Affiliation:
1Department of Biochemistry, University of Medicine and Pharmacy Craiova, 200349 Craiova, Romania
†These authors contributed equally to this work.
ORCID: https://orcid.org/0000-0002-3621-0566
Affiliation:
1Department of Biochemistry, University of Medicine and Pharmacy Craiova, 200349 Craiova, Romania
2Department of Psychiatry, University of Medicine and Pharmacy Craiova, 200349 Craiova, Romania
†These authors contributed equally to this work.
ORCID: https://orcid.org/0000-0003-3654-5275
Affiliation:
1Department of Biochemistry, University of Medicine and Pharmacy Craiova, 200349 Craiova, Romania
ORCID: https://orcid.org/0000-0002-8897-0241
Affiliation:
1Department of Biochemistry, University of Medicine and Pharmacy Craiova, 200349 Craiova, Romania
2Department of Psychiatry, University of Medicine and Pharmacy Craiova, 200349 Craiova, Romania
ORCID: https://orcid.org/0000-0003-0543-3596
Affiliation:
3Department of Neurology, University Hospital Essen, University of Duisburg-Essen, D-45122 Essen, Germany
ORCID: https://orcid.org/0000-0003-0198-3152
Affiliation:
4Stroke Pharmacogenomics and Genetics Group, Sant Pau Hospital Institute of Research, 08041 Barcelona, Spain
Affiliation:
5Department of Neurology, University of Gießen, 35390 Gießen, Germany
6Department of Neurology, University Medicine Göttingen, 37075 Göttingen, Germany
ORCID: https://orcid.org/0000-0002-1222-9211
Affiliation:
1Department of Biochemistry, University of Medicine and Pharmacy Craiova, 200349 Craiova, Romania
Email: roxana.surugiu@gmail.com
ORCID: https://orcid.org/0000-0003-4509-885X
Affiliation:
1Department of Biochemistry, University of Medicine and Pharmacy Craiova, 200349 Craiova, Romania
3Department of Neurology, University Hospital Essen, University of Duisburg-Essen, D-45122 Essen, Germany
Email: aurel.popa-wagner@geriatrics-healthyageing.com
ORCID: https://orcid.org/0000-0003-4574-8605
Explor Neurosci. 2023;2:27–47 DOI: https://doi.org/10.37349/en.2023.00010
Received: July 19, 2022 Accepted: October 11, 2022 Published: February 26, 2023
Academic Editor: Suyue Pan, Southern Medical University, China
The article belongs to the special issue Cerebral Ischemia, Genetics, Comorbidities, Risk Factors and New Therapeutic Options for Neurorestoration
Aim: Stroke is one of the leading causes of death and disability worldwide. Plasma biomarkers have long been used to evaluate physiological or pathological processes and to make predictions about the outcome of stroke patients. The current systematic review is focused on genetic plasma biomarkers as a new potential prognostic indicator for post-stroke recovery. The aim of the present systematic review is to assess the potential of genetic plasma biomarkers associated with stroke to predict post-stroke recovery.
Methods: The search strategy used PubMed and Web of Science databases to identified 166 studies that investigated genetic plasma biomarkers in patients with stroke between 2017 and 2021. However, only 21 of them met the inclusion criteria.
Results: The identified genetic biomarkers can be divided into: (i) serum/plasma circular RNA (circRNA) associated with stroke onset or recurrence (5; 23.80%), (ii) genetic polymorphisms associated with the atherosclerotic process and stroke recurrence (6; 28.57%), (iii) serum/plasma long non-coding RNA (lncRNA) levels involved in immunity/inflammatory processes (4; 19.04%), (iv) marker of DNA methylation associated with stroke onset and outcome (3; 14.28%), and (v) proteins and pathways of stroke identified by serum/ plasma proteomics/genomics analysis (3; 14.28%).
Conclusions: Overall, more than 100 potential biomarkers were found and the data suggest that combinations of plasma genetic biomarkers might be used as a better predictor for stroke.
Stroke is the second leading cause of death and disability in the European Union [1], the fifth in the USA [2], and the third leading cause of chronic disability worldwide [3] with over 795,000 people who experience a new or recurrent stroke every year [4]. Moreover, between 2008 and 2018, there was a 10.2% increase in the number of deaths caused by stroke [4]. Over 85% of all strokes are of the ischemic type.
There are several therapeutic options available during the acute phase of stroke. Thrombolysis using recombinant tissue plasminogen activator (r-tPA) and/or mechanical thrombectomy can be used to limit the consequences of acute occlusions of cerebral blood vessels. Clinical trials on patients that received r-tPA treatment within 3 h of the onset still reported functional deficits at 3 months and 12 months post-stroke, on a modified Rankin Scale 2–5 [5]. The motor deficit is the most frequent one encountered, however, disability-adjusted life years (DALYs) include cognitive, linguistic, optical, and sensorial deficits [5]. Numerous neuroprotectants have been tested with promising results in animal models. However, there is a difficult transition to human clinical trials, which may be the cause of slow progression in the field [6]. Some of the recently completed clinical trials have tested the therapeutic efficacy of human urinary kallidinogenase [7, 8], 3-methyl-1-phenyl-2-pyrazolin-5-one [9], nerinetide [10], and 3K3A-activated protein C, a recombinant variant of human activated protein C [11], with results of which still being evaluated.
Despite sustained efforts to develop clinically effective drugs, the complex mechanism underlying stroke recovery makes complete functional recovery unlikely. Therefore, research into the prevention and identification of biomarkers that could potentially improve the response to treatment is currently advisable [12]. By the Biomarkers Definitions Working Group [13] in 1998, a biological marker or biomarker refers to a vast category of indicators defined as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention”. More and more studies aim to identify useful plasma biomarkers associated with the prognosis of ischemic stroke (IS). Montellano et al. [14] reported a growth in the number of publications that evaluate the role of biomarkers associated with the prognosis of stroke between 2007–2018, while another recent review made a comprehensive systematic investigation of blood biomarkers associated with physical post-stroke recovery [15].
A search of PubMed and Web of Science databases as of May 5, 2022 identified 169 studies that investigated genetic plasma biomarkers in patients with stroke in the past 5 years (2017–2021). The search strategy used the following keywords: (i) genetic, (ii) biomarker, (iii) plasma, and (iv) stroke (Table S1). All results were filtered for English language, human patients, original work, and free access. Duplicate studies were removed from the list and the abstracts were read to assess the eligibility criteria. Finally, all eligible articles were fully read and only the relevant ones were included in the study. Articles that proved to be on animal models, studies on outcomes after a cardio-vascular event without a stroke, studies that included biomarkers from other tissues, and studies that focused on non-genetic biomarkers were excluded from the list. Data extracted from each study consisted of author information, year of publication, journal, impact factor (IF) in the publishing year, cohort data, biomarker with potential targeted pathway, and the REporting recommendations for tumor MARKer prognostic studies (REMARK) quality questionnaire [16].
Using the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) flow diagram [17], the search strategy identified 78 articles in PubMed and 90 articles in Web of Science. Two duplicate studies were found and from the remaining 76 PubMed articles, only 6 (7.89%) met the inclusion criteria. Forty-seven studies (61.84%) were excluded because they were done on patients without a stroke, 3 (3.94%) were not clinical trials, 9 (11.84%) identified non-genetic biomarkers, 10 (13.15%) used other tissue biomarkers, and 1 (1.31%) was an association of stroke with different types of active cancers. From the 90 Web of Science articles, 15 (16.66%) met the inclusion criteria, 29 (32.22%) were excluded because they were on patients without a stroke, 33 (36.66%) were not clinical trials, 5 (5.55%) identified non-genetic biomarkers, 7 (7.77%) used other tissue biomarkers, and 1 (1.11%) was an association of stroke with diabetes biomarkers (Figure 1).
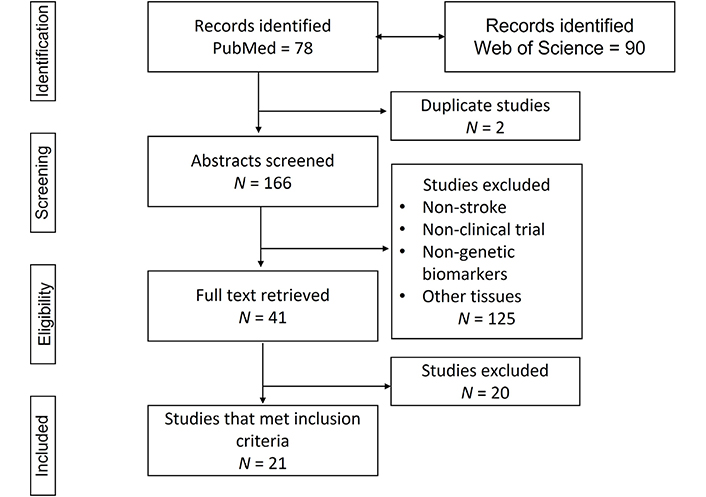
Flow diagram of the literature search
Note. Adapted from “The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration,” by Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, et al. BMJ. 2009;339:b2700 (https://www.bmj.com/content/339/bmj.b2700.long). CC BY-NC.
Studies were published between 2017–2021. The studies were conducted in China (42.85%), USA (28.57%), Japan (9.52%), Poland (4.76%), Turkey (4.76%), and Spain (9.52%), and the sample size varied from 6 patients to 2,763 patients. All studies enrolled patients with stroke and 11 studies (52.38%) were matched with a control cohort of healthy individuals.
The combined mean and standard deviation (SD) for the age of the stroke patients was 63.86 (SD = 13.98) and in the control group was 62.047 (SD = 12.21). Two of the studies (9.52%) did not provide the age of the patients enrolled and two (9.52%) provided only an age interval (49–80 and 64–85, respectively), for which statistical analysis could not be performed.
For the patients with stroke, the male cohort was larger than the females (55.43% vs. 44.56%). This was maintained for the studies that had a control group (57.29% vs. 42.70%). Two of the studies (9.52%) included samples from databases, for which characteristics of the patients were not provided.
In nine studies, the blood samples were collected during the acute phase (< 24 h from the onset) (42.85%). Two studies collected the blood samples in the first two weeks (9.52%), two in a time frame of 120 days (9.52%), one collected the blood samples at 6 months (4.76%), and seven did not provide a specific window (33.33%). Two studies were indexed in journals with an IF under 1 (9.52%), two were indexed in journals with an IF between 1 and 3 (9.52%), eleven were in journals with an IF between 3 and 5 (52.38%) and six were indexed in journals with an IF over 5 (28.57%). Over 100 biomarkers were identified in all the studies. The characteristics of the studies can be found in Table 1.
Characteristics of the study
| Item | Year | Authors | Link | Country | Journal | IF | Cohort data | Sample collection time |
|---|---|---|---|---|---|---|---|---|
| 1 | 2020 | Lu et al. [20] | https://www.webofscience.com/wos/woscc/full-record/WOS:000515868500001 | China | Frontiers in Neuroscience | 4.67 | Samples were matched for demographic and vascular risk factors as well as the history of previous use of anti-platelet medications. The study included 16 patients (M), with an age mean ± SD of 55.4 years ± 12.3 years in the stroke group and 54.11 years ± 10.1 years in the C. | 60 min to 280 min, with a mean time of 179 min from stroke symptoms onset. |
| 2 | 2020 | Ostolaza et al. [23] | https://www.webofscience.com/wos/woscc/full-record/WOS:000520976200005 | Spain | Cell and Bioscience | 7.13 | A cohort of 700 patients recruited from January 2015 to December 2016, 30 patients (19 M + 11 F) were included in the discovery cohort and 50 patients were included in the validation cohort. The age of patients in the discovery cohort was between 49 and 80. No healthy C. | Venous blood samples were drawn from acute stroke patients within 24 h after admission. |
| 3 | 2020 | Abe et al. [24] | https://pubmed.ncbi.nlm.nih.gov/32354168/ | Japan | International Journal of Molecular Sciences | 5.92 | The study included 45 patients (IS: 17 M + 6 F; C: 17 M + 5 F). Patients’ age (IS: 68.6 years ± 15.8 years; C: 69.2 years ± 16.4 years). | Blood samples were collected after the treatment. |
| 4 | 2018 | Vijayan et al. [26] | https://www.webofscience.com/wos/woscc/full-record/WOS:000438578900007 | USA | Human Molecular Genetics | 4.54 | The study included 45 patients (IS: 13 M + 21 F; C: 5 M + 6 F). Patients’ age (IS: 62.88 years ± 11.94 years; C: 62.63 years ± 6.6 years). | Not specified. |
| 5 | 2017 | Mick et al. [28] | https://www.webofscience.com/wos/woscc/full-record/WOS:000398207000017 | USA | Stroke | 6.33 | The study included 2,763 patients (IS: 1,264 M + 1,499 F). Age of patients: 66.3 years ± 8.9 years. No healthy C. | The blood samples were collected at the 8th exam from the patients that were included in Framingham Heart Study (FHS) off-spring cohort. |
| 6 | 2019 | Zhu et al. [29] | https://www.webofscience.com/wos/woscc/full-record/WOS:000457370300001 | China | Frontiers in Neurology | 3.55 | The study included 10 patients (IS: 4 M + 1 F; C: 4 M + 1 F). Patients’ age (IS: 61 years ± 11.3 years; C: 59 years ± 6.2 years). | Blood was collected within 24 h and 7 days after symptom onset. |
| 7 | 2020 | Cao et al. [31] | https://www.webofscience.com/wos/woscc/full-record/WOS:000513554900005 | China | International Journal of Molecular Medicine | 3.09 | Patients with IS had only one episode of stroke ≥ 6 months before blood collection, and controls could not have a family history of stroke. The study included 40 patients (IS: 10 M + 10 F; C: 10 M + 10 F). Patients’ age (IS: 60.2 years ± 10.6 years; C: 58.7 years ± 11.0 years). | The IS patients in the study suffered only one stroke episode ≥ 6 months before the blood collection. |
| 8 | 2022 | Wang et al. [33] | https://www.webofscience.com/wos/woscc/full-record/WOS:000711301400001 | China | Genes & Genomics | 1.83 | The study included 6 patients (IS: 3 M; C: 3 M). Patients’ age (IS: 51 years ± 0.81 years; C: 51.3 years ± 0.47 years). | Peripheral blood samples were collected the next morning after a fasting period of 10 h or overnight. |
| 9 | 2020 | Zhang et al. [38] | https://www.webofscience.com/wos/woscc/full-record/WOS:000575004300053 | China | Molecular Medicine Reports | 2.95 | The study included 168 patients divided into a stroke group and a non-stroke C. Patients’ age and sex are not specified. | Not specified. |
| 10 | 2020 | Zhu et al. [40] | https://www.webofscience.com/wos/woscc/full-record/WOS:000519997800015 | China | Genomics | 5.73 | The study included 610 patients (IS: 422 M + 188 F). Patients’ age (≤ 55 years old: 172; ≥ 55 years old: 438). No healthy C. | Samples were collected at the onset of IS. |
| 11 | 2018 | Li et al. [44] | https://www.webofscience.com/wos/woscc/full-record/WOS:000429737400001 | China | Lipids in Health and Disease | 0.95 | The study included 747 patients divided into an IS group and a non-stroke C. Patients’ age and sex are not specified. | Not specified. |
| 12 | 2020 | Xie et al. [45] | https://www.webofscience.com/wos/woscc/full-record/WOS:000601211900010 | China | Biomed Research International | 3.41 | The study included 34 patients (IS: 14 M + 20 F). Patients’ age: 71.94 years ± 14.61 years. No healthy C. | Samples were collected within 24 h from the known onset of symptoms and again at 24–48 h after onset. |
| 13 | 2019 | Shroff et al. [47] | https://www.webofscience.com/wos/woscc/full-record/WOS:000455396400002 | USA | Translational Stroke Research | 5.78 | The study included 155 patients (IS: 30 M + 13 F; C: 59 M + 53 F). Patients’ age (IS: 65.48 years ± 3.72 years; C: 63.83 years ± 2.41 years). | Venous blood draws were performed within 24 h of stroke onset. |
| 14 | 2017 | Williams et al. [48] | https://pubmed.ncbi.nlm.nih.gov/28495826/ | USA | Stroke | 6.33 | The study included 2,100 patients (IS: 1,335 M + 765 F). Patients’ age: 68.5 years ± 11.48 years. No healthy C. | Not specified. |
| 15 | 2021 | Davis Armstrong et al. [49] | https://pubmed.ncbi.nlm.nih.gov/33661917/ | USA | Plos One | 3.58 | The study included 44 patients (IS: 22 M + 22 F). Patients’ age: 63.56 years ± 10.59 years. No healthy C. | Participants were enrolled within 120 days of suffering a non-disabling cerebral infarction and followed for two years. |
| 16 | 2021 | Davis Armstrong et al. [51] | https://pubmed.ncbi.nlm.nih.gov/34252155/ | USA | Plos One | 3.58 | The study included 180 patients (IS: 103 M + 77 F). Patients’ age: 65.89 years ± 11.18 years. No healthy C. | Participants were enrolled within 120 days of suffering a non-disabling cerebral infarction and followed for two years. |
| 17 | 2018 | Soriano-Tárraga et al. [59] | https://pubmed.ncbi.nlm.nih.gov/29515201/ | Spain | Scientific Reports | 4.01 | The study included 594 patients (IS: 326 M + 268 F). Patients’ age: 76 (64–85) years. No healthy C. | DNA samples were extracted from whole peripheral blood collected in the acute phase of the stroke. |
| 18 | 2021 | Miao et al. [64] | https://www.webofscience.com/wos/woscc/full-record/WOS:000685052900002 | China | Neuropsychiatric Disease and Treatment | 0.77 | The study included 112 patients (IS: 58 M + 26 F; C: 13 M + 15 F). Patients’ age (IS: 61.94 years ± 8.03 years; C: 57.64 years ± 9.0 years). | Not specified. |
| 19 | 2019 | Sikora et al. [65] | https://www.webofscience.com/wos/woscc/full-record/WOS:000473756000246 | Poland | International Journal of Molecular Sciences | 4.55 | The study included 68 patients (37 M + 31 F). Patients’ age: 67.5 years ± 12.4 years. No healthy C. | Blood was drawn immediately after arrival at the hospital. |
| 20 | 2018 | Tufekci et al. [66] | https://www.webofscience.com/wos/woscc/full-record/WOS:000426733300001 | Turkey | Frontiers in Neurology | 3.55 | The study included 190 patients (IS: 57 M + 38 F; C: 47 M + 48 F). Patients’ age (IS: 69.65 years ± 8.12 years; C: 67.35 years ± 10.13 years). | Within 24 h after stroke onset followed by the 1st week, the 1st month, and the 6th month. |
| 21 | 2021 | Li et al. [68] | https://pubmed.ncbi.nlm.nih.gov/34188129/ | Japan | Scientific Reports | 4.54 | The study included 633 patients (IS: 253 M + 211 F; C: 105 M + 64 F). Patients’ age (IS: 70.12 years ± 10.90 years; C: 58.7 years ± 15.46 years). | Serum samples associated with it were obtained within 2 weeks after onset. |
M: male; F: female; C: control group (applicable to Table 1 only)
The REMARK criteria [15] are provided in Table S2, S3, and Figure 2. Overall, two studies (9.52%) met all the REMARK criteria, two (9.52%) met 87.5%, ten (47.61%) met 75%, five (23.80%) met 62.5%, and two (9.52%) were under 50%. All studies provided a precisely defined clinical outcome and the methods of measuring the biomarker, nineteen studies (90.47%) were prospective, eighteen (85.71%) provided characteristics of the patients, fifteen (71.42%) mentioned a defined time period for enrolment, five (23.80%) uses a blinded biomarker measurement, and only two studies (9.52%) provided a rational for sample size.
The genetic biomarkers associated with stroke are summarized in Table 2. Five of the studies investigated the levels of circular RNA (circRNA) in association with stroke onset or recurrence (23.80%), four focused on long non-coding RNA (lncRNA) involved in immunity/inflammatory processes (19.04%), six investigated the polymorphisms associated with the atherosclerotic process and stroke recurrence (28.57%), three inquired into DNA methylation (DNAm) processes, stroke onset and outcome (14.28%), and three studies did proteomics/genomics analysis to correlate levels of plasma proteins with pathways of stroke (14.28%).
Characteristics of biomarkers
| Item | Authors | Link | Biomarkers | Potential pathway | Results |
|---|---|---|---|---|---|
| 1 | Lu et al. [20], 2020 | https://www.webofscience.com/wos/woscc/full-record/WOS:000515868500001 | circPHKA2, circBBS2 | PHKA2 gene encodes PbK-subunit alpha involved in cellular energy metabolism.BBS2 gene is part of the BBS family reported to stimulate ciliary biosynthesis via GTPase Rab8. | Mean values of circPHKA2, circBBS2 were statistically significant in IS group vs. control group (P < 0.05). BBS2 function after stroke remains unclear. |
| 2 | Ostolaza et al. [23], 2020 | https://www.webofscience.com/wos/woscc/full-record/WOS:000520976200005 | hsa_circRNA_102488 | Six different miRNAs implicated in fatty acid biosynthesis, lysine breakdown, and ARVC have a binding site on hsa_circRNA_102488, encoded on chromosome 19. | The atherothrombotic group had decreased hsa_circRNA_102488 levels (P < 0.01), and UBA52 mRNA levels were also downregulated (P < 0.001). |
| 3 | Abe et al. [24], 2020 | https://pubmed.ncbi.nlm.nih.gov/32354168/ | miR-505-5p, miR-1255b-5p, miR-550b-2-5p, miR-4523, and miR-6795-3p | miRNAs downregulate WDR26, which is an apoptosis repressor factor. | The study identified five miRNAs with fluctuating patterns correlated with the onset and course of cerebral infarction: miR-505-5p, miR-1255b-5p, miR-550b-2-5p, miR-4523, and miR-6795-3p. Higher levels of miR-376a-3p and downregulation of miR-3184-5p were associated with the onset of cerebral infarction. |
| 4 | Vijayan et al. [26], 2018 | https://www.webofscience.com/wos/woscc/full-record/WOS:000438578900007 | PC-3p-57664, PC-5p-12969, hsa-miR-122-5p, hsa-miR-211-5p, hsa-miR-22-3p, PC-3p-32463, hsa-miR-30d-5p, hsa-miR-23a-3p | miRNAs play an essential role in the regulation of leukocyte gene expression. | Sixteen dysregulated miRNAs in the IS group vs. the control group, out of which eight were validated through RT-PCR: four upregulated [PC-3p-57664 (P = 0.01), PC-5p-12969 (P = 0.04), hsa-miR-122-5p (P = 0.01), and hsa-miR-211-5p (P = 0.03)] and four downregulated [hsa-miR-22-3p (P = 0.01), PC-3p-32463 (P = 0.0001), hsa-miR-30d-5p (P = 0.0009), and hsa-miR-23a-3p (P = 0.03)]. |
| 5 | Mick et al. [28], 2017 | https://www.webofscience.com/wos/woscc/full-record/WOS:000398207000017 | hsa-miR-877-5p, hsa-miR-124-3p, hsa-miR-320d, SNO1402, hsa-miR-656-3p, hsa-miR-3615, hsa-miR-656-3p, hsa-miR-941 | Exosome-derived ncRNAs are implicated in inflammation and fibrosis. | hsa-miR-877-5p [odds ratio, 0.18; 95% CI, 0.06–0.52; P = 0.002], hsa-miR-124-3p (odds ratio, 0.29; 95% CI, 0.11–0.72; P = 0.008), hsa-miR-320d (odds ratio, 0.33; 95% CI, 0.14–0.78; P = 0.01), and SNO1402 (odds ratio, 0.20; 95% CI, 0.07–0.52; P = 0.001). |
| 6 | Zhu et al. [29], 2019 | https://www.webofscience.com/wos/woscc/full-record/WOS:000457370300001 | SCARNA10, TERC, LINC01481, FLJ23867, H3F3AP6, TNPO1P1 | LncRNA levels in the pathway of antigen processing and presentation were elevated at 24 h, while lncRNA levels in TRP channels and GABAergic synapses were downregulated on day 7 following stroke. | SCARNA10, TERC, and LINC01481 showed significantly increased expression (P < 0.01), whereas FLJ23867, H3F3AP6, and TNPO1P1 showed significantly decreased expression (P < 0.05) in IS patient PBMCs compared to healthy controls. |
| 7 | Cao et al. [31], 2020 | https://www.webofscience.com/wos/woscc/full-record/WOS:000513554900005 | LncMRT | The PI3K/Akt signaling pathway was recognized by IS as a key route due to dysregulated LncMRTs. Improved cell survival is linked to the serine/ threonine kinase PI3K, and this impact is partially mediated by the activation of the serine/threonine kinase Akt. | The dysregulated LncMRT in IS (formed by PTEN, TFAP2A, and HOTAIR) suggests that PTEN is a key part of the PI3K/Akt signaling pathway by generating LncMRT. |
| 8 | Wang et al. [33], 2022 | https://www.webofscience.com/wos/woscc/full-record/WOS:000711301400001 | LNC_000015, LNC_001727 | Because it encourages stem cell development and inhibits erythrocyte apoptosis, lncRNA is also thought to be useful in the treatment of ischemic disorders. | Five differentially expressed lncRNAs (LNC_000015, LNC_001727, SNHG8, MIRLET7BHG, AF001548.5) were found in IS and were used to construct a pathway network that suggests their potential as biomarkers and therapeutic targets in IS linked with the inflammatory response. |
| 9 | Zhang et al. [38], 2020 | https://www.webofscience.com/wos/woscc/full-record/WOS:000575004300053 | MCM3AP-AS1, LINC01089, ITPK1-AS1, HCG27 | These hub genes were connected to signaling pathways relevant to inflammation, according to functional analysis. | Inflammatory associated DELs (MCM3AP-AS1/LINC01089/ hsa-miR-125a/FYN, ITPK1-AS1/ hsa-let-7i/RHOA/GRB2/STAT1, and HCG27/hsa-miR-19a/PXN) may function as a ceRNA network to accelerate the development of the genes. |
| 10 | Zhu et al. [40], 2020 | https://www.webofscience.com/wos/woscc/full-record/WOS:000519997800015 | ADIPOR2 rs12342 | Immunoinflammatory gene functional polymorphisms. | ADIPOR2 rs12342 was statistically significant for stroke recurrence (GG vs. AA: P = 0.025; GA vs. AA: P = 0.011; GG/GA vs. AA: P = 0.004), but not with the onset age. |
| 11 | Li et al. [44], 2018 | https://www.webofscience.com/wos/woscc/full-record/WOS:000429737400001 | Cx37 | The dysregulation of eNOS production and activity is brought on by the Cx37 C1019T polymorphism, which causes the transfer of proline to serine and also influences endothelial function and elasticity, leading to atherosclerosis. | The G allele of the Cx37 SNP rs1764390 was shown to be a risk factor for IS and the genotypes of the Cx37 SNP rs1764390 and rs1764391 are linked to an elevated risk of IS. |
| 12 | Xie et al. [45], 2020 | https://www.webofscience.com/wos/woscc/full-record/WOS:000601211900010 | RPS14, RPS15A, RPS24, FAU, RPL27, RPL31, RPL34, RPL35A, RSL24D1, EEF1B2 | The expression of sex-specific ribosomal differential genes during the early stages of stroke recovery may be the main underlying cause of sexually dimorphic diff in inflammatory responses. | They showed that there are 10 upregulated genes with mostly ribosomal and protein synthesis- related functions (RPS14, RPS15A, RPS24, FAU, RPL27, RPL31, RPL34, RPL35A, RSL24D1, and EEF1B2). |
| 13 | Shroff et al. [47], 2019 | https://www.webofscience.com/wos/woscc/full-record/WOS:000455396400002 | HDAC9 polymorphism rs2107595 | HDAC9 is a class IIa HDAC that is located at 7p21 and changes the chromatin state by removing acetyl groups from proteins. | Both the rs2107595 risk allele-positive and risk allele-negative LVAS groups, as well as the risk allele-positive and risk allele-negative VRFC groups, did not significantly differ from each other. |
| 14 | Williams et al. [48], 2017 | https://pubmed.ncbi.nlm.nih.gov/28495826/ | rs505922 | The production of thrombus during cerebrovascular injury is significantly influenced by vWF. | Their results suggest that high vWF levels associate with the risk of stroke recurrence in both unadjusted (P = 0.0007, HR = 1.26) and fully adjusted (P = 0.018, HR=1.19) models, with the most strongly associated SNP-rs505922. |
| 15 | Davis Armstrong et al. [49], 2021 | https://pubmed.ncbi.nlm.nih.gov/33661917/ | rs7025659, rs10757056, rs10118089, rs7020745, rs10125228, rs10757058, rs10118371 | Leukotriene B4 is a pro-inflammatory lipid mediator derived from arachidonic acid. | Sphingosine and sphinganine were nominally related with five common variants and one different variant [rs10757056 (sphingosine only), rs10118089, rs7020745, rs10125228, rs10757058, and rs10118371]. The most significant association included leukotriene B4 and rs7025659- SPTLC1 (P = 6.12e-07; post-hoc comparison between TG to GG genotypes P = 3.00E-07). |
| 16 | Davis Armstrong et al. [51], 2021 | https://pubmed.ncbi.nlm.nih.gov/34252155/ | cg03584380, which is located in an intron of ZDHHC6. | The ZDHHC6 gene, which makes the palmitoyltransferase ZDHHC6, contains this methylation site in its first intron. ZDHHC6 is required for the palmitoylation of numerous significant ER proteins, including calnexin and the ITPR1. | In the African population, cg03584380 [HR = 5.41 (2.91–10.06)] was significantly associated with the recurrence of stroke, but not in the European cohort. |
| 17 | Soriano- Tárraga et al. [59], 2018 | https://pubmed.ncbi.nlm.nih.gov/29515201/ | DNAm | DNAm affects the expression of genes and the structure of DNA, and fluctuates over the length of a human’s life. | Biological aging, an epigenetic biomarker calculated from DNAm values, outperforms chronological aging in predicting mortality three months after an IS. This connection is particularly important in the etiology of LAA stroke. |
| 18 | Miao et al. [64], 2021 | https://www.webofscience.com/wos/woscc/full-record/WOS:000685052900002 | Hypomethylation of the RIN3 | The RIN3 gene regulates the movement of the amyloid beta-protein and functions as an inducer to maintain the transit of GTP Rab5 to endosomes at the plasma membrane. This process interferes with the endocytosis of the amyloid protein, which is connected to cellular endocytosis. | In the TIA/MIS group, the RIN3 gene’s overall methylation level was considerably lower than in the control group (diff = 0.02, P = 0.001), a result that was maintained in the cognitive impairment vs. non-cognitive impairment group (diff = −0.013, P = 0.01). |
| 19 | Sikora et al. [65], 2019 | https://www.webofscience.com/wos/woscc/full-record/WOS:000473756000246 | APCS, APOM, C1qA, C4BPA, C4BP2, FBLN1, IGKV1D-12, KLKB1, SERPINF2, AMBP, APOA4, FCN3, ITIH4, LBP, PF4, APOL1, C5, GPX3, GSN, H2AFJ, IGK | The biological pathways of the significant proteins are linked with the clothing cascade (A2M, FBLN1, FGA, PLG, SERPIND1, F13B, etc.), the acute phase response (SAA1, CRP, SERPINA3, SERPINA1, LBP, ORM1, AHSG), and lipids metabolism (APOC1, APOC3, APOL1, etc.). | Nine proteins were unique to differentiate cardioembolic and large-vessel stroke (APCS, APOM, C1qA, C4BPA, C4BP2, FBLN1, IGKV1D-12, KLKB1, SERPINF2), six for differentiating large-vessel and lacunar stroke (APOL1, C5, GPX3, GSN, H2AFJ, IGK) and six for cardioembolic vs. large-vessel stroke (AMBP, APOA4, FCN3, ITIH4, LBP, PF4). |
| 20 | Tufekci et al. [66], 2018 | https://www.webofscience.com/wos/woscc/full-record/WOS:000426733300001 | TRAIL mRNA expression | TRAIL is linked to five different receptors, two of which, DR4 and DR5 that cause apoptosis and inflammation. | Serum TRAIL levels of stroke patients were statistically (P < 0.0001) lower at the time of admission than those of healthy controls.Surprisingly, the follow-up study revealed that serum TRAIL levels considerably raised after the first month following stroke onset (P = 0.002).In addition, the TRAIL mRNA expression in PBMC was substantially higher in the stroke patients than in the controls (P < 0.0001).When compared to the first week after the onset of the stroke, TRAIL mRNA expression in PBMC was observed to be decreased (P < 0.001). |
| 21 | Li et al. [68], 2021 | https://pubmed.ncbi.nlm.nih.gov/34188129/ | serum anti- AP3D1-antibody (s-AP3D1-Ab) | AP3D1 was the target antigen that serum IgG antibodies identified in the plasma of individuals with atherosclerosis. | s-AP3D1-Ab levels were substantially greater in AIS than in healthy donors. The levels of s-AP3D1-Ab were still considerably greater in patients with AIS than in healthy donors even after the subjects’ ages were matched to 65 years. The s-AP3D1-Ab-positive rates in healthy donors and patients with AIS and TIA were 2.4%, 10.1%, and 10.4%, respectively, at a cutoff value comparable to the average plus two SDs of the HD values. |
circPHKA2: circRNA phosphorylase kinase regulatory subunit alpha 2; circBBS2: circRNA Bardet-Biedl syndrome 2; PbK: phosphorylase b kinase; BBS2: Bardet-Biedl syndrome 2; miRNAs: microRNAs; ARVC: arrhythmogenic right ventricular cardiomyopathy; UBA52: ubiquitin A-52 residue ribosomal protein fusion product 1; mRNA: messenger RNA; WDR26: WD repeat domain 26; RT-PCR: reverse transcription polymerase chain reaction; CI: confidence interval; SCARNA10: small Cajal body-specific RNA 10; TERC: telomerase RNA component; LINC01481: long intergenic non-protein coding RNA 1481; H3F3AP6: H3 histone, family 3A, pseudogene 6; TNPO1P1: transportin 1 pseudogene 1; PBMCs: peripheral blood mononuclear cells; LncMRT: lncRNA-mediated regulatory triplet; PI3K: phosphatidylinositol 3-kinase; Akt: protein kinase B; PTEN: phosphatase and tensin homolog; TFAP2A: transcription factor (TF) AP-2 alpha; HOTAIR: HOX transcript antisense RNA; SNHG8: small nucleolar RNA (snoRNA) host gene 8; MIRLET7BHG: miRNA let-7b host gene; MCM3AP-AS1: minichromosome maintenance complex component 3 associated protein antisense 1; ITPK1-AS1: inositol-tetrakisphosphate 1-kinase-antisense RNA 1; HCG27: human leukocyte antigen complex group 27; DELs: DNA-encoded libraries; RHOA: Ras homolog family member A; GRB2: growth factor receptor bound protein 2; STAT1: signal transducer and activator of transcription 1; PXN: paxillin; ceRNA: competing endogenous RNA; ADIPOR2: adiponectin receptor 2; Cx37: CONNEXIN37; eNOS: endothelial nitric oxide synthase; SNP: single nucleotide polymorphism; RPS14: ribosomal protein S14; RSL24D1: ribosomal L24 domain containing 1; EEF1B2: eukaryotic translation elongation factor 1 beta 2; HDAC9: histone deacetylase 9; LVAS: large vessel atherosclerotic stroke; VRFC: vascular risk factor controls; vWF: von Willebrand factor; SPTLC1: serine palmitoyltransferase long chain base subunit 1; ZDHHC6: zinc finger DHHC-type palmitoyltransferase 6; ER: endoplasmic reticulum; ITPR1: inositol 1,4,5-trisphosphate receptor type 1; LAA: large artery atherosclerosis; RIN3: Ras and Rab interactor 3; GTP: guanosine triphosphate; TIA: transient ischemic attack; MIS: minor IS; diff: difference; APCS: serum amyloid P component; APOM: apolipoprotein M; C1qA: complement component 1q A chain; C4BPA: complement component 4 binding protein alpha; FBLN1: fibulin 1; IGKV1D-12: immunoglobulin kappa (IGK) variable 1D-12; KLKB1: kallikrein B1; SERPINF2: serpin family F member 2; AMBP: alpha-1-microglobulin/bikunin precursor; FCN3: ficolin 3; ITIH4: inter-alpha-trypsin inhibitor heavy chain 4; LBP: lipopolysaccharide binding protein; PF4: platelet factor 4; C5: complement component 5; GPX3: glutathione peroxidase 3; GSN: gelsolin; H2AFJ: H2A histone family member J; A2M: alpha-2-macroglobulin; FGA: fibrinogen alpha chain; PLG: plasminogen; F13B: coagulation factor XIII B chain; SAA1: serum amyloid A1; CRP: C-reactive protein; ORM1: orosomucoid 1; AHSG: alpha-2 Heremans Schmid glycoprotein; TRAIL: tumor necrosis factor-related apoptosis-inducing ligand; DR4: death receptor 4; AP3D1: adaptor-related protein complex 3 subunit delta 1; s-AP3D1-Ab: serum anti-AP3D1-antibody; IgG: immunoglobulin G; AIS: acute IS; PHKA2: phosphorylase kinase regulatory subunit alpha 2; TRP: transient receptor potential; GABA: gamma-aminobutyric acid; ncRNAs: non-coding RNAs
Cerebral ischemia causes a complex sequence of events, including neuroinflammation, reactive oxygen species, mitochondria dysregulations, impairment in neurotransmission, apoptosis, as well as epigenetic alterations [18]. Plasma biomarkers are very useful to predict post-stroke recovery and individualized treatment. In this study, it was found the following useful genetic biomarkers: (i) serum/plasma circRNA associated with stroke onset or recurrence (5; 23.80%), (ii) genetic polymorphisms associated with the atherosclerotic process and stroke recurrence (6; 28.57%), (iii) serum/plasma lncRNA levels involved in immunity/inflammatory processes (4; 19.04%), (iv) marker of DNAm associated with stroke onset and outcome (3; 14.28%), and (v) proteins and pathways of stroke identified by serum/plasma proteomics/ genomics analysis (3; 14.28%).
The role of ncRNA has been long discussed in the pathogenesis of different pathologies. Recent studies have been focusing on the function of circRNAs and their potential as biomarkers [19]. A comprehensive review of the biogenesis of circRNAs suggests that circRNAs can operate on protein scaffolds, interact with other RNA species, and modulate transcriptional silencing, translation, and degradation of certain mRNAs [19]. Thus, the circPHKA2 (hsa_circ_0090002) and circBBS2 (hsa_circ_0039457) mean values were statistically significantly increased in the IS group compared to the sham group (student’s t-test; P < 0.05) [20]. The PHKA2 gene encodes PbK-subunit alpha. PbK activates glycogen phosphorylase, which degrades glycogen when glucose is depleted, and hence plays a crucial role in cellular energy metabolism [21]. The BBS2 gene is a part of the BBS family, which is reported to stimulate ciliary biogenesis via GTPase Rab8 [22]. However, the function of BBS2 after ischemia in the brain remains unclear.
Another recent study focused on the potential of circRNAs with regard to stroke etiology. The authors reported over 200 diff in the expression of circRNAs between cardioembolic and atherothrombotic stroke and validated hsa_circRNA_102488, generated from pre-mRNA encoded by the UBA52 gene on chromosome 19. Indeed, the levels of hsa_circRNA_102488 were lower in the atherothrombotic group (P < 0.01) and the corresponding levels of UBA52 mRNA were also lower (P < 0.001) [23]. Functionally, hsa_circRNA_102488 has a binding site for six distinct miRNAs involved in fatty acids biosynthesis, lysine degradation, and ARVC. Another study focusing on circRNAs aimed to investigate the association between circRNA levels and the onset of stroke. The study identified five miRNAs with fluctuating patterns that correlated with the onset and time course of cerebral infarction: miR-505-5p, miR-1255b-5p, miR-550b-2-5p, miR-4523, and miR-6795-3p. Higher levels of miR-376a-3p and downregulation of miR-3184-5p were associated with the onset of cerebral infarction [24].
miRNAs are key post-transcriptional regulators that regulate target genes by binding to multiple mRNAs. In cases of AIS, miRNAs have also been discovered to be critically involved in leukocyte gene expression [25]. Vijayan et al. [26] reported 16 dysregulated miRNAs in the IS patients vs. controls, out of which eight were validated by RT-PCR: four upregulated [PC-3p-57664 (P = 0.01), PC-5p-12969 (P = 0.04), hsa-miR-122-5p (P = 0.01), and hsa-miR-211-5p (P = 0.03)] and four downregulated [hsa-miR-22-3p (P = 0.01), PC-3p-32463 (P = 0.0001), hsa-miR-30d-5p (P = 0.0009), and hsa-miR-23a-3p (P = 0.03)]. However, it was not clear if they act as biomarkers of stroke risk factors or are a consequence of cerebral ischemia.
Exosome-derived ncRNAs implicated in inflammation and fibrosis, notably miRNAs, have been found to modify organ-level phenotypes in animal models of stroke [27]. Because exosome-derived ncRNAs are released into circulation and may act as distant molecular mediators, there has been an increasing amount of research seeking to define a plasma exosome-derived ncRNA signature of systemic disorders. Thus, Mick et al. [28] found that exosome-derived ncRNAs were each independently associated with stroke incidence: hsa-miR-877-5p (odds ratio, 0.18; 95% CI, 0.06–0.52; P = 0.002), hsa-miR-124-3p (odds ratio, 0.29; 95% CI, 0.11–0.72; P = 0.008), hsa-miR-320d (odds ratio, 0.33; 95% CI, 0.14–0.78; P = 0.01), and snoRNA [SNO1402 (odds ratio, 0.20; 95% CI, 0.07–0.52; P = 0.001)]. In each case, individuals with a history of stroke at baseline were less likely to express the exosome-derived ncRNA at baseline than those without a history of stroke at baseline. Also, employing several Cox-proportional hazards models, they reported that hsa-miR-656-3p and hsa-miR-941 absolute expressions were independent of stroke risk and significantly associated with a higher risk of stroke [28]. Several studies found that serum/plasma lncRNA levels are associated with post-stroke immunity/inflammatory processes [29]. Thus, the involvement of lncRNA in the transition from acute to subacute stroke was investigated at 24 h and 7 days post-stroke by Gene Ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. The findings highlighted the role of peripheral immunity in AIS, with three lncRNAs (SCARNA10, TERC, and LINC01481) showing significantly increased expression (P < 0.01), whereas FLJ23867, H3F3AP6, and TNPO1P1 showed significantly decreased expression (P < 0.05) in PBMCs from IS patients compared to healthy controls [29].
A LncMRT is a functional network motif, made up of a lncRNA, a TF, and a gene that governs various aspects of life [30]. Cao et al. [31] used a global LncMRT network to identify their ability as new biomarkers and treatment targets for IS. Dysregulated LncMRTs in IS allowed the identification of the PI3K/Akt signaling pathway as a critical pathway associated with stroke onset and recurrence. The dysregulated LncMRT in IS patients (consisting of PTEN, TFAP2A, and HOTAIR) suggests that PTEN may play a role in the PI3K/Akt signaling pathway by generating LncMRT. Indeed, serine/threonine kinase PI3K is associated with improved cell survival, and this effect is mediated in part by the activation of the serine/threonine kinase Akt [32].
Another recent study aimed to identify the role of lncRNAs and mRNAs in a cohort of IS patients and healthy controls [33]. Five differentially expressed lncRNAs (LNC_000015, LNC_001727, SNHG8, MIRLET7BHG, AF001548.5) were found differentially expressed in IS vs. controls and used to construct a GO/KEGG pathway network that suggests their potential as biomarkers and therapeutic targets in IS-linked inflammatory response.
Several in vitro and in vivo investigations have already shown that lncRNAs associated with inflammation may be potential targets for the diagnosis and treatment of AIS [34–36]. By acting as ceRNAs for miRNAs, which are ncRNAs of about 20 nucleotides in length that negatively impact the expression of target genes at the post-transcriptional level, lncRNAs may play a part in the development of diseases [37]. Thus, Zhang et al. [38] identified multiple inflammatory associated lncRNAs, including DELs, MCM3AP-AS1, LINC01089, ITPK1-AS1, and HCG27, which may act as biomarkers for the diagnosis and treatment of AIS. These DELs (MCM3AP-AS1/LINC01089/hsa-miR-125a/FYN, ITPK1-AS1/hsa-let-7i/RHOA/GRB2/STAT1, and HCG27/hsa-miR-19a/PXN) may function as a ceRNA network to accelerate the expression of the genes. Contrary to other reports [39], HCG27 was found to be upregulated in IS. This finding may be explained by different sample selection (chronic vs. acute) and patient cohorts. The correlation of these lncRNAs’ interaction with miRNA-mRNA pairs and AIS suggests their participation in AIS. However, this study did not give any direct evidence to support it [39].
This study indicates that genetic polymorphisms associated with the atherosclerotic process and stroke recurrence account for 27.27% of serum genetic biomarkers. Thus, Zhu et al. [40] in a prospective investigation with 42 months follow-up aimed to investigate polymorphisms of five immunoinflammatory genes [C-X-C motif chemokine receptor 2 (CXCR2) rs1126579, Toll-like receptor 4 (TLR4) rs11536889, ADIPOR2 rs12342, matrix metalloproteinase-2 (MMP-2) rs7201, and MMP-9 rs1056628] in association with the recurrence of IS. They report that ADIPOR2 rs12342 was significantly associated with stroke recurrence (GG vs. AA: P = 0.025; GA vs. AA: P = 0.011; GG/GA vs. AA: P = 0.004), but not with the onset age [41].
Gap junction protein is encoded by the Cx37 gene (also known as GJA4). Cx37 C1019T polymorphism causes the conversion of proline to serine, which results in the dysregulation of endothelial nitric oxide synthase (eNOS) expression and activity [42]. Since Cx37 is mostly expressed in vascular endothelial cells, monocytes, and macrophages, the most significant cells in the process of arteriosclerosis, Cx37 C1019T polymorphism influences endothelial function and elasticity eventually leading to atherosclerosis [43]. Indeed, Li et al. [44] revealed that Cx37 SNP rs1764390 and rs1764391 genotypes are associated with an increased risk of IS, and the G allele of rs1764390 was also proven to be a risk factor for IS. Another study tested the hypothesis that sex-specific genes in peripheral blood are associated with the progression of IS and can serve as potential biomarkers to predict outcome and sex-specific peripheral immunological response [45]. They showed that there are 10 upregulated genes with mostly ribosomal and protein synthesis-related functions. The downstream translation level is more significantly affected by the changes in the transcription level of ribosome-related genes. Therefore, ribosome profiling was used in an in vitro assay to investigate how oxygen and glucose deprivation (OGD) can immediately impact the transcription and translation of the genes in brain cells and they came to the conclusion that OGD had a larger effect on translation than transcription.
Thus, the prognostic ribosomal biomarkers RPS14, RPS15A, RPS24, FAU, RPL27, RPL31, RPL34, RPL35A, RSL24D1, and EEF1B2 could become novel therapeutic targets for IS. Of note, women are more likely to experience stroke-induced immunodeficiency syndrome (SIDS), and the potential mechanism could be the sex diff in the inflammatory response of ribosomes in stroke-induced peripheral immunosuppression [45].
HDAC9 found on chromosome 7p21, is a class IIa HDAC that affects the chromatin state by removing acetyl groups [46]. Shroff et al. [47] hypothesized that vascular atherosclerosis, which is an important cause of IS, is associated with HDAC9 polymorphism rs2107595. However, neither rs2107595 risk allele-positive and risk allele-negative variants, as well as the risk allele-positive and risk allele-negative VRFC groups, did not significantly differ from each other.
Recurrent pathways involving interleukin-6 (IL-6) signaling, extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) signaling, liver X receptors (LXR)/retinoid X receptor (RXR) activation, and the function of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis were identified using pathway analysis for the 155 genes associated with rs2107595 polymorphism. Indeed, leukocyte recruitment, chronic inflammation, immunological response of macrophages, cholesterol efflux, and platelet aggregation were all connected to SNP rs2107595 and the risk of LVAS. However, this risk allele did not change significantly the levels of HDAC9 expression. LVAS is not induced by the risk allele rs210759 either. Thus, many individuals who carry the risk allele do not suffer a stroke. Nevertheless, the HDAC9 polymorphism may interact with other factors to increase the risk of stroke. A polymorphic allele at some other locus, epigenetic regulation, or external factors could all have an impact on the HDAC9 polymorphism’s incomplete penetrance [47].
Williams et al. [48] analyzed the involvement of plasma vWF in the recurrence of IS using expression quantitative trait locus (eQTL) analysis. Their results suggest that high vWF levels associate with the risk of stroke recurrence in both unadjusted (P = 0.0007; HR = 1.26) and fully adjusted (P = 0.018; HR = 1.19) models, with the most strongly associated SNP rs505922 [48].
A metabolomic-epigenetics study investigated the stroke recurrence in an African American population [49]. Thus, the alkaline ceramidase 2 (ACER2) locus encoding ACER2, sphingosine, and sphinganine were associated with five common variants and one different variant [rs10757056 (sphingosine only), rs10118089, rs7020745, rs10125228, rs10757058, and rs10118371]. The most significant association included leukotriene B4 and rs7025659-SPTLC1 (P = 6.12e-07; post-hoc comparison between TG to GG genotypes, P = 3.00E-07) [49]. Further, individuals with IS and high leukotriene B4 levels were linked to less successful functional recovery [50].
In a follow-up study, Davis Armstrong et al. [51] investigated DNAm loci associated with stroke recurrence in a cohort of African descendants and European descendants. They found that cg03584380 [HR = 5.41 (2.91–10.06)] polymorphism was significantly associated with the recurrence of stroke in the African population, but not with the European cohort. This methylation site is located in the first intron of the ZDHHC6 gene, which encodes palmitoyltransferase ZDHHC6 and is necessary for the palmitoylation of several important ER proteins, such as calnexin and the ITPR1 [52].
DNAm is an epigenetic pathway that influences the expression of genes and greater-order DNA structure. At a cytosine-phosphate-guanine (CpG) locus, the heritable but also reversible addition of a methyl group to the 5-carbon position of cytosine is linked with gene suppression [53]. DNAm levels fluctuate over the length of a human’s life and are affected by both genetic and lifestyle variations [54–56]. Age-related changes in DNAm are commonly accepted and two recent studies used methylation measurements at several CpGs loci across the genome to determine chronological age (C-age) in humans [57, 58]. Thus, Soriano-Tárraga et al. [59] found that an epigenetic biomarker calculated from DNAm values outperformed chronological aging in predicting mortality three months after an IS. This connection is particularly important in the etiology of LAA stroke [59]. Their findings are consistent with earlier research on many disorders, which has demonstrated that this biomarker accurately predicts death regardless of lifestyle, genetic factors, or health condition [60, 61].
A number of neurodegenerative disorders are significantly influenced by DNAm [41] and RIN3 gene is overexpressed and hypomethylated in the blood of patients with Alzheimer’s disease [62, 63]. Miao et al. [64] looked for the dysregulation of RIN3 gene methylation in association with cognitive impairment secondary to transitory or mild cerebral ischemia. In the TIA/MIS group, the RIN3 gene’s overall methylation level was considerably lower than in the control group (diff = 0.02; P = 0.001), the result that was maintained in the cognitive impairment vs. non-cognitive impairment group (diff = −0.013; P = 0.01), concluding that methylation levels of RIN3 could be used as a predictor factor for early cognitive impairment in IS.
Another proteomic study aimed to identify diff between stroke subtypes and cystathionine β-synthase (CBS) deficiency those innated deficit is associated with thromboembolism [65]. Regarding stroke subtypes, nine proteins uniquely differentiated cardioembolic and large-vessel stroke (APCS, APOM, C1qA, C4BPA, C4BP2, FBLN1, IGKV1D-12, KLKB1, SERPINF2). Another six were specific for large-vessel and lacunar stroke (APOL1, C5, GPX3, GSN, H2AFJ, IGK), and six differentiated cardioembolic and large-vessel stroke (AMBP, APOA4, FCN3, ITIH4, LBP, PF4). Furthermore, Ingenuity Pathway Analysis software (IPA; Ingenuity Systems; Mountain View, CA, USA) was used to identify proteins and biological pathways linked to the clothing cascade (A2M, FBLN1, FGA, PLG, SERPIND1, F13B, etc.), the acute phase response (SAA1, CRP, SERPINA3, SERPINA1, LBP, ORM1, AHSG), and lipid metabolism (APOC1, APOC3, APOL1, etc.).
Tufekci et al. [66] aimed to verify if serum TRAIL protein levels and mRNA expression in PBMCs of stroke patients were linked with stroke prognosis throughout the course of 6 months follow-up.
It is known that TRAIL is linked to five different receptors, two of which, DR4 and DR5 causing apoptosis and inflammation [67]. On the onset, blood levels of TRIAL protein were lower in the stroke cohort (P = 0.0001), but with a higher TRAIL mRNA expression in PBMC. In the follow-up period, the authors reported a down-regulation of the protein levels, while TRAIL mRNA expression in PBMC was found to be upregulated. However, no statistical significance was seen when blood TRAIL protein levels and PBMC TRAIL mRNA expression were compared among stroke subtypes, concluding that levels of serum TRIAL protein or PBMC TRAIL mRNA expression could not be used as a potential diagnostic biomarker.
Path analysis [68] indicated a strong correlation between anti-AP3D1-antibody levels and maximum intima-media thickness, suggesting that this marker accurately captured the initiation of atherosclerosis. It was discovered that the target antigen recognized by serum IgG antibodies in the plasma of atherosclerosis patients was AP3D1. The findings demonstrated that s-AP3D1-Ab levels were substantially greater in patients with IS than in patients with healthy donors. The levels of s-AP3D1-Ab were still considerably greater in patients with AIS than in healthy donors even after the subjects’ ages were matched to 65 years. The s-AP3D1-Ab-positive rates in healthy donors and patients with AIS and TIA were 2.4%, 10.1%, and 10.4%, respectively, at a cutoff value comparable to the average plus two SDs of the HD values.
A strength of this systematic review is the search for genetic biomarkers, which allowed us to investigate the less known potential biomarkers of stroke. Moreover, further classification of types of genetic biomarkers shall give a more clear overview of what is the main focus of recent research. On the other hand, the results could have been impacted negatively by unknown biomarker patterns and/or inconsistent blood collection times within and between experiments. Future prospective studies on the expression of biomarkers after IS should take these issues into consideration. Another drawback is the small sample sizes in some of the studies and the rationale of the sample size calculation was not taken into account, which inevitably weakens the statistical strength. Also, the study did not specifically take into account the recovery association of all the biomarkers, it was found to be associated with stroke incidence and functional recovery, which means that the data available are mixed between potential recovery prognostic genetic biomarkers and the onset of stroke.
In conclusion, more than 100 potential biomarkers were found and the data suggest that combinations of plasma genetic biomarkers might be used as a better predictor for stroke.
To the best of our knowledge, this is the first systematic review to focus only on genetic biomarkers. More than 100 biomarkers were significantly associated with IS, classified in circRNAs, lncRNA, polymorphisms, and genetic/proteomic networks. The data found suggest that genetic plasma biomarkers might be a useful tool with which to assess the risk of stroke onset and to allow prognosis of recovery in the clinic. However, more studies are needed with wider sample sizes and more homogeneous results to allow the identification of reliable plasma genetic factors unambiguously associated with stroke.
ADIPOR2: adiponectin receptor 2
AIS: acute ischemic stroke
Akt: protein kinase B
AMBP: alpha-1-microglobulin/bikunin precursor
AP3D1: adaptor-related protein complex 3 subunit delta 1
APCS: serum amyloid P component
APOM: apolipoprotein M
BBS2: Bardet-Biedl syndrome 2
C1qA: complement component 1q A chain
C4BPA: complement component 4 binding protein alpha
C5: complement component 5
ceRNA: competing endogenous RNA
CI: confidence interval
circBBS2: circular RNA Bardet-Biedl syndrome 2
circPHKA2: circular RNA phosphorylase kinase regulatory subunit alpha 2
circRNA: circular RNA
Cx37: CONNEXIN37
DELs: DNA-encoded libraries
diff: difference
DNAm: DNA methylation
DR4: death receptor 4
EEF1B2: eukaryotic translation elongation factor 1 beta 2
F: female
FBLN1: fibulin 1
FCN3: ficolin 3
GPX3: glutathione peroxidase 3
GSN: gelsolin
H2AFJ: H2A histone family member J
H3F3AP6: H3 histone, family 3A, pseudogene 6
HCG27: human leukocyte antigen complex group 27
HDAC9: histone deacetylase 9
IF: impact factor
IGK: immunoglobulin kappa
IGKV1D-12: immunoglobulin kappa variable 1D-12
IS: ischemic stroke
ITIH4: inter-alpha-trypsin inhibitor heavy chain 4
ITPK1-AS1: inositol-tetrakisphosphate 1-kinase-antisense RNA 1
KLKB1: kallikrein B1
LBP: lipopolysaccharide binding protein
LINC01481: long intergenic non-protein coding RNA 1481
LncMRT: long non-coding RNA-mediated regulatory triplet
lncRNA: long non-coding RNA
LVAS: large vessel atherosclerotic stroke
M: male
MCM3AP-AS1: minichromosome maintenance complex component 3 associated protein antisense 1
miRNAs: microRNAs
mRNA: messenger RNA
ncRNAs: non-coding RNAs
PbK: phosphorylase b kinase
PBMCs: peripheral blood mononuclear cells
PF4: platelet factor 4
PI3K: phosphatidylinositol 3-kinase
PTEN: phosphatase and tensin homolog
REMARK: REporting recommendations for tumor MARKer prognostic studies
RIN3: Ras and Rab interactor 3
RPS14: ribosomal protein S14
RSL24D1: ribosomal L24 domain containing 1
s-AP3D1-Ab: serum anti-adaptor-related protein complex 3 subunit delta 1-antibody
SCARNA10: small Cajal body-specific RNA 10
SD: standard deviation
SERPINF2: serpin family F member 2
SNP: single nucleotide polymorphism
TERC: telomerase RNA component
TIA: transient ischemic attack
TNPO1P1: transportin 1 pseudogene 1
TRAIL: tumor necrosis factor-related apoptosis-inducing ligand
UBA52: ubiquitin A-52 residue ribosomal protein fusion product 1
vWF: von Willebrand factor
ZDHHC6: zinc finger DHHC-type palmitoyltransferase 6
The supplementary material for this article is available at: https://www.explorationpub.com/uploads/Article/file/100610_sup_1.pdf.
MAR, DB, RS, and APW: Writing—review & editing. AMC and MA: Investigation. RS and APW: Funding acquisition. DMH, IFC, and TRD: Supervision.
The authors declare no conflicts of interest related to this study.
Not applicable.
Not applicable.
Not applicable.
Data will be made available upon request (aurel.popa-wagner@geriatrics-healthyageing.com).
APW received funding from the EU Horizon 2020 Research and Innovation Programme [263/16.11.2020]. RS received funding from the EU Horizon 2020 Research and Innovation Programme [PN-III-P1-1.1- PD-2021-0360]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Chaitanya Sanghadia ... Brandon Lucke-Wold
Vivian Molina Cuevas, Ambar Oyarzábal Yera
Divya Sharma, Rahul Kumar
Lorena Dellagnesi Depieri ... Lorena Souza Viana
Ana-Maria Dobri-Nicoară ... Cristiana Tanase
