Affiliation:
1Department of Genetics, Cytology and Bioengineering, Voronezh State University, Voronezh 394018, Russia
ORCID: https://orcid.org/0009-0002-7294-9525
Affiliation:
1Department of Genetics, Cytology and Bioengineering, Voronezh State University, Voronezh 394018, Russia
ORCID: https://orcid.org/0009-0007-6743-3766
Affiliation:
1Department of Genetics, Cytology and Bioengineering, Voronezh State University, Voronezh 394018, Russia
ORCID: https://orcid.org/0009-0000-1069-9108
Affiliation:
1Department of Genetics, Cytology and Bioengineering, Voronezh State University, Voronezh 394018, Russia
ORCID: https://orcid.org/0009-0001-1193-9203
Affiliation:
2Laboratory of Metagenomics and Food Biotechnology, Voronezh State University of Engineering Technology, Voronezh 394036, Russia
ORCID: https://orcid.org/0000-0002-5881-0845
Affiliation:
1Department of Genetics, Cytology and Bioengineering, Voronezh State University, Voronezh 394018, Russia
2Laboratory of Metagenomics and Food Biotechnology, Voronezh State University of Engineering Technology, Voronezh 394036, Russia
Email: gureev@bio.vsu.ru
ORCID: https://orcid.org/0000-0003-3562-5329
Explor Neuroprot Ther. 2025;5:1004122 DOI: https://doi.org/10.37349/ent.2025.1004122
Received: June 17, 2025 Accepted: October 22, 2025 Published: November 17, 2025
Academic Editor: Marcello Iriti, Università degli Studi di Milano, Italy
The article belongs to the special issue Natural Products in Neurotherapeutic Applications
Aim: Systemic inflammation is a key factor in cognitive decline and neurodegenerative diseases. Polyphenols, such as curcumin, resveratrol, and salidroside, exhibit neuroprotective effects, but their low bioavailability raises questions about their mechanism of action. The gut-brain axis, mediated by microbiome modulation, may play a critical role in their cognitive benefits. This study investigated whether polyphenols (curcumin, resveratrol, and salidroside) improve cognitive function in mice with lipopolysaccharide (LPS)-induced gut inflammation by modulating the gut microbiome and reducing neuroinflammation.
Methods: C57BL/6 mice were divided into five groups: control, LPS, and LPS + polyphenol treatments (curcumin, resveratrol, or salidroside). LPS was administered intraperitoneally to induce inflammation, while polyphenols were given orally for three weeks. Cognitive performance was assessed using the Morris water maze. Gut microbiome composition (16S rRNA sequencing), mitochondrial DNA (mtDNA) damage, and gene expression in brain regions were analyzed.
Results: LPS impaired spatial memory, but resveratrol and salidroside significantly mitigated these deficits. Polyphenols restored beneficial bacteria (e.g., Alloprevotella, Eubacterium) and suppressed pathogenic taxa (e.g., Peptostreptococcales). They also reduced pro-inflammatory markers in the cortex and hippocampus. Curcumin showed weaker effects. No significant mtDNA damage was detected.
Conclusions: Polyphenols, particularly resveratrol and salidroside, improve cognition during systemic inflammation by remodeling the gut microbiome and attenuating neuroinflammation. These findings highlight the gut-brain axis as a therapeutic target for inflammation-driven cognitive disorders.
Systemic inflammation is a well-established contributor to cognitive decline across neurodegenerative disorders, including Alzheimer’s [1, 2] and Parkinson’s diseases [3, 4], as well as age-related neurodegeneration [5, 6]. Particularly relevant are inflammation-associated memory impairments [7, 8], highlighting the urgent need for dietary compounds that can modulate neuroinflammatory pathways. Polyphenols represent promising candidates due to their dual anti-inflammatory properties and suitability for dietary incorporation, offering a practical preventive approach against inflammation-driven cognitive deterioration.
Polyphenols represent a class of ubiquitously distributed, bioavailable dietary compounds demonstrating significant neuroprotective properties [9]. These neuroprotective effects of polyphenols, particularly in age-related brain diseases, have been demonstrated in various experimental models. For example, in vivo studies have shown anti-aging activity of resveratrol [10], apigenin [11], and salidroside [12]. Collectively, studies report that polyphenols exert various beneficial effects on brain functions, including improved cognition, learning, and memory [10, 13–15], as well as reduced risk of dementia [15]. Among the various mechanisms proposed for their neuroprotective effects, the most prominent involve their antioxidant and anti-inflammatory properties. Another important effect of polyphenols that has garnered significant interest is their immunomodulatory capacity [16–18] and microbiota-remodeling properties [19]. However, the pharmacological potential of polyphenols is substantially limited by their poor water solubility, chemical instability, and rapid metabolism [20–22].
Research on polyphenol bioavailability has raised questions about their efficacy as direct antioxidant compounds [23]. However, emerging evidence demonstrates that polyphenols can exert biological activity following gut microbiota-mediated chemical modifications. Specifically, intestinal microbial enzymes facilitate three key transformations: deglycosylation, dehydroxylation, and ademethylation of polyphenols, generating small catabolic products with enhanced intestinal absorption [24].
Polyphenols directly influence the growth of specific bacterial populations. Notably, they exhibit prebiotic-like effects by shifting the Firmicutes/Bacteroidetes ratio toward increased Bacteroidetes abundance [25]. These dietary phenolic compounds contribute to neuroprotection [26] through their ability to regulate neuronal signaling pathways [27], particularly the nuclear factor erythroid 2-related factor 2/antioxidant response element (Nrf2/ARE) pathway that governs both synaptic plasticity [27, 28] and mitochondrial homeostasis. Collectively, these mechanisms position polyphenols as promising candidates for combating cognitive decline and neurodegenerative diseases [29]. We hypothesize that polyphenol-induced microbiome modulation primarily acts through three key signaling pathways: nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), sirtuin 1 (SIRT1), extracellular signal-regulated kinase (ERK), and Nrf2, ultimately regulating both antioxidant gene expression and pro-inflammatory cytokine production. Given their antioxidant properties and Nrf2/ARE activation capacity, we further investigated whether polyphenols could protect against lipopolysaccharide (LPS)-induced mitochondrial DNA (mtDNA) damage.
Using three structurally distinct polyphenols—curcumin (linear diarylheptanoid with potent NF-κB inhibition), resveratrol (stilbenoid activating SIRT1), and salidroside (phenylethanoid glycoside modulating Nrf2/ARE pathway)—this study investigates their bidirectional gut-brain axis interactions to determine how these specific compounds mitigate LPS-induced neuroinflammation and cognitive deficits, thereby identifying the most promising candidates for dietary interventions in inflammation-driven neurodegeneration.
The study was conducted on three-month-old C57BL/6 mice divided into five groups: control (n = 10, 5 males and 5 females), LPS (n = 9, 5 females and 4 males), LPS + curcumin (n = 10, 5 males and 5 females), LPS + resveratrol (n = 9, 5 females and 4 males), and LPS + salidroside (n = 10, 5 males and 5 females). The two-way ANOVA analysis revealed no significant sex differences between experimental groups for the studied factors (p = 0.295), while significant differences were observed depending on polyphenol treatment (p = 0.008). Control animals received a standard diet, while treatment groups were pretreated for three weeks with respective polyphenols: curcumin (50 mg/kg/day), resveratrol (20 mg/kg/day), or salidroside (30 mg/kg/day), administered orally. These drug concentrations were selected based on previously obtained experimental data [30–32].
Neuroinflammation was induced by daily intraperitoneal injections of LPS (0.375 µg/kg) for 7 days in all experimental groups, with polyphenol supplementation continuing throughout LPS exposure and subsequent behavioral testing. Spatial memory was assessed using the Morris water maze during a 12-day testing period. Following behavioral tests, mice were deeply anesthetized via intraperitoneal injection of a combination of tiletamine (12.5 mg/kg), zolazepam (12.5 mg/kg), and xylazine (7.5 mg/kg) [33]. Euthanasia was then performed by cervical dislocation in strict accordance with the Guide for the Care and Use of Laboratory Animals and approved by the Institutional Ethical Committee of Voronezh State University (Protocol No. 42-03). Tissue samples were subsequently collected for molecular analysis.
The study utilized three-month-old male and female C57BL/6 mice acquired from an approved breeding facility (Stolbovaya Nursery, Russia). Animals were housed in a controlled vivarium environment maintained at 25 ± 1°C with 40–60% relative humidity and standardized 12:12 light-dark cycles. All mice received ad libitum access to pure water and standard laboratory chow. Mice were randomly assigned to treatment groups using a coded identification system prior to behavioral testing.
To assess the cognitive abilities of mice, we conducted a Morris water maze according to the protocol for assessment of spatial long-term memory [34].
Additionally, we calculated the learning coefficient for each individual mouse. To do this, we took the sum of the time values for 4 attempts within one training day (control days were not taken into account), in chronological order, for the entire period of the Morris water maze. And thus, we tracked the dynamics of the memorization process, based on the value of the coefficient k, which we looked at from the linear dependence formula:
Where, y: time, x: corresponding training day.
Thus, the lower the value of this coefficient, the better the performance, that is, it spends less time searching for a platform every day.
mtDNA damage quantification was conducted using quantitative long-range polymerase chain reaction (PCR) on a Bio-Rad CFX96TM Real-Time PCR System (# 184-5096, Hercules, USA) using Encyclo Plus PCR Kit (# PK101, Evrogen, Russia) with 1× SYBR Green Master Mix (BioDye, Russia), and gene-specific primer pairs. Thermal cycling parameters included: initial denaturation (95°C, 3 min) followed by 35 cycles of denaturation (95°C, 30 s), primer annealing (59°C, 30 s), and extension (72°C, 4.5 min). Primer sequences targeting long mtDNA fragments were previously designed using Primer-BLAST [35]. DNA lesion frequency was normalized to 10,000 base pairs using the following calculation:
Where, Δlong = Cq control − Cq experiment for the long fragment and Δshort = Cq control − Cq experiment for the short fragment (Cq: quantification cycle).
Total RNA was isolated using the Extract RNA Kit (# BC032 Evrogen, Russia) following the manufacturer’s protocol, with subsequent cDNA synthesis performed using the REVERTA-L reverse transcription system (# K3-4-100 AmpliSens, Russia) on an Eppendorf Mastercycler personal thermal cycler (Eppendorf, Germany). Quantitative real-time PCR analysis was conducted on a Bio-Rad CFX96TM Real-Time PCR System (Hercules, USA) using Encyclo Plus PCR Kit (Evrogen, Russia) with 1× SYBR Green Master Mix (BioDye, Russia), and gene-specific primer pairs. The thermal profile consisted of initial denaturation at 95°C for 3 min, followed by 38 cycles of denaturation (95°C, 30 s), annealing (61°C, 30 s), and extension (72°C, 30 s), with a final extension at 72°C for 5 min and melting curve analysis from 65 to 95°C. Gene-specific primers were designed using Primer-BLAST (Table 1), and relative expression levels were calculated from fluorescence data (RFU) following normalization to reference genes.
Primers that were used to analyze gene expression.
| Gene | The sequence of the forward primer 5′ to 3′ | The sequence of the reverse primer 5′ to 3′ |
|---|---|---|
| Nfe2l2 | CTCTCTGAACTCCTGGACGG | GGGTCTCCGTAAATGGAAG |
| Bdnf | AAGGACGCGGACTTGTACAC | CGCTAATACTGTCACACACGC |
| Mtor | AGATAAGCTCACTGGTCGGG | GTGGTTTTCCAGGCCTCAGT |
| Akt1 | TGATCAAGATGACAGCATGGAGTG | GATGATCCATGCGGGGCTT |
| p62 | GCCAGAGGAACAGATGGAGT | TCCGATTCTGGCATCTGTAG |
| Pink1 | GAGCAGACTCCCAGTTCTCG | GTCCCACTCCACAAGGATGT |
| Txnrd2 | GATCCGGTGGCCTAGCTTG | TCGGGGAGAAGGTTCCACAT |
| Prdx3 | TGGCTTGATCGTAGGGGACT | GTGGTTTGGGCCACATGAAC |
| Gpx | AGTCCACCGTGTATGCCTTCT | GAGACGCGACATTCTCAATGA |
| Gclc | GCAGCTTTGGGTCGCAAGTAG | TGGGTCTCTTCCCAGCTCAGT |
| Il1b | TTGCGGACCCCAAAAGATG | AGAAGGTGCTCATGTCCTCA |
| Il6 | CGGAGAGGAGACTTCACAGAG | CATTTCCACGATTTCCCAGA |
| Tnf | TATGGCTCAGGGTCCAACTC | GGAAAGCCCATTTGAGTCCT |
| Cat | CGGCACATGAATGGCTATGGATC | AAGCCTTCCTGCCTCTCCAACA |
| Ptgs2 | AGTCCGGGTACAGTCACACTT | TTCCAATCCATGTCAAAACCGT |
| Gfap | CAACGTTAAGCTAGCCCTGGACAT | CTCACCATCCCGCATCTCCACAGT |
| Gapdh (reference) | GGCTCCCTAGGCCCCTCCTG | TCCCAACTCGGCCCCCAACA |
The gut microbiota composition was characterized through high-throughput sequencing of the V3 hypervariable region of the 16S rRNA gene using the Ion Torrent PGM platform. DNA amplification was performed with 337F/518R universal primers and 5× ScreenMix-HS Master Mix (# PK143S Evrogen, Russia) under the following thermal conditions: initial denaturation at 94°C for 4 min, followed by 37 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 5 min. Amplification products were purified using AMPureXP magnetic beads (# 21169900, Beckman Colter, USA) and processed into sequencing libraries with the NEBNext Fast DNA Library Prep Kit (# E6270L, New England Biolabs, USA), including barcode ligation and quality verification. Emulsion PCR amplification and sequencing were conducted using the OneTouch 2 System and Ion PGM Hi-Q View Sequencing Kit (# A30044, Thermo Fisher Scientific, USA) according to manufacturer protocols. Raw sequence data in BAM format were converted to FASTQ files and analyzed through a bioinformatics pipeline implemented in RStudio. Sequence processing included demultiplexing, quality filtering (maximum expected error cutoff of 1.0), and read trimming using VSEARCH v.2.8.2. Operational taxonomic units (OTUs) were identified using the UNOISE2 algorithm, with taxonomic classification performed through the DADA2 package employing a Naive Bayes classifier against the SILVA database (version 132) at 100% amplicon sequence variant identity.
Statistical analysis was carried out using the Statistica 12 software package (StatSoft, USA). Results are presented as means ± SEM. The normality of the data distribution was assessed using the Shapiro-Wilk test. Since the data distribution differed from normal, we used non-parametric statistical methods. The statistical significance of differences between groups was assessed by the Kruskal-Wallis test. Statistical significance was considered to be p < 0.05.
During the Morris water maze testing, we evaluated drug effects on memory parameters during the 6th and 12th day probe trials, and assessed learning performance during acquisition training (days 1–5) and reversal training (days 7–11). Neither LPS injections nor polyphenol administration significantly altered the time spent searching for the platform during the 6th day [H (4, N = 48) = 1.886, p = 0.757] or 12th day [H (4, N = 48) = 2.905, p = 0.574] probe trials (Figure 1).
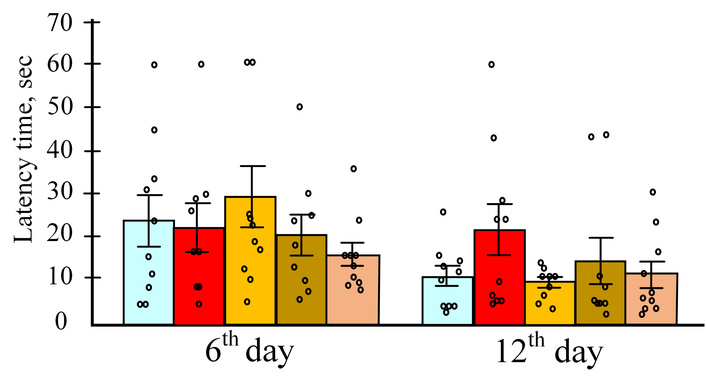
The time that the mice spent on the control days of the test. The results are expressed as means ± SEM. Control, n = 10; LPS, n = 9; LPS + curcumin, n = 10; LPS + resveratrol, n = 9; LPS + salidroside, n = 10. LPS: lipopolysaccharide.
Analysis of the acquisition training phase revealed that LPS injections were associated with increased platform search time, though the Kruskal-Wallis test failed to reach statistical significance for this phase. Mice in the LPS group showed a 1.25-fold increase in search time relative to controls. All polyphenol treatments attenuated this LPS effect, though curcumin showed limited efficacy, failing to demonstrate significant improvement over LPS-only controls (Figure 2).
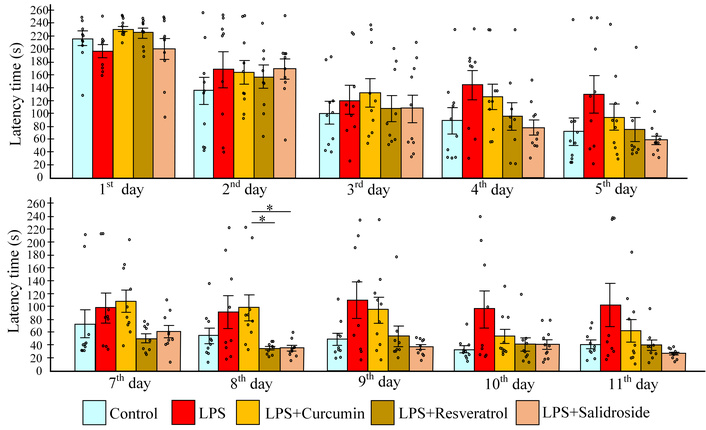
Learning performance during acquisition training and reversal training. Time in acquisition (upper row), reverse learning (lower row). The results are expressed as means ± SEM. Control, n = 10; LPS, n = 9; LPS + curcumin, n = 10; LPS + resveratrol, n = 9; LPS + salidroside, n = 10. *p < 0.05-comparison of the control group and experimental groups using the Kruskal-Wallis test. LPS: lipopolysaccharide.
All experimental groups demonstrated improved performance during acquisition training, as indicated by negative learning coefficients ranging from –13.6 (LPS group) to –35.2 (LPS + salidroside group), though intergroup differences were non-significant. While resveratrol treatment showed a trend toward enhanced learning (lower coefficient vs. LPS group; p = 0.07), this did not reach statistical significance. During reversal training, only the LPS group exhibited a positive learning coefficient (53.3 ± 33.7), reflecting impaired relearning capacity. In contrast, all polyphenol-treated groups maintained negative coefficients, with the curcumin group showing the lowest value (–17.3 ± 3.5), approximately 4-fold lower than LPS controls, though this difference trended toward but did not reach significance (p = 0.056) (Figure 3).
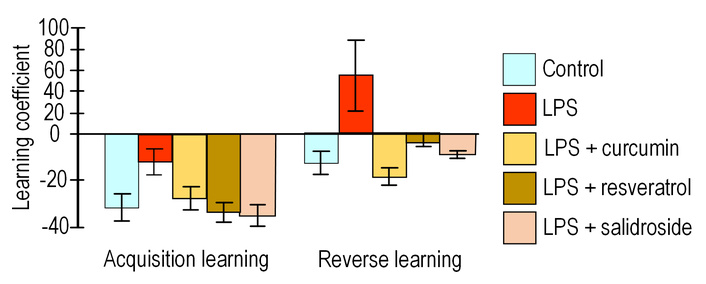
Learning coefficient. The results are expressed as means ± SEM. Control, n = 10; LPS, n = 9; LPS + curcumin, n = 10; LPS + resveratrol, n = 9; LPS + salidroside, n = 10. Comparison of the control group and experimental groups using the Kruskal-Wallis test. LPS: lipopolysaccharide.
Clustering analysis revealed two distinct gene expression patterns (Figure 4). The first cluster comprised antioxidant-related genes, including peroxiredoxin pathway components (Prdx3, Txnrd2), glutathione metabolism regulators (Gpx, Gclc), catalase (Cat), and the master antioxidant regulator Nfe2l2. Notably, the pro-inflammatory cytokine Il1b unexpectedly grouped within this antioxidant cluster. The second cluster contained canonical inflammatory markers: cytokines (Il6, Tnf), cyclooxygenase Ptgs2, astrocyte activation marker Gfap, and autophagy-related genes (Mtor, Pink1, p62).
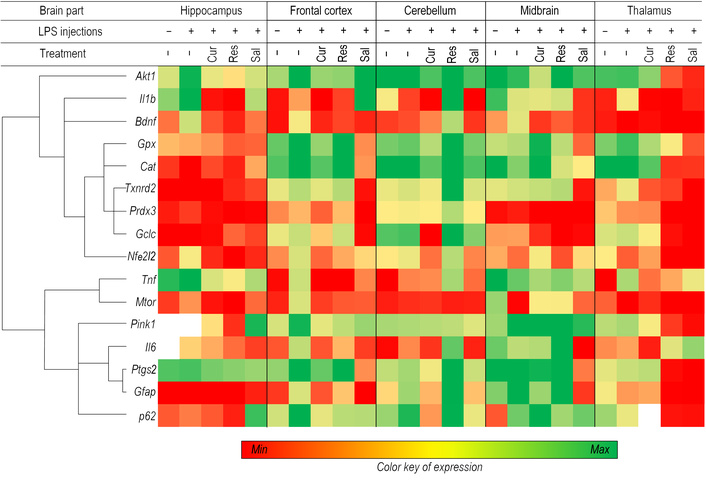
A heat map representing genes based on expression values in different parts of the brain is divided into two main clusters. Control, n = 10; LPS, n = 9; LPS + curcumin, n = 10; LPS + resveratrol, n = 9; LPS + salidroside, n = 10. LPS: lipopolysaccharide.
LPS administration triggered robust pro-inflammatory responses across brain regions, with striking regional specificity. The frontal cortex showed the most pronounced activation, with 8.7-fold Tnf upregulation, 4.4-fold Il6 increase, and 4.1-fold Gfap elevation. The thalamus exhibited extreme Tnf induction (14-fold), while the cerebellum demonstrated moderate but significant increases in Il6 (3-fold) and Tnf (4.8-fold).
All tested polyphenols counteracted LPS-induced inflammation, though with compound- and region-specific patterns. Salidroside demonstrated the most consistent anti-inflammatory profile, particularly in the cortex, where it reduced Il6 (5.6-fold), Tnf (3.1-fold), and Gfap (13-fold) expression. Its effects extended to other regions, including the hippocampus (2.6-fold Tnf reduction) and midbrain (11-fold Il6 decrease). Curcumin showed exceptional potency against Tnf (> 20-fold suppression in cortex) but more variable effects on other markers, with moderate Il6 reduction (4.7-fold in cortex) and limited cerebellar activity. Resveratrol exhibited region-dependent effects, significantly lowering Tnf (3.5-fold) and Ptgs2 (9-fold) in the thalamus while paradoxically increasing several inflammatory markers in the cerebellum.
We quantified mtDNA damage levels in the hippocampus, cortex, cerebellum, midbrain, and thalamus. No statistically significant increase in mtDNA damage was observed in any of these regions following LPS administration. Thus, LPS exposure did not induce significant mtDNA damage in key brain structures associated with long-term memory formation and storage (Figure 5).
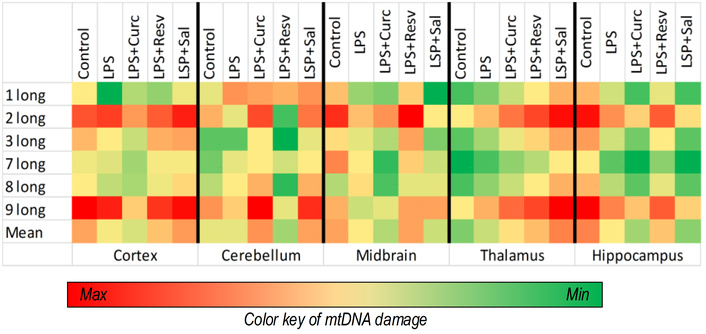
A heat map showing the amount of mtDNA damage in different parts of the brain. Control, n = 10; LPS, n = 9; LPS + curcumin, n = 10; LPS + resveratrol, n = 9; LPS + salidroside, n = 10. mtDNA: mitochondrial DNA; LPS: lipopolysaccharide.
However, the salidroside-treated group exhibited higher mtDNA damage in the thalamus compared to other experimental groups, with an average increase of 27% relative to controls across all measured fragments (all p < 0.05). Furthermore, salidroside administration was associated with significantly elevated cortical damage for long fragments 1, 3, and 9 (all p < 0.01) compared to the LPS group. Additionally, both curcumin (fragment 1, p < 0.05) and resveratrol (fragment 3, p < 0.05) treatments resulted in greater cortical damage relative to LPS controls (Figure 5).
Analysis of the gut microbiome composition across all experimental groups revealed a consistent profile dominated by the phyla Bacteroidetes and Firmicutes. Within these phyla, the most abundant classes were Bacilli, Bacteroidia, and Clostridia. Overall, no major differences in bacterial taxonomic diversity were observed between the experimental groups (Figure 6).
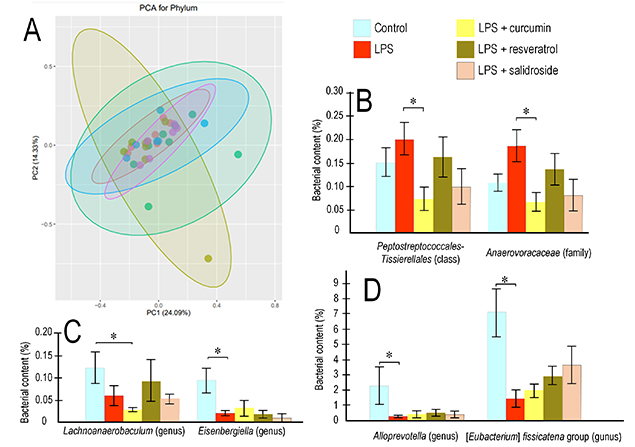
Bacterial composition of the gut microbiome. PCA graph for the microbiome of mice that received polyphenols against the background of induced inflammatory processes (A). Changes in the level of bacteria of the Peptostreptococcales-Tissierellales class and the Anaaerovoraceae family (B); genera Lachnoanaerobaculum and Eisenbergiell (C); as well as Alloprevotella and [Eubacterium] fissicatena group (D). Control, n = 10; LPS, n = 9; LPS + curcumin, n = 10; LPS + resveratrol, n = 9; LPS + salidroside, n = 10. Reliability of differences between groups: *p < 0.05 (Kruskal-Wallis criterion). LPS: lipopolysaccharide; PCA: principal component analysis.
Polyphenol consumption significantly altered the abundance of bacteria belonging to the classes Clostridia and Bacteroidia. Among pathogenic representatives of Clostridia, statistically significant differences were observed for the order Peptostreptococcales-Tissierellales, which was significantly enriched in mice receiving LPS alone compared to those supplemented with curcumin. Specifically, the abundance of this order was 2.8-fold higher in the LPS group than in the curcumin-treated group (p < 0.05). A similar trend was observed for the family Anaerovoracaceae (within the same order), which showed a 2.7-fold increase in the LPS group relative to the curcumin group (p < 0.05) (Figure 7).
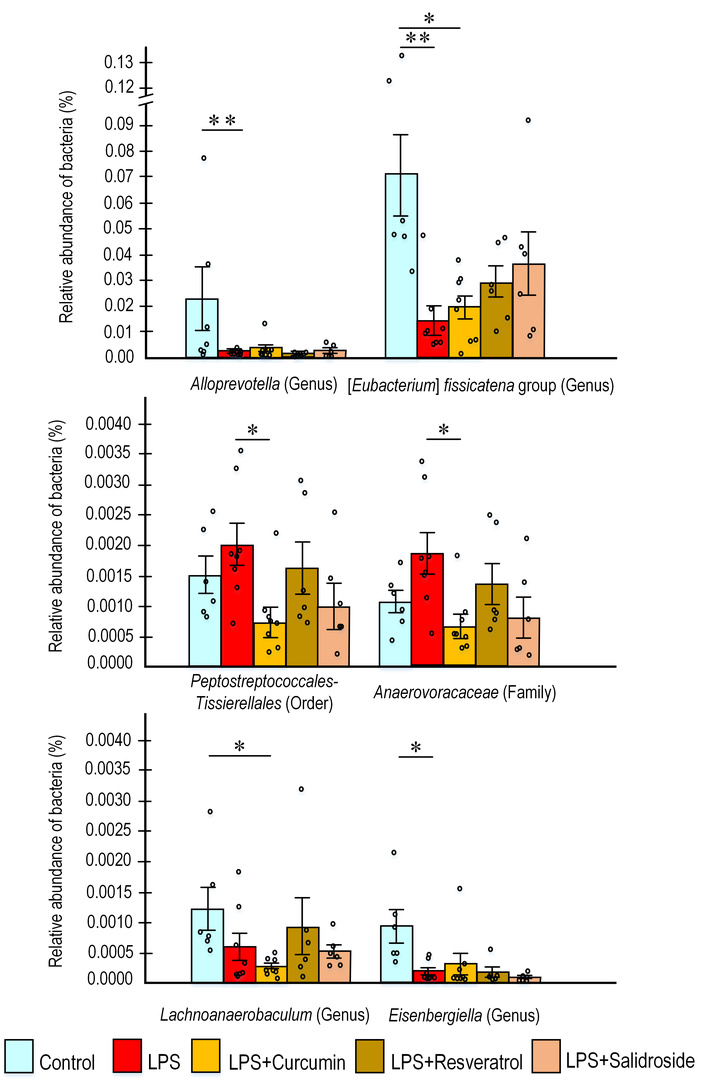
The content of the predominant bacteria in the gut microbiome. The results are expressed as means ± SEM. Control, n = 10; LPS, n = 9; LPS + curcumin, n = 10; LPS + resveratrol, n = 9; LPS + salidroside, n = 10. *p < 0.05, **p < 0.01 comparison of the control group and experimental groups using the Kruskal-Wallis test. LPS: lipopolysaccharide.
Among beneficial representatives of the Clostridia class—which support intestinal barrier and protective functions through short-chain fatty acid (SCFA) production—we observed statistically significant changes in several genera of the Lachnospiraceae family, specifically Eubacterium, Eisenbergiella, and Lachnoanaerobaculum. These genera generally showed reduced abundance in LPS-treated groups, whereas polyphenol supplementation partially restored their levels. Specifically, the abundance of Eisenbergiella was 6-fold lower in the LPS group compared to controls (p < 0.05), but 2-fold higher in the curcumin group compared to the LPS group. Eubacterium abundance was significantly lower in the LPS group relative to controls (p < 0.05); similarly, the resveratrol group showed reduced levels compared to controls. However, both curcumin and salidroside counteracted this effect, increasing Eubacterium abundance 2-fold compared to the LPS group. Conversely, the curcumin group exhibited 4.5-fold fewer Lachnoanaerobaculum compared to controls (p < 0.05), while resveratrol restored near-control levels of this genus (Figure 7).
Among representatives of the Bacteroidia class, we observed significant changes in the abundance of Alloprevotella, a genus known for its anti-inflammatory properties and enhancement of intestinal barrier function. LPS administration resulted in a 5-fold reduction in Alloprevotella abundance compared to controls (p < 0.01). In contrast, polyphenol supplementation prevented this LPS-induced depletion, with no significant reduction observed in these treatment groups (Figure 7).
Analysis of the Shannon diversity index revealed no significant differences between groups, indicating that LPS exposure did not alter overall microbial diversity but specifically affected certain bacterial taxa. However, evaluation of the Bacteroidetes/Firmicutes ratio showed that curcumin treatment restored a near-control ratio of approximately 2, compared to the LPS group (ratio = 1.4). Resveratrol and salidroside also increased this ratio (1.5 and 1.2, respectively), though these changes did not reach statistical significance.
Correlation analysis assessed relationships between identified gut microbiota bacterial groups and parameters reflecting memory function. Notably, a positive correlation was identified between the acquisition learning coefficient and groups of bacteria from the Negativicutes class and genus Veillonella (both rs = 0.35, p < 0.05), which are classified as potential pathobionts. A negative correlation was observed between the acquisition learning coefficient and bacteria that favorably affect the intestine, producing SCFAs and improving the protective function of the intestine. Among such groups, we isolated bacteria of the genus Ileibacterium (rs = –0.37, p < 0.05), Alloprevotella (rs = –0.35, p < 0.05), Odoribacter (rs = –0.36, p < 0.05), as well as the families Erysipelotrichaceae (rs = –0.36, p < 0.05) and Marinifilaceae (rs = –0.36, p < 0.05). Odoribacter (rs = –0.4, p < 0.05), Marinifilaceae (rs = –0.4, p < 0.05), and Marvinbryantia (rs = –0.34, p < 0.05) also negatively correlated with the coefficient of reverse learning (Figure 8).
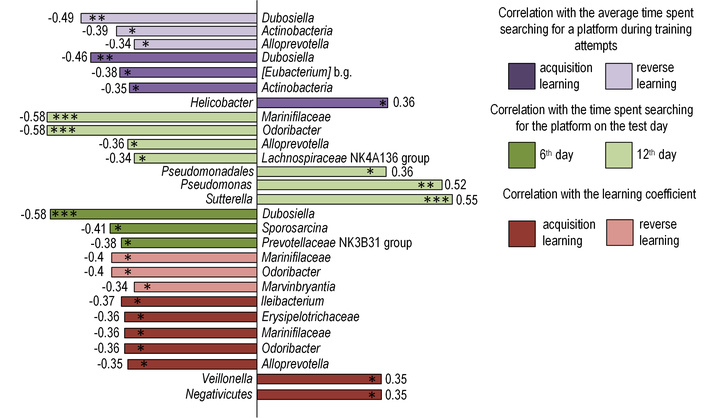
Results of correlation analysis. Correlation analysis was performed using Spearman’s rank correlation coefficient (rs). Statistical significance was considered to be *p < 0.05, **p < 0.01, ***p < 0.001.
Among the main data that we obtained during the Morris water maze, we examined the average values of the time that mice spent searching for a platform during five training days during acquisition and reverse training. Bacteria of the genus Dubosiella (rs = –0.49, p < 0.01 for reverse learning and rs = –0.46, p < 0.01 for acquisition learning) and the class Actinobacteria (rs = –0.39, p < 0.05 for reverse learning and rs = –0.35, p < 0.05 for acquisition learning) correlated negatively with these parameters. [Eubacterium] brachy group was negatively correlated with the total average time during acquisition training (rs = –0.38, p < 0.05), and a positive correlation was observed with the pathogenic genus Helicobacter (rs = 0.36, p < 0.05). The total training time during the reverse learning is negatively correlated with Alloprevotella (rs = –0.34, p < 0.05) (Figure 8).
We also considered the time that the mice spent searching for the platform in the control attempt on the 6th day of the Morris water maze. Dubosiella (rs = –0.58, p < 0.001), Prevotellaceae NK3B31 group (rs = –0.38, p < 0.05), and Sporosarcina (rs = –0.41, p < 0.05) negatively correlate with this parameter (Figure 8). Alloprevotella (rs = –0.36, p < 0.05), Odoribacter (rs = –0.58, p < 0.001), Marinifilaceae (rs = –0.58, p < 0.001), and Lachnospiraceae NK4A136 group (rs = –0.34, p < 0.05) negatively correlate with the time of the control attempt during reverse training. Pathogenic Pseudomonas (rs = 0.52, p < 0.01), Sutterella (rs = 0.55, p < 0.001), and Pseudomonadales (rs = 0.36, p < 0.05) correlate positively with the time of the control run during reverse training (Figure 8).
Systemic inflammation represents a key pathogenic factor in numerous diseases, contributing significantly to their development and progression. Substantial evidence links systemic inflammation to memory deterioration and cognitive impairment [36, 37], highlighting its detrimental role in neural function. Consequently, identifying strategies to modulate inflammatory processes and mitigate their adverse effects on memory has emerged as a critical research objective.
In our study, we investigated the effects of polyphenols on cognitive function in mice following LPS injections. The use of LPS as a chemical model of inflammation is often used in many studies, including as a neuroinflammation model to study the cognitive abilities of experimental animals. Substantial accumulated evidence demonstrates that various LPS administration methods induce central nervous system alterations, as reflected by impaired performance across multiple physiological tests. For example, LPS administration has been shown to cause cognitive deficits in mice tested in the Morris water maze [38–40], in the passive avoidance test [39], and the “Y-maze” test [41], with similar cognitive impairments observed in rats [42].
By conducting the Morris water maze, we found that resveratrol and salidroside mitigated the effect of LPS injection, which was primarily manifested in the learning process (Figure 2). There were impairments in long-term memory in the experimental group receiving only LPS injections, taking significantly longer to switch between spatial strategies, compared to mice receiving polyphenolic supplements. The mice from the LPS group exhibited no systematic search pattern, which was the reason for the increase in their search time. At the same time, the resulting trajectories of other experimental groups demonstrate a decrease in the distance traveled to the location of the platform; that is, they moved in order to get to the platform, which in turn indicates a better memory consolidation (Figure 9). Additionally, the learning coefficient shows a chronological decrease in platform search time from day to day in the learning process for mice that consumed polyphenols (Figure 3).
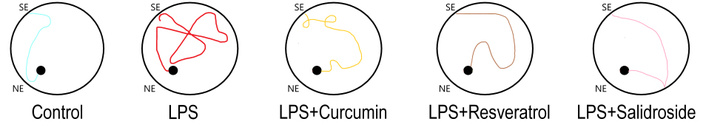
Representative trajectories of mice in the Morris water maze test. SE: start point; NE: end point; LPS: lipopolysaccharide.
Analysis of gene expression levels revealed that LPS administration predominantly increased expression of inflammatory markers across multiple brain regions, with the most pronounced effects observed in the frontal cortex (Figure 4). These findings align with existing studies reporting cortical inflammation following LPS exposure [43, 44]. While some evidence suggests potentially greater LPS sensitivity in the hippocampus compared to the cerebral cortex [45], the characteristic neuroinflammatory response—including microglial activation and upregulation of pro-inflammatory markers—consistently involves both regions [46, 47].
One primary mechanism of LPS action on the cells of the nervous system is the activation of astrocytes, which in turn participate in the immune response of the brain, and are able to support chronic inflammation and progressive neurodegeneration due to the overexpression of cytokines, growth factors, and chemokines [48]. Key inflammatory indicators include interleukins. In our study, we analyzed expression changes of Il1b, Il6, and Tnf genes, which serve as markers of the pro-inflammatory process [49]. The Gfap gene encodes one of the main proteins of intermediate filaments of mature astrocytes [50]. In turn, Ptgs2 is an induced form of Cox-2 and catalyzes the conversion of arachidonic acid into prostaglandins. Cox-2 is expressed by inflammatory cells, such as macrophages, and can be induced by TNF. It is involved in pathological processes, such as acute and chronic inflammatory conditions [51]. Reduced expression of these genes in mice treated with polyphenols compared to the LPS group indicates attenuated inflammatory responses.
Curcumin [52–54], resveratrol [55–57], and salidroside [41, 58, 59] inhibit the production of pro-inflammatory interleukins such as IL-1β, IL-6, and TNF. Curcumin also inhibits the translocation of LPS-induced NF-κB activated by interleukins, astrocyte growth factors, and phosphorylates mitogen-activated protein kinase [60]. Curcumin can suppress pro-inflammatory pathways associated with most chronic diseases and block both TNF production and TNF-mediated cellular signaling in various cell types. Curcumin can also be a TNF blocker in in vitro and in vivo studies by direct binding to TNF [61]. In addition, other studies have reported that salidroside reduces the production of LPS-induced proinflammatory cytokines and mediators and weakens acute lung damage caused by LPS by inhibiting the JAK2-STAT3 signaling pathway, and it also reduces phosphorylation of NF-κB, ERK, and p38 [62]. Recent data have shown that salidroside provides neuroprotection by modulating mitochondrial biogenesis and microglial polarization [63, 64]. There is also evidence of a relationship between different signaling pathways, such as NF-κB, ERK, and Nrf2, which influence each other by coordinating antioxidant and inflammatory responses, determining the cellular response to oxidative stress, and providing protection of cells from free radicals [65, 66]. However, our data demonstrate that although polyphenol treatment reduced pro-inflammatory cytokine expression, it did not enhance the expression of antioxidant genes, despite their putative antioxidant properties.
The primary limitation of polyphenols is their low bioavailability; upon ingestion, they undergo rapid metabolism, resulting in insufficient concentrations to exert neuroprotective effects in brain regions critical for long-term memory formation [67]. Currently, there are results showing that polyphenols can exert their biological effects after chemical modifications performed by the gut microbiota. Intestinal microbiota enzymes can perform deglycosylation, dihydroxylation of polyphenols, which leads to the formation of small catabolic products that can be easily absorbed during intestinal transit. These catabolites can fall into two classes: some have higher biological activity compared to the “parent” compound, while others lose biological activity [68]. For example, resveratrol is metabolized by hepatic, intestinal, and microbial processes [69]. A recent study has shown that the gut microbiota promotes the metabolism of resveratrol precursors into resveratrol and may also increase the bioavailability of resveratrol [70]. It has been reported that dihydroresveratrol, 3,4'-dihydroxybibenzyl, and 3,4'-dihydroxy-trans-stilbene are the main metabolites of resveratrol derived from microbiota [71]. Additionally, resveratrol has been shown to modulate gut microbiota composition in bowel diseases under conditions of severe oxidative stress [72].
Indeed, the gut microbiota plays a crucial role in determining polyphenol bioavailability, as the majority of these compounds are poorly absorbed in the small intestine [73]. Most dietary polyphenols transit to the colon, where they are metabolized into low molecular weight phenolic acids and related compounds [74]. These microbial metabolites exhibit enhanced absorption across the intestinal barrier [75] and can have beneficial effects, including weakening the adhesion of monocytes to activated TNF endothelial cells [76], reducing IL-1β secretion [77]. One study demonstrated that the variability of the microbiota, especially the type of Proteobacteria, positively correlates with Il6 expression [78]. Furthermore, inhibition of Il1a was shown to reshape the gut microbiome and attenuate inflammation and tissue damage in a murine model of Crohn’s-like ileitis [79]. Microbial metabolites directly influence the immune system, which affects brain function through circulating cytokines [80]. Notably, many beneficial effects of gut bacteria on learning and memory are associated with reduced levels of pro-inflammatory cytokines [41, 81, 82].
It can be assumed that changes in cognitive parameters in experimental groups are mediated through the modulation of the bacterial composition of the intestine by polyphenols, while the microbiota, in turn, enhances their bioavailability in the intestine. This assumption is indirectly confirmed by the data of the correlation analysis. Thus, a positive correlation was observed between cognitive deficits and bacteria with pathogenic potential. Among such bacteria, we were able to identify the class Negativicutes, which has pathogenic properties [83], and the genus Veillonella, which is referred to as a potential pathobiont. Bacteria of the genus Veillonella were found in large numbers in the microbiome of organisms suffering from depression, and as a result, with a reduced level of cognitive abilities [84], as well as a positive correlation was observed with the pathogenic genus Helicobacter, which regulates innate host reactions by injection of one of its main virulence factors, cytotoxin-associated gene A [85]. Positively correlates pathogenic Pseudomonas, Sutterella, and Pseudomonadale with the time of the control run during reverse training. Sutterella species are often associated with human diseases such as autism, Down syndrome, and inflammatory bowel disease [86]. However, Sutterella does not appear to cause significant inflammation; rather, this genus has the ability to destroy IgA [87]. Environmental and clinical strains of Pseudomonadales can produce mucus composed of glycolipoprotein and LPS [88].
Conversely, negative correlations were observed with beneficial bacteria that enhance intestinal health through SCFA production and improved barrier function. These included the genus Ileibacterium [89], Alloprevotella (a butyrate producer [90]), Odoribacter [91], as well as the families Erysipelotrichaceae and Marinifilaceae. Erysipelotrichaceae is linked to a healthy microbiome and may strengthen the intestinal mucus barrier [92], while Marinifilaceae (Bacteroidetes phylum) degrades plant polysaccharides, produces propionate, and enhances barrier function by reducing inflammation and oxidative stress [93]. The genus Marvinbryantia, which also produces SCFAs [94], showed negative correlations with cognitive deficits. Similarly, Dubosiella (SCFA producer [95]), the class Actinobacteria, the [Eubacterium] brachy group, and Alloprevotella were negatively associated with escape latency during training. Notably, Alloprevotella abundance was 5-fold lower in LPS-treated mice versus controls (p < 0.01), an effect prevented by polyphenol supplementation. Negative correlations were also found with Prevotellaceae NK3B31 group, Sporosarcina, and Prevotellaceae species are key regulators of rectal microbiota composition and correlate strongly with rectal SCFA production and serum IgG levels [96]. Other studies confirm a positive relationship between Prevotellaceae abundance and propionate/total SCFA concentrations [96]. Additionally, Sporosarcina exhibits traits suitable for probiotic development [97].
We observed statistically significant changes in several genera of the Lachnospiraceae family, including Eubacterium, Eisenbergiella, and Lachnoanaerobaculum. LPS administration significantly reduced the abundance of these Clostridia class genera compared to controls (Figure 7). Polyphenol treatment partially restored their levels: curcumin increased Eisenbergiella and Eubacterium abundance, resveratrol enhanced Lachnoanaerobaculum, and salidroside counteracted the LPS-induced reduction in Eubacterium. These microorganisms support essential intestinal functions. For instance, Lachnospiraceae NK4A136 and Odoribacter produce butyrate, which serves as an energy source for colonocytes, suppresses inflammatory cytokine release, and upregulates tight junction proteins to enhance epithelial barrier integrity [98]. The Eubacterium fissicatena group metabolizes dietary carbohydrates into butyrate—the predominant colonic SCFA—and may modulate colonic inflammatory responses. Although poorly characterized, this group can cleave riboflavin to hydroxyethylflavin [99]. Eisenbergiella species produce butyrate and have been linked to Th2 immune responses [100], while Lachnoanaerobaculum generates butyrate, acetate, H2S, and NH3 as major metabolic products [101, 102]. Butyrate itself provides nutrients to colonic tissues and maintains mucosal integrity.
It is also worth noting that LPS administration significantly increased the abundance of the order Peptostreptococcales-Tissierellales (Clostridia class). The LPS group showed a 2.8-fold higher abundance than in mice from the curcumin group (p < 0.05). Within this order, we identified the family Anaerovoracaceae, which exhibited similar trends to the overall order. Limited literature exists on the functional role of these bacteria, though some members are associated with pathogenicity. For example, certain taxa can exacerbate intestinal damage through alpha-toxin production [103], while others in the Anaerovoracaceae family appear to inhibit straight-chain SCFA production while promoting branched-chain SCFA accumulation [104]. Decreased SCFA levels are an integral part of dysbiosis and the development of a wide range of diseases associated with it, including autoimmune diseases [105].
The observed microbiome modifications exert bidirectional effects along the gut-brain axis, both preventing and reversing neuroinflammatory processes. The molecular mechanisms involve bacterial molecules that act directly on neurons, affecting their excitability, or indirectly on non-neuronal cells, inducing changes in the production of proinflammatory or anti-inflammatory mediators [106]. This is evidenced by reduced expression of key pro-inflammatory genes in polyphenol-treated mice.
The microbiota enzymatically metabolizes unabsorbed polyphenols in the colon, generating metabolites with enhanced antioxidant activity [75, 107, 108]. Conversely, polyphenols, namely curcumin in particular, show an inhibitory effect on some pathogenic bacterial species. One study showed that curcumin inhibited the growth of Helicobacter pylori bacteria on agar plates and eradicated the bacteria in mice, respectively. The bactericidal effect of curcumin appears to occur by inhibiting bacterial cell division, which leads to improper assembly of the bacterial protofilament. In addition, some studies have demonstrated antimicrobial activity of curcumin against a number of common pathogenic gram-negative and gram-positive bacteria [109]. However, the pharmacological potential of curcumin is widely limited due to its poor solubility in water, chemical instability, and rapid metabolism. In addition, the bioavailability of curcumin is very low after oral administration [110]. These physicochemical properties may explain its reduced efficacy in our study compared to resveratrol and salidroside, which exhibit higher stability and absorption. Furthermore, curcumin’s microbial metabolism differs significantly from other polyphenols: while resveratrol and salidroside are converted into active metabolites by specific gut bacteria, curcumin undergoes more complex transformations that often reduce its activity. These differences in bioavailability, metabolic fate, and microbial interactions likely contribute to the differential neuroprotective outcomes observed.
While our study demonstrates significant associations between polyphenol intake, microbial shifts, and cognitive improvement, we acknowledge that the causal pathways within the gut-brain axis may involve alternative mechanisms. Specifically, polyphenols may exert their beneficial effects not only through microbiota remodeling but also via direct systemic immunomodulation [111]. Given their known anti-inflammatory properties, polyphenols could directly attenuate LPS-induced neuroinflammation, leading to improved cognitive function independently of microbial changes. Further studies using germ-free models or fecal microbiota transplantation are needed to dissect the precise contributions of each pathway (Figure 10).
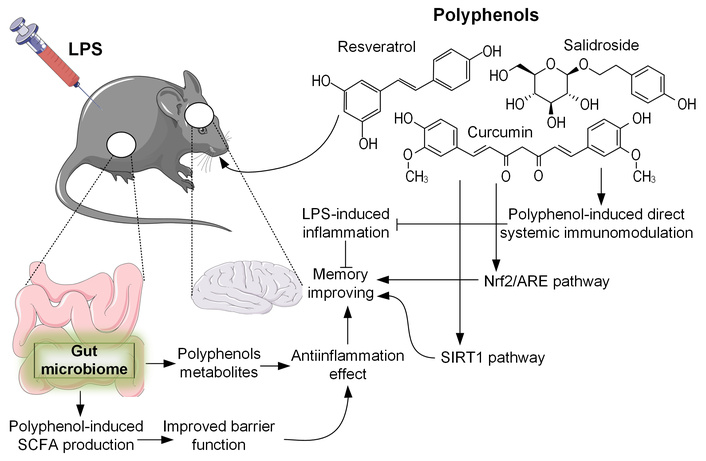
An overview of the multifaceted biological activities of polyphenols. Polyphenols are metabolized by the gut microbiome, leading to increased production of short-chain fatty acids (SCFAs) and an improvement in the intestinal barrier function, which reduces the systemic impact of lipopolysaccharides (LPSs). These gut-mediated effects contribute to systemic immunomodulation, potent anti-inflammatory properties, and memory-enhancing capabilities. Additionally, polyphenols can directly influence immune function and activate cytoprotective pathways such as the Nrf2/ARE and SIRT1 signaling pathways. Nrf2/ARE: nuclear factor erythroid 2-related factor 2/antioxidant response element; SIRT1: sirtuin 1. All icons were sourced from Bioicons (https://bioicons.com), a library providing free open-source icons under Creative Commons (CC 3.0, CC BY-SA) and MIT licenses.
Dietary phenolic compounds are associated with beneficial health effects and are considered partially responsible for their neuroprotective activity [26]. A number of these compounds are able to modulate the Nrf2/ARE pathway and thus represent a potential tool for preventing cognitive decline and neurodegenerative diseases [29]. The Nrf2/ARE pathway is closely related to the pathogenesis of neurodegenerative diseases, being a potential neuroprotective factor and a target for modulation of neuroinflammation [112]. Some examples of dietary phenolic compounds that are known to have a neuroprotective effect through Nrf2 include the yellow pigment in curcumin [29].
Several mechanisms may explain the influence of polyphenolic substances on cognitive function. Since the mechanism of memory impairment as a whole is based on the processes of oxidative stress, which cause corresponding reactions along the signaling pathways associated with Nrf2/ARE, then a logical step would be to consider damage in mtDNA. Since polyphenols, being powerful antioxidants, can activate the Nrf2/ARE signaling pathway responsible for maintaining mitochondrial homeostasis, we assumed a protective effect of these substances on mtDNA when LPS acts on it, but it turned out that LPS injections did not cause disturbances in the structure of mtDNA of the brain regions responsible for the formation of long-term memory. The absence of significant mtDNA damage in our model may be attributed to the specific experimental conditions, including the LPS dosage, administration protocol, or the time point of analysis. Alternatively, compensatory mechanisms such as enhanced DNA repair or antioxidant defenses may have mitigated mtDNA damage in our experimental setting. This finding is consistent with reports by Zhan et al. (2021) [113], where it was found that LPS stimulates the biogenesis of extranuclear DNA and its subsequent release into the cytoplasm, which is a pathological process; however, an increase in the level of markers of oxidative DNA damage was not detected. However, it has been demonstrated that LPS-induced functional and structural injury of the mitochondria in the nigrostriatal pathway [114].
It is classically believed that resveratrol is able to activate SIRT1, which, by deacetylating target proteins, plays an important role in metabolic processes. It is well-established that resveratrol activates SIRT1, which deacetylates target proteins and plays a crucial role in metabolic regulation. Resveratrol-mediated activation of both Nrf2/ARE and SIRT1 has been demonstrated in aging mouse models [115]. Similarly, salidroside can activate SIRT1 to exert antioxidant effects [62]. Recent evidence further indicates a link between SIRT1 and the modulation of inflammatory processes. Additionally, literature reports describe resveratrol’s ability to inhibit cyclooxygenase activity [116], a finding consistent with our own data. However, quantitative PCR analysis revealed that LPS injections increased the expression of not only neurogenesis-related genes in the cortex but also genes associated with the antioxidant system. This observed upregulation may reflect a compensatory mechanism activated to counteract reactive oxygen species (ROS)-mediated oxidative stress.
While this study provides evidence for polyphenol-mediated improvements in cognition through gut microbiome modulation, several limitations should be acknowledged. First, the experimental design does not fully establish causality in the gut-brain axis interactions. Although we observed significant correlations between specific bacterial taxa and cognitive performance, the absence of fecal microbiota transplantation or antibiotic depletion experiments prevents definitive conclusions about whether microbiome changes are necessary or sufficient for the observed effects. Future studies should incorporate these approaches to validate the mechanistic role of the microbiota. Second, our memory assessment focused exclusively on spatial memory using the Morris water maze, which provides robust measures of hippocampal-dependent learning but does not evaluate non-spatial memory domains such as recognition or associative memory. This narrow focus was chosen to maintain experimental consistency with established LPS-induced cognitive deficit models, but it limits the generalizability of our findings to broader cognitive functions. Finally, while LPS-induced inflammation is a widely used model for studying neuroinflammation, it represents an acute, non-physiological challenge that may not fully replicate the chronic, low-grade inflammation observed in age-related cognitive disorders. The translational relevance of our findings to human conditions should therefore be interpreted with caution, and future work should explore complementary models such as high-fat diet-induced systemic inflammation or genetic models of chronic gut barrier dysfunction.
In conclusion, our study expands the current understanding of how dietary polyphenols modulate the gut-brain axis, highlighting their potential role in mitigating inflammation-associated cognitive impairment. While confirming the importance of antioxidant mechanisms, we provide evidence that microbiota remodeling and immunomodulation significantly contribute to the neuroprotective effects of resveratrol and salidroside. These findings complement existing paradigms and emphasize the multifactorial nature of polyphenol-mediated neuroprotection. Our results support the exploration of polyphenol-rich diets as complementary strategies for maintaining cognitive health. The observed compound-specific effects highlight the importance of personalized nutritional approaches.
ERK: extracellular signal-regulated kinase
LPS: lipopolysaccharide
mtDNA: mitochondrial DNA
NF-κB: nuclear factor kappa-light-chain-enhancer of activated B cells
Nrf2/ARE: nuclear factor erythroid 2-related factor 2/antioxidant response element
PCR: polymerase chain reaction
SCFA: short-chain fatty acid
SIRT1: sirtuin 1
EPK: Conceptualization, Methodology, Writing—original draft, Writing—review & editing. PIB: Investigation, Visualization. VVN: Formal analysis, Investigation. ADT: Software, Validation. IYB: Data curation. APG: Conceptualization, Methodology, Writing—review & editing, Funding acquisition, Project administration. All authors have read and approved the submitted version.
The authors declare that they have no conflicts of interest.
The animal study protocol was approved by the Institutional Ethical Committee of Voronezh State University (section of animal Care and Use, 94 Protocol No. 42-03 of October 8, 2020).
Not applicable.
Not applicable.
The data generated and analyzed during the current study are available from the corresponding author upon reasonable request.
The study was supported by the Ministry of Science and Higher Education of the Russian Federation within the framework of the State task for universities in the field of scientific activity (project No. FZGU-2023-0009). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 2018
Download: 21
Times Cited: 0
Leonel Pereira, Ana Valado
Sharon Smith ... David Heal
Prerna Sarup ... Sonia Pahuja
Susmita Das ... Shylaja Hanumanthappa
Sneha Bagle ... Sadhana Sathaye
Luis Antonio Ramirez-Contreras ... Andrés Frausto de Alba
Agustina Lulustyaningati Nurul Aminin ... Muhammad Ajmal Shah
