIC) and allogeneic cell products (e.g., HPC cord blood-derived treatments for inherited or acquired hematopoietic disorders) have broadened the scope of gene-based medicine. These modalities collectively demonstrate the clinical potential of both autologous and allogeneic approaches, employing vectors derived from AAVs, oncolytic herpes simplex viruses, or lentiviruses to confer durable therapeutic benefits. Many of these products also exemplify the transition toward personalized medicine, custom-tailored fronts in which patient-derived cells and tissues are central to the treatment process.
Taken together, the FDA-listed therapies highlight the diversity of targets and delivery systems in modern gene and cell-based interventions. From scaffold-based tissue-engineered products to genetically reprogrammed T cells, they suggest a future where an even wider range of diseases, beyond cancer and inherited disorders, can be treated with safe, effective, and personalized approaches. These advancements not only open novel avenues of treatment but also set the stage for integrating precision diagnostics and emerging technologies like AI into the development of next-generation therapies.
Specific examples include the FDA approvals of gene therapies targeting inherited retinal diseases and β-thalassemia, indicating that rigorous clinical trials can validate the safety and efficacy of gene-based interventions and drive new therapeutic avenues [26]. Meanwhile, in the field of oncology, CAR T-cell therapies have demonstrated compelling results, and ongoing research and clinical trials are expanding their application to multiple cancer types, including solid tumors [27]. These successes underscore both the feasibility of gene-based therapies and the vibrant pipeline of emerging candidates moving through preclinical and clinical phases. One illustrative example is the use of the one-time gene therapy onasemnogene abeparvovec-xioi (Zolgensma) for the treatment of pediatric patients suffering from spinal muscular atrophy type 1. This is a lethal hereditary condition caused by mutations in the survival motor neuron 1 (SMN1) gene. In pivotal clinical trials, infants not older than 2 years who received this AAV9-based gene therapy demonstrated significant improvements in motor milestones, including head control and the ability to sit unsupported, which are typically unattainable with standard care [28]. These outcomes were also correlated with prolonged survival and reduced respiratory support requirements. Despite the remarkable efficacy of the therapy, the high cost and potential liver toxicity or adverse immune responses highlight the need for ongoing long-term surveillance [28]. Nevertheless, Zolgensma represents a breakthrough in treating a previously intractable neuromuscular disorder, showcasing how gene therapies can provide pronounced clinical benefits while underscoring the importance of addressing safety, long-term benefits, and accessibility concerns in new clinical trials.
Another compelling case study involves ornithine transcarbamylase deficiency (OTCD), an X-linked urea cycle disorder characterized by life-threatening hyperammonemia. In early clinical investigations, gene therapy efforts primarily employed adenoviral vectors to introduce a functional OTC gene into hepatocytes, thereby restoring partial enzymatic activity [29]. Although these initial trials underscored the potential for gene replacement to reduce ammonia levels, adverse events also highlighted the need for more refined vector design and improved safety protocols. Ongoing research now focuses on using AAV-based approaches, which generally offer lower immunogenicity and longer-term gene expression [29]. While still in development, these next-generation therapies aim to mitigate the severe metabolic complications associated with OTCD and provide a durable, potentially curative option for patients otherwise reliant on strict dietary regimens and chronic ammonia-lowering medications. Nevertheless, the experience made in the first patient treated in the UK with the liver-directed editing treatment with ECUR-506, delivering a gene encoding the editing enzyme and an OTC gene, is highly encouraging [30].
Nevertheless, recent developments underscore that gene therapy can pose significant safety risks. According to a July 18, 2025, communication from the FDA, three patients experienced fatal acute liver failure following treatment with AAVrh74 gene therapy from Sarepta. Two cases occurred in non-ambulatory pediatric males with Duchenne muscular dystrophy treated with ELEVIDYS, while the third involved an adult with limb girdle muscular dystrophy who died 80 days post-treatment after presenting with elevated liver enzymes. These incidents prompted the FDA to request a suspension of ELEVIDYS distribution and to place clinical trials involving the implicated gene therapy products on hold, highlighting the need for ongoing vigilance and stringent safety monitoring in the rapidly evolving field of gene-based medicine [31]. Therefore, well-designed clinical trials, along with a careful assessment of which gene therapies are ethically and scientifically defensible, remain crucial in minimizing risks and ensuring patient safety.
Moreover, discussions of gene editing methodologies include ethical, legal, and economic dimensions that extend well beyond familiar concerns about the generation of “designer babies” and equitable access. Real-world examples, such as the controversial use of CRISPR in human embryos by a researcher in China, illustrate the risks of unapproved germline manipulation and have prompted governments worldwide to revisit and tighten their regulatory frameworks [32, 33]. In the European Union, therapies must meet stringent standards laid out in directives on advanced therapy medicinal products, while the FDA closely monitors clinical trials and post-market surveillance. Moreover, in the US the use of federal funds to finance genetic modification experiments in gametes and embryos is prohibited [34].
Beyond regulatory oversight, high therapy costs, often exceeding hundreds of thousands of dollars, spark intense debate over financial accessibility, insurance reimbursement, and the broader societal implications of gene editing [35]. These factors, coupled with growing concerns about data privacy and the ownership of genetic information, highlight a complex landscape that requires robust public engagement, international collaboration, and transparent governance to balance innovation with responsibility.
Despite inspiring breakthroughs and success stories, gene-based medicine faces substantial hurdles. These challenges encompass a broad spectrum, from the technical details of ensuring safe and efficacious therapies to the larger ethical and social considerations that arise whenever healthcare delves into the genetic domain.
Safeguarding patient well-being is paramount, and gene-editing tools must surmount issues related to off-target effects, long-term stability, and immune responses. Regulatory bodies across the globe weigh both the promise and the risks associated with gene-editing tools, often grappling with inadequate precedents and evolving guidelines and special regulations for researchers, clinicians, and industry [36, 37]. As the field matures, regulators must find a balance between enabling technological innovation and maintaining rigorous standards that protect patients from unforeseen complications. Collaboration among academic institutions, industry, and regulatory authorities is imperative to craft agile yet robust frameworks for approving and monitoring gene-based therapies [36, 37].
Ethical debates surrounding genetic manipulation span a spectrum that includes germline editing, equity in access, and the broader societal impact of modifying the human genome [35]. Intervening in germline cells, where changes can be inherited by subsequent generations, sparks particularly heated controversy, fueling both hope for disease eradication and fears of “designer babies” [38, 39]. Public engagement and transparent dialogue are essential to ensure that ethical guidelines remain both scientifically informed and socially responsive. By actively involving patient advocates and community representatives, researchers and policymakers can work to preserve public trust while fostering responsible innovation.
Scaling up gene-based therapies from proof-of-concept studies to large patient populations involves complexities in manufacturing, quality control, and distribution [40]. Issues such as batch-to-batch variability, costs associated with complex production processes, and the stringent conditions required to maintain product efficacy all challenge the feasibility of widespread deployment. Delivery constraints remain a fundamental barrier as well. Many gene-based therapies rely on tissues or cells being transduced at high efficiency while minimizing exposure to off-target areas, which can result in unintended genetic modifications at sites closely resembling the desired target sequence [32, 41]. Incorporating regulatory elements for tissue-specific expression in viral vectors can also be an effective means to reduce off-target effects [42]. Moreover, diverse experimental and in silico methods, as well as bioinformatics algorithms, have emerged that are also helpful in identifying off-target effects and their consequences [43, 44]. Concerted efforts in research and development devoted to vector engineering, manufacturing protocols, and supply-chain logistics will be critical in overcoming these bottlenecks. Examples show that standardized protocols can reduce production time, generation costs, and process risk [45].
Even when scientific and regulatory aspects are overcome, high prices can limit the reach of gene-based therapies to only a small subset of patients. The cost burden comes from specialized materials, complex manufacturing processes, and significant investment in research and development. Insurance frameworks and reimbursement policies vary widely across countries, creating ethical dilemmas and economic disparities [46]. To prevent a scenario where only wealthy nations or patients benefit from gene-based advances, stakeholders must explore novel pricing models, public-private partnerships, and global collaboration to distribute responsibility and risk more equitably.
Although gene-based therapies offer unparalleled potential for treating previously intractable conditions, high development and production costs often result in prohibitively expensive treatments for patients. For example, some advanced therapies come with price tags in the hundreds of thousands or even millions of dollars, raising concerns about equitable access and reimbursement models. While in certain regions, such as parts of the European Union, national healthcare systems help subsidize or negotiate the cost of these treatments, significant disparities persist globally [35]. Cost-control measures for gene and cell therapies tend to be stricter in Europe compared to the US. For instance, Roctavian therapy is priced at around US$2.9 million but only US$1.5 million in Germany, while Lenmeldy is priced at US$4.25 million in the USA but a slightly more reasonable £2.8 million (about US$3.8 million) in the UK. Companies seem to have significant freedom in shaping their therapies, highlighting the need for greater transparency in pricing [39].
Unfortunately, in some low- and middle-income countries, insurance frameworks are limited or nonexistent, making many gene therapies inaccessible to most patients. Visual comparisons of therapeutic costs by region, along with transparent assessments of both direct and indirect financial burdens on patients and healthcare systems, can shed light on how pricing structures impact widespread adoption and long-term sustainability. As a result, there is a rising demand for innovative financing mechanisms, such as installment payment plans or outcomes-based pricing, to ensure that advances in gene-based medicine benefit patients worldwide in a fair manner [47].
The outlook for gene-based medicine is undoubtedly bright as researchers refine current methods and discover next-generation solutions. However, sustainable progress requires ongoing innovation, careful governance, and interdisciplinary partnerships that integrate the needs and insights of diverse stakeholders.
Refinements to CRISPR-Cas9, along with emerging technologies like base editing, aim to reduce off-target edits and expand the genetic changes that can be made. Scientists are also exploring more controlled inducible systems that activate gene editing only in response to specific signals [48]. These advances are expanding the scope from single-gene monogenic disorders to more complex, polygenic diseases, unlocking therapeutic possibilities for once-deemed intractable conditions.
Recent advances in gene editing technology go far beyond the groundbreaking CRISPR-Cas9 approach. New, more refined and precise systems have been developed that can achieve single-nucleotide modifications or complex genomic alterations. As discussed, base editors, for example, use a “nickase” variant of Cas9 fused to a cytosine or adenine deaminase, allowing for targeted nucleotide conversions without causing double-stranded breaks. Another innovative technique is prime editing, which combines Cas9 nickase with a reverse transcriptase module and specially designed pegRNAs to enable flexible editing, including insertions, deletions, and substitutions. It is now crucial to improve the accuracy of all these tools and establish conditionally activated enzymes to minimize off-target activity and enhance therapeutic potential. The advancement of these tools, including base editors, prime editors, and advanced CRISPR variants, will further expand the scope and precision of genome manipulation. This progress paves the way for novel precision therapies in inherited diseases, oncology, and beyond.
Encouraged by promising clinical results in oncology and early-phase trials for neurological and cardiovascular diseases, many researchers anticipate broader use of gene-based therapies or exosome-based vesicles that can effectively cross the blood-brain barrier for conditions with substantial unmet clinical needs [49, 50]. As more patients undergo these treatments and sustained investment in research, growing data sets will help define safety parameters, optimal dosing, and long-term benefits, continually enhancing delivery vector construction and manufacturing, as well as treatment algorithms, including ethical considerations [50–52]. Over time, gene-based medicine, including nanoparticle-based genomic medicines, may prove transformative for disorders that have remained elusive under conventional therapeutic paradigms, ultimately reshaping standards of care in multiple clinical specialties [53].
There are also some important challenges regarding clinical translation when using different delivery systems. Although AAV vectors have shown robust gene delivery and sustained expression, pre-existing neutralizing antibodies may limit their efficacy. In contrast, LNPs and exosomes exhibit lower immunogenicity and can facilitate multiple dosing, but can face challenges in achieving high transduction rates and consistent targeting (Table 2), emphasizing the importance of tailoring delivery strategies to balance efficacy, safety, and long-term viability in clinical applications. As the field continuously evolves, these parameters may shift over time with ongoing research and technological advancements.
The concept of “multi-omics” emphasizes the integration of genomic, transcriptomic, proteomic, and metabolomic datasets, providing a comprehensive view of human disease mechanisms [54]. Combining gene-based therapies with advanced diagnostics that synthesize these large data streams could refine patient selection and predict therapeutic outcomes more accurately. Genome-wide association studies (GWAS), combined with transcript-level analyses or transcriptome-wide association studies (TWAS), can identify essential regulatory networks, highlighting pathways amenable to gene-based interventions [55]. Through this holistic approach, clinicians can make data-driven decisions that enhance both efficacy and safety.
A more comprehensive view of gene therapy requires recognizing that regulatory pathways vary widely across regions, influencing both the pace and scope of clinical translation. For instance, the European Medicines Agency (EMA) applies a stringent approval process under regulations specific to advanced therapy medicinal products, while the FDA oversees a structured pathway that includes expedited designations for promising therapies [56, 57]. In China, the National Medical Products Administration (NMPA) has introduced recent reforms to accelerate innovative treatments with advanced therapy medical products, but ethical controversies, such as unauthorized germline interventions, have spurred tighter oversight [58]. Against this complex backdrop, it is also important to acknowledge the perspectives of skeptics, patients who have experienced off-target effects, and advocacy groups that champion more patient-centric and ethically responsible application of these technologies from philosophical, theological, public, scientific, and clinical perspectives [59, 60]. By engaging these diverse viewpoints and regulatory frameworks, researchers and policymakers can develop a balanced, globally informed strategy for advancing gene-based medicine.
Fostering broader success in gene-based medicine requires a strong ecosystem of collaboration. Partnerships range from academia and industry-driven research consortia to regulatory agencies and patient advocacy groups. Large-scale public-private collaborations have already accelerated vaccine development through pooled resources, shared data repositories, and coordinated clinical trial networks, as seen in the vaccine development during the COVID-19 pandemic [61]. A similar model could streamline gene therapy development, where transparent communication and shared objectives reduce redundancies and promote ethical, efficient progress. Crucially, fostering strong links between scientific innovation and ethical oversight will be vital to maintaining public confidence and ensuring that gene-based therapies uphold societal values [62]. Despite international and national legal regulations that need to be considered, human genome editing research requires a close network of collective scientific self-regulation, regardless of whether it is conducted in public institutions, private sectors, or in public-private partnerships [63].
Gene-based medicine represents an unprecedented leap forward in the quest to combat human disease at its foundational genetic level. Buoyed by breakthroughs in gene editing, sophisticated delivery systems, and personalized therapeutics, the field shows immense promise for treating both rare and widespread disorders. Nevertheless, these transformative possibilities are accompanied by equally weighty challenges. Technical limitations in delivery, ethical considerations surrounding the sanctity of the genome, and economic barriers that restrict access all warrant thoughtful, multifaceted solutions. As gene-based therapies move more firmly into clinical practice, the collective focus must remain on responsible innovation, global collaboration, and patient-centricity. By balancing robust scientific advances with vigilant regulation and community engagement, gene-based medicine could revolutionize healthcare, ushering in a future where targeted, prophylactic, and even curative interventions become reality for patients in need.
AAV: adeno-associated virus
AI: artificial intelligence
CAR: chimeric antigen receptor
Cas: clustered regularly interspaced short palindromic repeat-associated
CRISPR: clustered regularly interspaced short palindromic repeat
FDA: Food and Drug Administration
gRNA: guide RNA
GWAS: genome-wide association studies
LNPs: lipid nanoparticles
OTCD: ornithine transcarbamylase deficiency
pegRNA: prime editing guide RNA
TWAS: transcriptome-wide association studies
The supplementary table for this article is available at: https://www.explorationpub.com/uploads/Article/file/1008124_sup_1.pdf.
The author is grateful to Sabine Weiskirchen (IFMPEGKC, RWTH Aachen University) for preparing the figures and graphical abstract for this article. During the preparation of this work, the author used the RWTHgpt to improve readability and language. After using the tool, the author reviewed and edited the content as needed and takes full responsibility for the content of the publication.
RW: Conceptualization, Funding acquisition, Writing—original draft, Writing—review & editing.
Ralf Weiskirchen who is the Associate Editor of Exploration of Drug Science had no involvement in the decision-making or the review process of this manuscript.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
The study was supported by grants from the German Research Foundation [WE2554/17-1]; the Deutsche Krebshilfe [70115581]; and a grant from the Interdisciplinary Centre for Clinical Research within the Faculty of Medicine at the RWTH Aachen University [PTD 1-5]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 4497
Download: 133
Times Cited: 0
