Affiliation:
Institute of Microbiology, University of Agriculture Faisalabad, Faisalabad 38000, Pakistan
ORCID: https://orcid.org/0009-0002-2989-9809
Affiliation:
Institute of Microbiology, University of Agriculture Faisalabad, Faisalabad 38000, Pakistan
ORCID: https://orcid.org/0009-0009-2334-7668
Affiliation:
Institute of Microbiology, University of Agriculture Faisalabad, Faisalabad 38000, Pakistan
Email: aamir.aslam.dr@gmail.com
ORCID: https://orcid.org/0000-0002-0958-4749
Explor Drug Sci. 2025;3:1008133 DOI: https://doi.org/10.37349/eds.2025.1008133
Received: August 31, 2025 Accepted: October 21, 2025 Published: November 12, 2025
Academic Editor: Cheorl Ho Kim, Sungkyunkwan University, Samsung Advances Institute of Health Science and Technology (SAIHST), Republic of Korea
The escalating threat of antibiotic resistance and its advancing mechanisms for resistance development underscore the imperative need for alternative approaches to treat life-threatening infections. Consideration of bacteriophages, as well as antimicrobial peptides (AMPs) that can specifically target and eliminate particular bacteria, is gaining prominence for the improved treatment of infections. The effectiveness of bacteriophages and AMPs has been known for a long time, and their combined use is being investigated recently. Studies have shown that the use of phages or phage-derived enzymes (endolysins) in combination with AMPs has shown promising results in combating multidrug resistant bacteria. Bacteriophages lyse bacteria by hijacking the bacterial cell’s metabolic machinery, leading to the production of phage virus inside it and finally bursting the bacteria, while AMPs act by disrupting the bacterial cell membrane or affecting intracellular targets after penetration. In this review, we discuss previous studies on the combined use of both phages or phage-derived enzymes and AMPs, demonstrating their synergistic effects for combating multidrug resistant pathogens. Their mechanisms of action, and possible mechanisms of synergy and development of bacterial resistance to these, are discussed. Approaches, including genetic engineering, for improving their efficacy have been discussed. Safety and ethical issues regarding their use in human subjects are discussed. In summary, this review emphasizes the need for further research on the combined use of AMPs and bacteriophages to tap their potential effectiveness for treating antimicrobial-resistant infections.
A pressing concern and public health crisis is the alarming rise in infections caused by antibiotic-resistant bacteria of this age. The World Health Organization (WHO) declared a post-antibiotic era in 2014, stating that antibiotics will be largely ineffective in the 21st century [1, 2]. Due to this increase in antibiotic resistance, there is a need to find other alternatives for treating bacterial infections. Phages and antimicrobial peptides (AMPs, described below) are being considered as one of the alternatives.
The bacteriolytic action of phage on the bacterial cell starts with its adsorption onto the bacterial surface through the binding of receptor-binding proteins (RBPs) on the tail fiber of phage to specific receptors on the bacterial cell. This interaction is very specific, making the phage treatment distinct in nature as it mostly targets specific bacteria, but some phages can be polyvalent, meaning that these can target more than one species or genus. The examples of adsorption receptors on bacterial surfaces include OmpA, OmpC, and polysaccharides [3, 4]. Following adsorption, the phage injects its nucleic acid (enclosed in its capsid) into the cytoplasm of the bacterial cell after piercing a hole in the cell membrane and cell wall through enzymatic hydrolysis of the cell envelope using enzymes called virion-associated peptidoglycan hydrolases and depolymerases. After inserting its genetic material, the phage takes control of the metabolic machinery of the bacterial cell, and expression of phage genes occurs, leading to synthesis of phage components. Hundreds of new copies of phage DNA are synthesized, which are packed into newly formed capsids. Finally, the newly synthesized phage tails are attached to capsids, creating a fully infectious phage virion. To release these phages from infected bacteria, phages synthesize two proteins, i.e., holin and endolysin. Holins insert themselves into the inner bacterial cell membrane, creating holes in the membrane. Endolysins are muralytic enzymes that access the cell wall through these pores and enzymatically digest the cell wall, thus leading to bacterial cell lysis and release of phage progeny (see Figure 1) [5, 6]. The newly released phages find other bacterial cells and again start the lytic cycle. Strictly lytic phages are preferred for therapeutic purposes. Another type of cycle displayed by phages is the lysogenic cycle. In this cycle, phages integrate their genome inside the bacterial genome, remain dormant and are called prophages, and replicate alongside their host. In response to a specific trigger, dormant prophages excise themselves from the bacterial genome and burst into a lytic cycle [7]. Phages with lysogenic properties are not preferred for therapeutic purposes as these do not always kill bacteria and also lead to the transfer of toxin genes to host bacteria [8].
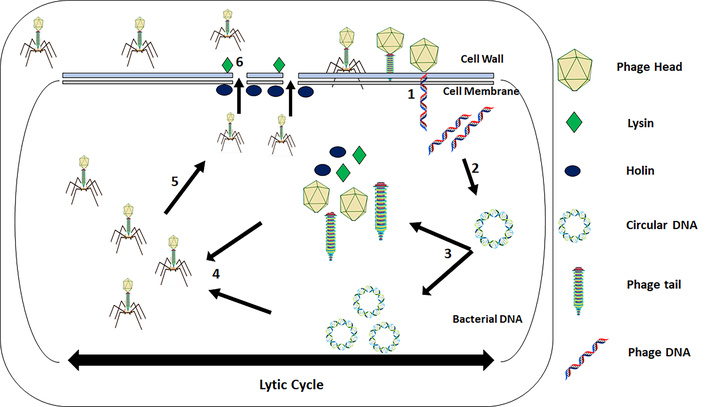
Schematic illustration of phage-induced bacteriolysis through the lytic cycle. (1) Adsorption of phage to the bacterial body and DNA injection. (2) Circularization of phage DNA to avoid enzymatic degradation. (3) Replication of phage DNA as well as transcription of phage DNA to produce phage proteins. (4) DNA packaging and assembly of the head and tail into a complete phage. (5) Disruption of the cell wall through the endolysin-holin system. (6) Release of the phage progeny.
In eukaryotes, AMPs are a part of the innate immune system in a wide variety of organisms. Furthermore, besides direct killing of pathogens, AMPs interact with the immune system through certain mechanisms in mammals, e.g., involvement in the neutrophils’ chemoattraction, stimulation of Toll-like receptors, T-cells, and dendritic cells activation, and enhancing the activity of phagocytosis [9]. AMPs are mainly classified into two broad categories: (1) membranolytic AMPs, (2) non-membranolytic AMPs [10].
Membranolytic AMPs interact with the lipid-peptide bonds in the bacterial cell membrane, resulting in its lysis. Multiple models have been presented to interpret this mechanism, such as the toroidal-pore model, barrel-stave model, and carpet model. In the toroidal pore model, AMPs are inactively inserted in the lipid-peptide layer of the membrane, and upon exceeding a threshold limit, they activate themselves, bend the membrane, and disrupt it by producing pores in the membrane, leading to cell death. The barrel-stave model explains cell death by forming a bundle in a barrel-like shape, inserting into the lipid bilayer, and forming transmembrane channels, leading to cell death. In the carpet model, AMPs arrange themselves in a parallel manner (sheet-like) on the membrane’s surface. Upon reaching the threshold limit, AMPs disintegrate the membrane due to electrostatic interaction between positively charged AMPs and negatively charged phospholipid membrane, leading to cell death [11].
Besides the lysis of the cell membrane, non-membranolytic AMPs target internal organelles such as proteins, enzymatic complexes, nucleic acids, or processes of the cell, such as protein folding, replication of DNA, and RNA synthesis [11]. There are countless examples of that type of AMPs, such as in dolphins, where Tur1A (an AMP) interacts with the ribosomes of the pathogens by blocking translation of mRNA to proteins. Another trp-rich AMP has been proven to kill Pseudomonas aeruginosa by inhibiting the genes involved in its DNA replication. Tridecaptin AMP interrupts bacterial ATP synthesis and is also effective against multidrug resistant (MDR) and colistin-resistant Enterobacterales. Moreover, in arthropods, thanatin AMP kills the bacterial cell by targeting its lipopolysaccharide (LPS) [12].
Previous studies have focused on phage-antibiotic combinations for treating resistant infections, whereas research on phage-AMP synergy is still emerging, with few direct investigations. According to Rothong et al. (2024) [13], three peptides were identified, particularly PE04-1(NH2), PE04-2, and PE04-1, encoded from phage (vB_AbaAut_ChT04) endolysin. The sequence alignment of these peptides revealed their similarity with mammalian cathelicidin AMPs. These three peptides showed strong activity against extensively drug resistant and MDR bacteria, particularly Acinetobacter baumannii, and effectively inhibited biofilm formation. These peptides also improved survival in Galleria mellonella infection models with no cytotoxicity in human cell lines, supporting their potential therapeutic safety. In another study, Zhang et al. (2023) [14] showed that a combination of AMP, cathelicidin LL-37, and endolysin Ply2660 was superior in eliminating the biofilm of Enterococcus faecalis as well as for enhancing the survival rates of diseased mice in in vivo animal models.
Research was carried out by Gouveia et al. (2022) [15]. It was found that Staphylococcus aureus pre-treated with R8K (a modified AMP derived from cathelicidin SMAP-29) exhibited a markedly increased sensitivity to the endolysin Lys11 from phage ϕ11. This increased sensitivity led to fast and extensive Lys11-mediated bacterial lysis, even at low levels. A related study by Mirski et al. (2019) [16] showed that phages selectively eliminated specific bacteria, whereas AMPs can damage bacterial membranes and prevent biofilm development.
Similarly, Duc et al. (2020) [17] investigated S. aureus in both planktonic and biofilm forms on various surfaces, including LB broth, stainless steel surfaces, polystyrene plates, and pasteurized milk. They found that using phage SA46-CTH2 in combination with nisin (a natural AMP) was more effective than using either treatment alone. In another study by Tyagi et al. (2024) [18], it was shown that a combination of T7 endolysin (T7L) with polymyxin B and colistin displayed synergistic effects for the eradication of biofilm of P. aeruginosa. This study also demonstrated that a combination of T4 endolysin (T4L) with nisin displayed synergistic effects for the eradication of biofilm of S. aureus. Another study showed that a synergistic effect was achieved on the inhibition of growth of polymyxin B-resistant Salmonella Typhimurium, resulting in a decrease in minimum inhibitory concentration (MIC) values of FK13 and FK16 (AMPs) when combined with LysPB32 endolysin [19].
A previous study showed that genetically engineered T7 phage, which expressed AMP 1018, called engineered phage 1018, was able to display superior efficacy in planktonic cell lysis as well as biofilm eradication [20]. From the results of all these previous studies, it is apparent that the synergistic potential of AMPs and phages can be utilized for the eradication of MDR bacteria (see Table 1 for a summary of these AMPs and phage combinations).
Summary of results from previous studies combining bacteriophage/bacteriophage endolysins and AMPs.
| AMP + bacteriophage/lysin combination | Target bacteria | Outcome | Reference |
|---|---|---|---|
| FK13, FK16 (AMPs) + endolysin LysPB32 | Polymyxin B-resistant Salmonella Typhimurium | The combination decreased the MIC values against the test bacteria. Enhanced reduction of bacterial growth in broth culture. | [19] |
| Pseudomonas aeruginosa, Staphylococcus aureus | All three combinations exhibited synergism to eradicate the biofilms of the respective bacteria. | [18] |
| R8K (AMP) + endolysin Lys11 (phage ϕ11) | Staphylococcus aureus | Increased bacterial killing due to increased binding of lysin to the bacterial cell wall. | [15] |
| Nisin + bacteriophage, SA46-CTH2 | Staphylococcus aureus | Better bacterial killing in broth and milk. Enhanced biofilm eradication. | [17] |
| LL-37 + endolysin Ply2660 | Enterococcus faecalis | Enhanced biofilm eradication. | [14] |
| Genetically engineered T7 phage expressed the AMP (1018) | Escherichia coli | Enhanced bacterial killing and biofilm eradication. | [20] |
AMPs: antimicrobial peptides; MIC: minimum inhibitory concentration.
Recent research has shown that peptides derived from endolysins hold strong potential as new antimicrobial agents, especially for targeting gram-negative bacteria. Endolysins mainly kill the bacterial cells by enzymatic hydrolysis of the peptidoglycan layer of the bacterial cell wall. But the access to the bacterial cell wall is hindered by the cell membrane. Some of the endolysins, especially from those of gram-negative bacteria, are known to possess segments in the endolysin structure that help in accessing the cell wall by damaging and permeabilizing the cell membrane. These segments are known to possess a positive charge in C-terminal segments in cases of endolysins; from T4 [21], RL-2015 [22], phage 53 [23], PhiKo [24], and JG004 phages [25, 26], and also from another endolysin, PlyPa01 [27]. Another AMP named P30/Intestinalin was derived from the N-terminal region of the endolysin of LysC derived from phage against the bacteria Clostridium intestinale. Antimicrobial properties of this peptide have been shown to surpass the full-length enzyme [24]. These peptides have also been shown to be effective in in vivo murine models of infections, and these usually show low toxicity to human cells [28].
Different possible mechanisms of synergy between AMPs and phages have been proposed. For example, AMPs can enhance the binding of bacteriophage endolysins [15]. A combination of AMPs’ (nisin) ability to kill bacteria, along with bactericidal effects of phages, enhanced the overall bacterial elimination [17]. Both AMPs and phages have the ability to disrupt bacterial biofilms by disruption of bacterial membranes and digestion of capsular polysaccharides or exopolysaccharides (EPSs), respectively. AMPs and phages, or endolysins, each kill bacterial cells by engaging independent targets in bacterial cells, and their mechanisms of action can augment each other; furthermore, this ensures that resistance against one will not affect the action of the other.
AMPs can augment the killing ability of endolysins by making disruptions in cell membranes, making the accessibility of the cell wall easy for the enzymatic digestion of the cell wall by endolysins [29]. This mechanism of synergy is further reinforced by the fact that certain AMPs have been derived from phage endolysins of gram-negative bacteria (described in the previous section), which have been shown to possess positively charged peptide moieties that interact with negatively charged cell membranes to cause cell membrane perturbations.
Several studies have demonstrated that resistance to AMPs can develop, often as a result of changes in the surface charge of the bacterial cell wall or cell membrane. The outer membrane of gram-negative bacteria limits permeability as their natural defense. Negatively charged LPS attracts cationic AMPs, while outer membrane proteins further contribute to resistance via their physiological functions. Since LPS shields bacteria from both hydrophilic and hydrophobic molecules, enzymes like Lpx involved in its synthesis, along with modifications regulated by PhoPQ and PmrAB systems, are vital for AMPs resistance in many gram-negative bacteria [30]. Alarmingly, A. baumannii can develop colistin resistance through the total elimination of LPS from its outer membrane [31].
Further, mutations in two-component systems such as ParRS, ColRS, and CprS in P. aeruginosa can trigger their continuous activation, which causes overexpression of genes that modify LPS. In species lacking capsules, the O antigen chains determine surface characteristics and serve as a protective layer that blocks AMPs from penetrating the LPS. O-antigens shield K. pneumoniae and S. flexneri from the harmful effects of histones, which are found in neutrophil extracellular traps and act as AMPs. Another example belongs to AMPs resistance, it is the outer membrane proteins OmpU and OmpT, such as in V. cholerae, these proteins play a partial role in intrinsic resistance to cationic AMPs [30, 32].
Bacteria can also generate extracellular molecules that bind and trap AMPs, e.g., the SIC protein, staphylokinase from S. aureus, and various M protein types made by Streptococcus pyogenes [31]. They bind to AMPs with high affinity, stopping these peptides from reaching and damaging the host cell membrane and interior. Bacteria also develop resistance by modifying their efflux pumps or forming capsules, mainly in gram-negative bacteria [33]. Further, bacteria can develop resistance to bacteriocins (AMPs produced by bacteria) by mimicking producer defenses, altering their membranes, or using enzymes, similar to antibiotic resistance. For instance, resistance to nisin may involve enzymes like dehydropeptide reductase or nisinase that inactivate the bacteriocin [31, 34].
The clinical use of AMPs faces several obstacles, especially their tendency to be broken down easily due to bacterial protease activity. This enzymatic degradation reduces their effectiveness as therapeutic agents. Different strategies like adding D-amino acids, N-acetylation, lipidation, cyclization, C-amidation, and PEGylation have been shown to improve AMPs’ stability but often reduce antibacterial activity or increase toxicity. For example, lipidation can increase toxicity, while D-amino acids may lower hydrophobicity and effectiveness. However, homoarginine (hArg), a non-standard amino acid, has shown potential to enhance AMPs’ stability and improve resistance to protease degradation [35]. Further, selective fluorination of the AMPs, such as buforin and magainin, improves protease resistance and enhances or maintains their antibacterial activity [36].
To enhance the bioavailability of AMPs without altering their structure, two main strategies can be used: enzyme inhibition and sustained release systems. The first involves co-administering enzyme inhibitors to improve absorption, such as via the oral route [37]. For example, cathelin-like proteins, part of the mammalian innate immune system, help protect host tissues by inhibiting microbial cysteine proteases and also possess direct antimicrobial properties against pathogens [38]. The second uses delivery systems like ethosomes, liposomes, cubosomes, transferosomes, solid lipid nanoparticles, and nanostructured lipid carriers. Biodegradable polymers like polylactic acid, polylactic glycolic acid, emulsions, or cyclodextrin derivatives are also commonly employed [37]. Other approaches include conjugating AMPs with nanoparticles such as metal nanoparticles, lipid-based nanoparticles, and polymer-based nanostructures [39].
Only using strictly lytic phages can minimize the chances of the spread of toxin or antibiotic resistance genes among microbial communities. Genetic engineering allows phages to be modified for removing toxin or antibiotic resistance genes present in the phage genome, allowing for an increase in their targetable host range by modifying tail fiber phage proteins [40, 41]. AMPs can be expressed from the phage genome, minimizing side effects associated with AMPs [20]. It can be used to alter properties of both phages and AMPs to extend their half-life in blood circulation and reduce the rate of their action to minimize side effects occurring due to the release of bacterial toxins, due to rapid bacterial lysis [10].
The adoption of lytic phages to treat ailments is known as phage therapy. Lytic phages completely destroy bacteria, unlike bacteriostatic antibiotics that merely inhibit growth [42]. Other advantages are that phages are self-replicating and inherently low in toxicity [43]. Their application is versatile, usable in liquids, creams, or embedded in solids. Moreover, they can disrupt biofilms [44] and there is less propensity of developing resistance against phages and their lytic enzymes, as these target indispensable elements in bacterial cells, such as conserved chemical bonds in peptidoglycan linkages in the bacterial cell wall.
Limitations of phage therapy include being highly sensitive to environmental conditions like pH, temperature, and moisture, which affect their survival and efficacy. Bacteria can also develop resistance through receptor changes, biofilm barriers, or CRISPR defenses and inhibition of adsorption of phages [45]. High specificity of phages becomes a disadvantage when infection is caused by multiple bacterial populations; phage therapy targeting only one bacterium can favor other bacteria, which can multiply rapidly. Phage cocktails can be used to treat mixed infections. During bacterial lysis, toxins and cell wall components can be released, potentially triggering harmful immune responses, especially in immunocompromised patients [46, 47]. The host’s immune system often clears phages rapidly, reducing their therapeutic effectiveness and complicating pharmacokinetics [48], but other studies showed these effects to be minimal [49]. Phages struggle to treat intracellular infections and may unintentionally spread antimicrobial resistance (AMR) via gene transfer [48]. Additionally, the complex composition of phage preparations makes it challenging for dosage calculation and quality assessment. A lack of standardized protocols and regulatory policies further hinders their clinical application. More time is required for new phage isolation, testing its interaction with bacteria in the laboratory, and phage-DNA sequencing for the purpose of excluding lysogeny or virulence genes, which makes the application of phage therapy more suitable for patients suffering from chronic infections [49].
Since AMPs target the highly conserved and vitally important targets in bacterial cells, such as cell membranes, there is very little likelihood of developing resistance against AMPs, which is a clear advantage as compared to antibiotics [10, 20, 50]. AMPs are also known to have potential limitations due to cytotoxicity, resulting in nephrotoxicity and hemolysis. The results of AMPs on bacterial populations can vary under in vivo conditions due to attack by proteases and other enzymes present in the mammalian body [51]. Furthermore, these can also provoke the immune system and lead to the production of neutralizing antibodies. Other limitations regarding their clinical applications include being unstable in the gastrointestinal tract and other bodily fluids, displaying poor absorption, distribution, rapid metabolic degradation, and excretion, causing limited bioavailability [52, 53], high production costs, and large-scale production challenges [54].
Regarding the use of phages in clinical settings, many trials have been initiated either by investigators or commercial companies covering diverse clinical departments and infections caused by widely different bacteria, such as Acinetobacter, Enterobacter, Staphylococcus, Enterococcus, Shigella, Pseudomonas, Salmonella, Vibrio, Burkholderia, Serratia, Neisseria gonorrhoeae, Mycobacteria, and other common clinical bacteria [55]. The results from these studies have generally shown phages to be effective for bacterial eradications with promising clinical results and indicated that phages are generally safe for use under clinical settings and given a GRAS status, i.e., generally recognized as safe [56].
Though phage therapy has been practiced for a very long time in Eastern Europe and Russia [57], many countries are recently rediscovering their interest in this due to rapidly emerging AMR. Only recently have different regulations been introduced in different countries regarding the use of phages, since the use of phages is at its nascent stage in many Western countries. Though the regulations regarding the use of phages vary across different countries, the majority of countries practice the “compassionate” use of phage therapy, wherein phages are used as an unapproved drug for the benefit of patients when all other drugs and antibiotics have failed to treat infections. Phage cocktails for therapeutic purposes can be purchased without a prescription in Georgia and Russia [58]. In the United Kingdom, phage therapy is allowed under compassionate use when phages are prepared following GMP, i.e., good manufacturing practice [59]. Similarly, France and Belgium also practice the compassionate use of phages. Additionally, Belgium has also started to allow the magistral phage preparations, which allow a pharmacist to produce medicinal products based on a physician’s prescription for each patient following pharmaceutical standards [60]. In Australia, phage therapy is only being administered to patients through a special access scheme at Phage Australia center after being referred by a family doctor or infectious disease specialist [61]. In the USA, phages are classified as biological products, and their manufacturing and use for therapy must follow standards like GMP, preclinical research, and clinical trials [62], but the Food and Drug Administration (FDA) allows the compassionate use of phage therapy under exceptional situations when patients can not be enrolled for clinical trials [63].
The FDA has approved seven AMPs, whereas nearly 4,000 AMPs have been registered in the peptide database [10]. Phages are characterized as biological entities, whereas AMPs are regulated as peptides or small-molecule drugs [10]. When a protein molecule, whether it is an AMP or an endolysin, is expressed as a recombinant molecule through recombinant DNA technology using a host cell such as E. coli, it has specific requirements that need to be met according to criteria set out by the European Medicines Agency (EMA), the regulatory authority for medical products in Europe. These requirements are listed under 2001/83/EC and EC regulation 726/2004 [64]. Regulatory requirements for the use of phages are less stringent than compared for endolysins and AMPs. When using any of these treatment modalities, either alone or in combination, they may need to meet these regulatory criteria independently. Since the research regarding the combination of phages/phage-derived enzymes and AMPs is in its initial phase, with none of the combined therapies progressing into clinical trials, and furthermore, the regulatory distinction regarding the nature of these therapies, it makes it difficult to comment regarding the regulatory requirements of these combination therapies.
Progress is being made on the use of AMPs and phages for the treatment of bacterial infections. Yet, very few studies have been performed on the combined use of both of these to combat infections. The results from these few studies are promising. More future studies on clinical trials, efficacy, and safety are required. Regulatory requirements need to be worked out, but results from compassionate use of phages and magistral phage preparations of phages for patients set the stage for a regulatory framework for different governments across the world. The results for using these therapies for combating MDR bacteria are promising and set the stage for further research.
AMPs: antimicrobial peptides
AMR: antimicrobial resistance
FDA: Food and Drug Administration
GMP: good manufacturing practice
LPS: lipopolysaccharide
MDR: multidrug resistant
SN: Conceptualization, Investigation, Writing—original draft. KI: Writing—original draft. MAA: Conceptualization, Investigation, Writing—original draft, Writing—review & editing, Supervision. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 2470
Download: 109
Times Cited: 0
