Affiliation:
1Current address: Radiodiagnosis, Sree Balaji Medical College and Hospital, Chennai 600003, Tamil Nadu, India
2Radiodiagnosis, Bernad Institute of Radiodiagnosis, Madras Medical College, Chennai 600003, Tamil Nadu, India
†
ORCID: https://orcid.org/0000-0001-5640-2656
Affiliation:
3Department of Computer Science, St. Peter’s Institute of Higher Education and Research (Deemed to be University), Chennai 600054, Tamil Nadu, India
†
ORCID: https://orcid.org/0009-0006-5827-1649
Affiliation:
4Multidisciplinary Research Unit (MRU), Department of Health Research, Madras Medical College, Chennai 600003, Tamil Nadu, India
†
ORCID: https://orcid.org/0000-0002-6665-5312
Affiliation:
5Barnard Institute of Radiology, Madras Medical College, Chennai 600003, Tamil Nadu, India
Email: kapudr@gmail.com
ORCID: https://orcid.org/0000-0003-3825-3145
Explor Med. 2025;6:1001341 DOI: https://doi.org/10.37349/emed.2025.1001341
Received: January 17, 2025 Accepted: April 21, 2025 Published: July 01, 2025
Academic Editor: Xiaofeng Wang, Cleveland Clinic Lerner College of Medicine of Case Western Reserve University, USA
Aim: Lung cancer is a leading cause of cancer-related deaths globally, where early and accurate diagnosis significantly improves survival rates. This study proposes an AI-based diagnostic framework integrating U-Net for lung nodule segmentation and a custom convolutional neural network (CNN) for binary classification of nodules as benign or malignant.
Methods: The model was developed using the Barnard Institute of Radiology (BIR) Lung CT dataset. U-Net was used for segmentation, and a custom CNN, compared with EfficientNet B0, VGG-16, and Inception v3, was implemented for classification. Due to limited subtype labels and diagnostically ambiguous “suspicious” cases, classification was restricted to a binary task. These uncertain cases were reserved for validation. Overfitting was addressed through stratified 5-fold cross-validation, dropout, early stopping, L2 regularization, and data augmentation.
Results: EfficientNet B0 achieved ~99.3% training and ~97% validation accuracy. Cross-validation yielded consistent metrics (accuracy: 0.983 ± 0.014; F1-score: 0.983 ± 0.006; AUC = 0.990), confirming robustness. External validation on the LIDC-IDRI dataset demonstrated generalizability across diverse populations.
Conclusions: The proposed AI model shows strong potential for clinical deployment in lung cancer diagnosis. Future work will address demographic bias, expand multi-center data inclusion, and explore regulatory pathways for real-world integration.
Lung cancer remains the leading cause of cancer-related mortality, accounting for over 18% of all cancer deaths globally as of 2020 (GLOBOCAN) [1]. Adenocarcinoma is the most prevalent histopathological subtype. Accurate evaluation of pulmonary nodules is critical for predicting malignancy and determining prognosis, as emphasized in the 8th edition of the TNM classification of lung cancer [2, 3]. Pulmonary part-solid nodules with a solid component larger than 5 mm are often considered malignant, as the extent of the solid component strongly correlates with invasive adenocarcinoma (IVA) [4–6]. These nodules typically require aggressive management, such as surgical resection, unless regression is evident in follow-up imaging. Early and precise assessment of invasive components significantly impacts treatment strategies and improves patient outcomes [7, 8].
Despite its clinical importance, accurately assessing invasive components on CT scans remains a challenge, owing to variability in nodule morphology and the subjective nature of radiological interpretation. Recent advancements in artificial intelligence (AI), particularly convolutional neural networks (CNNs), have revolutionized medical imaging, offering promising solutions to these challenges [9]. CNN-based systems are widely applied in medical diagnostics for tasks such as detecting pulmonary nodules and differentiating between benign and malignant lesions [10–16]. This study focuses on lung nodule segmentation using CT images. A modified U-Net, combined with Respath in the proposed ResNodNet model, achieves 98.6% accuracy in segmenting and classifying lung nodules [17]. These models utilize deep learning techniques to extract intricate image features, enhancing diagnostic accuracy, supporting treatment planning, and improving prognostic predictions [18, 19]. The author discussed the advancements in AI, enhanced lung nodule detection, and classification using CT scans, addressing the critical need for early lung cancer diagnosis. Using a deep-learning model, the proposed CNN achieved promising accuracy in malignancy detection. This non-invasive approach supports early diagnosis, personalized treatment, and reduced morbidity, offering significant real-world healthcare implications. The study highlights AI’s role in improving patient outcomes and advancing digital healthcare [20].
In the realm of lung cancer imaging, CNNs have shown significant potential for automating feature extraction and analyzing extensive datasets. They have demonstrated superior diagnostic performance in detecting invasive pulmonary adenocarcinoma, thereby providing critical support to radiologists. Building on these advancements, this study introduces a novel AI-driven framework aimed at addressing the limitations of existing diagnostic methods. The proposed framework incorporates advanced CNN architectures for automated lung nodule segmentation and classification. Specifically, the framework integrates U-Net architecture, enhanced with Gaussian and bilateral filters, to achieve precise nodule segmentation. For classification, it employs ClassyNet, a novel CNN model designed to differentiate benign from malignant nodules with high accuracy. Feature engineering techniques are incorporated to exclude non-cancerous nodules, enhancing the model’s robustness. The study utilizes the Barnard Institute of Radiology (BIR) Lung Dataset, comprising proprietary CT scans from the BIR, and applies rigorous preprocessing steps such as data augmentation and normalization to ensure the models’ generalizability. Comparative analyses with pre-trained networks, including Inception v3, VGG Net, and EfficientNet B0, highlight the superior performance of the proposed framework. The objectives of this study are to enhance segmentation accuracy using U-Net integrated with Gaussian and bilateral filters. To develop a novel CNN architecture for the precise classification of benign and malignant nodules. To reduce diagnostic ambiguity and improve early detection outcomes through AI-driven tools. By achieving these objectives, the study seeks to make a significant contribution to AI-assisted diagnostics in lung cancer, where early and accurate detection can profoundly impact patient survival. This research aims to bridge the gap between imaging and diagnosis, offering an innovative solution to the complex challenges of pulmonary nodule analysis.
The dataset used in this research article is ethically approved by the Institutional Ethics Committee, Madras Medical College, Chennai, “Ec.No.02122021” titled “An Automatic Nodule Point Detection and Classification of Lung Mass by HRCT”. The initial version 1 dataset of three classes is Benign, Malignant, and Normal lung CT images, is available online with the following link https://doi.org/10.34740/KAGGLE/DSV/8288306. The other cancer types will be hosted online in the next version release. The dataset description is shown in Figure 1.
This clinical study included CT imaging data from 388 individuals who underwent chest CT scans at the BIR, Madras Medical College, Chennai. The inclusion and exclusion criteria were meticulously established to ensure the accuracy and relevance of the study’s findings.
Patients with a CT diagnosis of lung pathologies referred for biopsy, with Lesion size ranging from 8 mm to 20 cm. Lesion characteristics: solid nodules, part-solid nodules, cavitatory lesions, non-resolving pneumonias. A pathological diagnosis of non-mucinous adenocarcinoma based on the 2015 World Health Organization (WHO) classification of lung tumors.
Patients allergic to contrast agents, patients who were uncooperative during the study, patients with contraindications to CT-guided biopsies, cases with inconclusive histopathological examination (HPE) results, patients without preoperative thin-section CT images or with images that could not be analyzed due to artifacts or image noise, patients with prior treatment to the lungs cases with pathological specimens deemed inadequate for diagnosis under the 2015 WHO classification, individuals under the age of 18, pregnant women, patients who don’t provide consent to be part of study.
A proprietary dataset, referred to as the BIR Lung Dataset, was developed using CT scans of 388 cases, resulting in a total of 16,172 images. Scans were pre-processed to anonymize patient information, and annotations were performed by a radiologist to ensure precise image training. Both plain and contrast-enhanced CT imaging were conducted using a Siemens 32-slice CT scanner. Imaging parameters included: KVp: 130, mAs: average (80), slice thickness: 5 mm, reconstruction interval: 1.5 mm.
Ethical approval for this study was obtained from the Institutional Ethics Committee Review Board of Madras Medical College, Chennai (Ec.No.02122021). The need for informed consent was waived for this retrospective review of patient records, imaging data, and biomaterials. All CT data and pathological specimens were provided by the host institution.
Two independent pathologists reviewed and diagnosed all the lung specimens stained with hematoxylin–eosin and/or elastic van Gieson stain according to the 2015 WHO classification of lung tumors. The histological diagnoses of adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), or IVA were confirmed by consensus decisions. The data of the 388 patients used for 3D-CNN model construction comprised AIS (n = 248), MIA (n = 47), and IVA (n = 93).
The CT image data were acquired with three types of multidetector-row CT scanners: Discovery CT750 HD (GE Healthcare), Aquilion PRIME (Canon Medical Systems), and LightSpeed VCT (GE Healthcare). The protocols used with each of the three scanners are summarized in Table 1. All targeted lung CT images were reconstructed using a 200–230 mm field of view from thin-section CT images reconstructed with a high spatial-frequency algorithm.
Transfer learning architecture and its features
| Model | Architecture | Features with ImageNet weights | Applications |
|---|---|---|---|
| VGG-16 | 16 weight layers (13 convolutional + 3 fully connected). | Robust feature extraction for classification tasks. | Transfer learning, object detection. |
| Small 3 × 3 convolutional kernels with 2 × 2 max-pooling layers. | Pretrained on ImageNet, provides generalizable features. | ||
| Requires significant memory due to its size. | |||
| Inception v3 | Modular architecture with inception blocks (1 × 1, 3 × 3, 5 × 5 convolutions). | Highly efficient and accurate for hierarchical feature extraction. | Image classification, segmentation, and image captioning. |
| Uses auxiliary classifiers to combat vanishing gradients. | Pretrained weights reduce the need for large datasets. | ||
| Dimensionality reduction within inception modules. | Optimized for efficiency without sacrificing performance. | ||
| EfficientNet B0 | Compound scaling balances network depth, width, and resolution. | High accuracy with minimal resources when pre-trained on ImageNet. | Edge computing, facial recognition, and anomaly detection. |
| Utilizes MBConv (mobile inverted bottleneck blocks) and squeeze-and-excitation layers. | Scales effectively to larger EfficientNet variants for higher accuracy. | ||
| Lightweight and resource-efficient, ideal for deployment on low-power devices. |
First, without using the 3D-CNN model, three chest radiologists from our institute who have sub-specialization in chest radiology evaluated the cases, i.e., with the grade of junior level to senior level, were independently assessed the CT findings. Each radiologist’s findings were then pooled into a common finding and compared to the 3D-CNN. The results were expressed in terms of percentage.
The model was initially developed using the BIR Lung Dataset, which originates from a single medical center. This introduces potential dataset bias due to the lack of demographic diversity and institutional variability. Furthermore, the dataset lacks access to patient-specific metadata such as age, gender, ethnicity, and smoking history, making it difficult to assess and mitigate demographic and population-level biases.
To partially address this limitation, external validation was conducted using the publicly available LIDC-IDRI dataset, which includes scans from multiple centers. This step was taken to evaluate the model’s generalizability across different populations. Additionally, future work will focus on incorporating demographically diverse, multi-center datasets with annotated patient profiles to enable fairness-aware model training and evaluation.
In order to train the deep learning models, high-performance computing systems that are equipped with graphics processing units (GPUs) were used. The code was written in Python, and numerous deep learning packages, including TensorFlow and Keras, were used in this process.
Pre-processing: The image processing techniques used to enhance CT images during the preliminary processing phase. To optimize the robustness of the model, image clarity is most important. This phase includes image normalization and feature enhancement to remove noise in the images.
Model development: The U-Net architecture was implemented to segment lung nodules. The classification of nodules as benign or malignant was accomplished via the development of a custom-designed neural network.
Transfer learning: In order to identify malignant cells, pre-trained models (Inception v3, VGG Net, and Efficient B0 Net) were used in order to exploit the information that was obtained from the ImageNet dataset.
Training and validation: In order to guarantee a high level of accuracy, the models were trained with the use of the BIR Lung Dataset and a rigorous cross-validation technique. There were three criteria that were used to evaluate the performance: accuracy, sensitivity, and specificity.
Evaluation: In order to evaluate the dependability and efficiency of the automated system, the findings of the models were compared to the evaluations that were conducted by radiologists from the Barnard Institute of Radiology (see Figure 2).
The image enhancement is carried out to highlight the inner features of the lung region to identify the nodule. The original greyscale images are transformed to 32-bit color, quantize applied to segment the lung region. The preprocessed images are fed for training. The lung nodule segmentation using the U-Net architecture. First, the CT images are pre-processed to enhance image quality and normalize the intensity values, ensuring consistency across the dataset. The pre-processed images are then fed into the U-Net model, which consists of an encoder-decoder structure shown in Figure 2. The encoder extracts multi-scale features through successive convolution and pooling layers, capturing both local and global contextual information. Skip connections are used to transfer high-resolution features from the encoder to the decoder, enabling precise segmentation by combining spatial and semantic information. The decoder reconstructs the segmentation map by progressively upsampling the encoded features and merging them with corresponding features from the encoder. This step ensures that the model retains fine-grained details crucial for delineating small and irregularly shaped lung nodules. During training, the model is optimized using a loss function, typically a combination of cross-entropy or Dice loss, to maximize segmentation accuracy. Data augmentation techniques are applied to increase the variability of training data and improve model generalization.
Once trained, the model predicts binary segmentation masks for input CT images, highlighting the boundaries of the lung nodules. Post-processing steps, such as morphological operations, may be applied to refine the segmentation and reduce noise. The resulting segmentation masks are then used for quantitative analysis, including measuring nodule size, shape, and volume, and can aid in further diagnostic and treatment planning tasks. The segmented nodules are fed to a transfer learning model for lung cancer classification.
In this study, we limited the classification task to distinguishing between benign and malignant nodules due to the presence of suspicious cases in the dataset and the lack of sufficient labeled examples for specific cancer subtypes. These suspicious cases, based on histopathology outcomes, represent diagnostically uncertain scenarios and were treated with caution during model training and evaluation. As a result, the current model prioritizes robust binary classification to ensure clinical reliability. These specific cases are handled as a validation set to test the prediction accuracy.
The U-Net architecture is a deep learning model built for semantic segmentation tasks. It is especially useful in medical image analysis because of its ability to collect spatial and contextual elements. This design is made up of two major components: an encoder (contracting path) and a decoder (expanding path), which are linked by skip connections to retain spatial information shown in Figure 3.
The model starts with an input layer that provides CT pictures of lung nodules. These photos are usually pre-processed to guarantee size and intensity uniformity, such as shrinking to 256 × 256 pixels and normalizing intensity ranges.
The encoder extracts hierarchical information from input pictures using repeated convolutional and downsampling techniques. It includes the following steps:
Convolutional layers: Each block has two or three convolutional layers with tiny 3 × 3 filters, followed by ReLU activation. These layers extract spatial characteristics while minimizing information loss.
Max pooling: After each convolutional block, max pooling is used to downsample the feature maps by halving their spatial dimensions (from 256 × 256 to 128 × 128).
The number of feature channels doubles with each level, beginning with 64 and progressing to 128, 256, 512, and beyond, depending on the model depth.
The bottleneck is the deepest region of the U-Net model, where the feature mappings are most compressed and abstract. This layer comprises of extra convolutional layers that improve the previously learnt features without using spatial pooling. The bottleneck connects the encoder and decoder.
The decoder reconstructs the spatial resolution of the feature maps, gradually upsampling them until they match the original input dimensions. It contains the following:
Upsampling layers: The decoder starts with transposed convolutional layers or upsampling operations, which double the spatial dimensions (from 16 × 16 to 32 × 32).
Concatenation with skip connections: The feature maps from the respective encoder layers are concatenated with the upsampled maps to preserve fine-grained spatial information.
Convolutional layers: After each upsampling step, two or three convolutional layers are used to enhance the segmentation, much as the encoder.
Skip connections connect each encoder block to the associated decoder block. These links convey high-resolution characteristics from the encoder to the decoder, maintaining spatial information and allowing for accurate localisation of microscopic objects like lung nodules.
The final layer produces a binary segmentation mask with the same spatial dimensions as the input picture (256 × 256). Each pixel in the mask indicates whether it belongs to the lung nodule or the backdrop (see Figure 3).
The U-Net model is used to segment the lung nodules. The segmented lung region is further reduced to the lung nodule by applying a color threshold integrating the Gaussian and bilateral filters that enhance the image outcome and make the machine learning model better understand how to classify the cancer types, such as adenocarcinoma, squamous cell carcinoma, large cell carcinoma, and small cell carcinoma. The segmented nodule is processed with a Gaussian and bilateral filter. Figure 4 shows different sigma values and the outcome. Figure 4A original input image, Figure 4B sigma = 0.8, Figure 4C sigma = 0.7, Figure 4D sigma = 0.5, range of outcome. The various lung nodule obtained is stored as a feature subset to feed the input for the transfer learning model for classification.
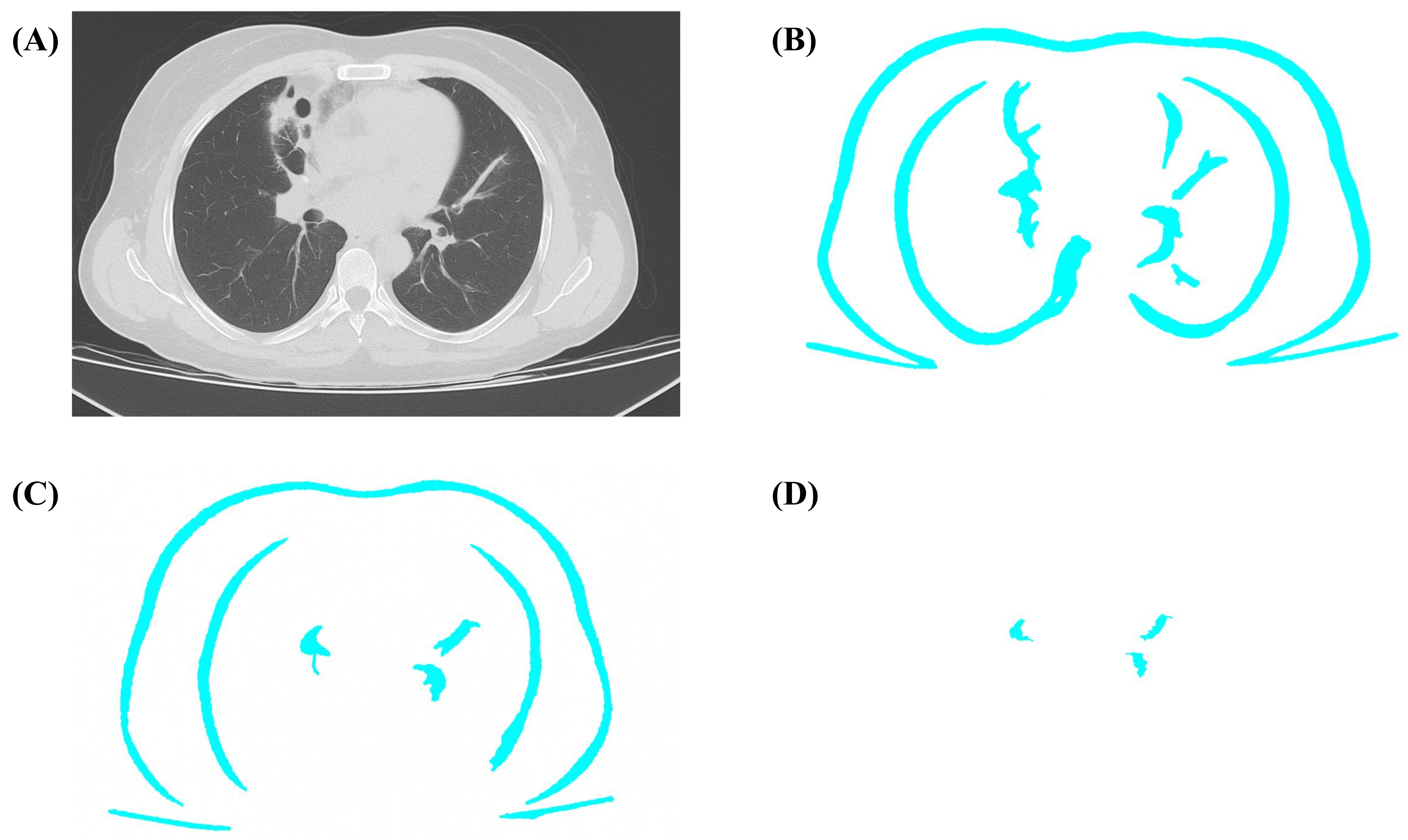
The segmented lung region segmentation with various sigma ranges. (A) Original input image; (B) segmented lung region of sigma range = 0.8; (C) segmented lung region of sigma range = 0.7; (D) segmented lung nodule of sigma range = 0.5
The enhanced U-Net model in the proposed method attains 97% accuracy, as shown in Figure 5. Thus, the model is better at identifying nodules when compared with the traditional identification.
In this research article, the transfer learning model is employed for the classification of the segmented nodule. The models employed for the study are VGG-16, Inception v3, and Efficient B0 Net. The architecture and its features are shown in Table 1. The assessment of the deep learning model in this research used recognised performance criteria, including accuracy, precision, recall, and F1-score, to enable an extensive evaluation of the model’s predictive abilities. The proposed method demonstrated noteworthy classification accuracy, prevailing over the baseline model, Efficient B0 Net, with a significant score of 98.3%. The accuracy and recall scores highlight the ability of the model to accurately distinguish between cancerous and benign nodules while reducing false positives and negatives.
Cross-validation enhanced robustness and generalisability across varied datasets, whereas confusion matrix analysis offered insights into particular categorisation difficulties. The model’s exceptional performance compared to leading architectures, such as VGG-16, Inception v3, and EfficientNet, further confirms its effectiveness. The use of sophisticated methods like colour transformation and transfer learning improved feature extraction and classification accuracy, enhancing its applicability in lung cancer detection.
The experimental evaluation of VGG-16, Inception v3, and EfficientNet B0 for classifying lung nodules into benign and malignant categories reveals significant findings. The results, summarized in Tables 2 and 3, highlight the comparative efficacy of these models based on accuracy, precision, recall, and F1-score metrics. Figures 6 and 7 provide detailed visualizations of the models’ training accuracy, validation accuracy, training loss, and validation loss over 100 epochs. The model was trained using the Adam optimizer with a learning rate of 0.0001, batch size of 32, and a maximum of 100 epochs with early stopping (patience = 10). The dropout rate was set to 0.5, and L2 regularization (λ = 0.001) was applied to prevent overfitting.
Model evaluation metrics
| Metric | Description |
|---|---|
| Accuracy | Measures the proportion of correct predictions to the total predictions. |
| Precision | Proportion of true positive predictions out of all positive predictions. |
| Recall (sensitivity) | Proportion of true positive predictions out of all actual positives. |
| F1-score | Harmonic means of precision and recall are used to balance both metrics. |
A clear and concise overview of each model’s performance metrics and key remarks
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1-score (%) | Remarks |
|---|---|---|---|---|---|
| VGG-16 | 96.0 | 94.0 | 92.0 | 93.0 | Strong feature extraction but higher computational demands and lower recall, risking false negatives. |
| Inception v3 | 97.8 | 96.5 | 95.2 | 95.8 | Modular architecture and multi-scale processing offer balanced precision-recall performance. |
| EfficientNet B0 | 99.3 | 98.9 | 99.0 | 98.9 | Top performance with high accuracy and computational efficiency due to compound scaling. |
EfficientNet B0 demonstrated superior performance, achieving ~99.3% training accuracy and ~97% validation accuracy. Its high accuracy and low loss metrics across both datasets underline its robustness and efficiency as a diagnostic tool. The minimal gap between training and validation metrics suggests effective generalization with negligible overfitting. In contrast, VGG-16 and Inception v3 displayed slightly lower accuracies and higher losses, emphasizing EfficientNet B0’s advanced feature extraction and architecture optimization.
The learning curves for all models showed consistent training loss reduction, with EfficientNet B0 converging near zero by the 100th epoch. Validation loss followed a similar trend, with minor fluctuations stabilizing at slightly higher values than the training loss. These fluctuations reflect potential variations in nodule characteristics across validation samples.
EfficientNet B0’s architecture enabled a balanced trade-off between model depth, width, and resolution, leading to superior diagnostic precision. The model consistently outperformed VGG-16 and Inception v3 in reducing overfitting and ensuring reliable predictions, as depicted in Figure 7 and Table 4. Despite high initial metrics raising concerns about overfitting, implementation of stratified 5-fold cross-validation and regularization techniques such as dropout, L2 weight decay, early stopping, and data augmentation helped mitigate these concerns.
Comparison and summary of the AI-based model CNN with radiology imaging results
| Category | Radiologist 1 | Radiologist 2 | Radiologist 3 | EfficientNet B0 (AI) | Pooled radiologist finding | Proposed AI model CNN finding |
|---|---|---|---|---|---|---|
| Total cases with nodules | 100 | 100 | 100 | 100 | - | - |
| Benign nodules | 73 | 70 | 65 | 72 | 69.3% | 72% |
| Malignant nodules | 22 | 25 | 20 | 28 | 22.3% | 28% |
| Suspicious cases | 5 | 5 | 15 | 0 | 8.4% | 0 |
AI: artificial intelligence; CNN: convolutional neural network; -: not applicable
We further addressed class imbalance through advanced strategies. Data augmentation was applied more extensively to the minority class (MIA), and class-weighted loss functions were used to penalize misclassification of the minority class. Monitoring per-class metrics throughout training ensured balanced model performance.
The validation performance comparison between the private and LIDC-IDRI datasets demonstrates strong generalizability and high classification accuracy of the proposed model. On the private dataset, the model achieved superior scores across all key metrics: accuracy of 0.983, precision of 0.981, recall of 0.985, F1-score of 0.983, and AUC of 0.990. When evaluated on the external LIDC-IDRI dataset, the performance remained robust with slightly lower but still impressive scores: accuracy of 0.957, precision of 0.948, recall of 0.962, F1-score of 0.955, and AUC of 0.970. These results suggest that while the model performs optimally on the internal data, it also maintains high predictive capability on diverse imaging protocols and patient populations, validating its potential for real-world deployment in clinical scenarios (see Figures 8 and 9).
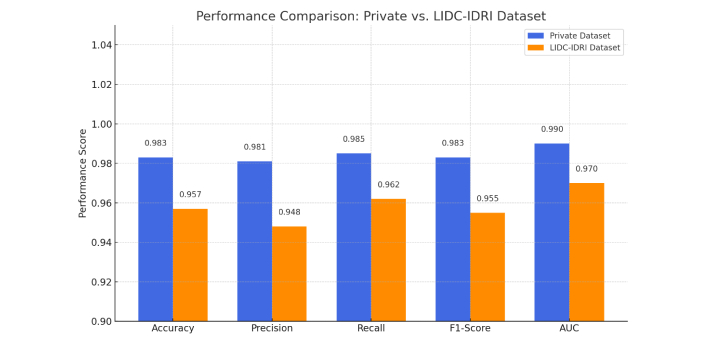
The performance comparison of the Barnard Institute of Radiology (BIR) Lung Dataset and the LIDC-IDRI dataset
In a 100-case classification challenge, the AI model outperformed three radiologists: AI Model: 72% accuracy in benign classification, 28% in malignant, 0% in suspicious cases. Radiologists (combined): 69.3% accuracy for benign, 22.3% for malignant, 8.4% for suspicious cases. This performance highlights the AI’s consistency and reduction in diagnostic ambiguity.
This study demonstrates the effective application of transfer learning models—VGG-16, Inception v3, and EfficientNet B0—for classifying lung nodules as benign or malignant using CT imaging. Among these, EfficientNet B0 achieved the highest performance, with ~99.3% training accuracy and ~97% validation accuracy. Its superior performance is attributed to compound scaling, which uniformly adjusts network depth, width, and resolution, enabling optimized model complexity and better generalization with fewer parameters [21–23].
Our results highlight EfficientNet B0’s ability to outperform both VGG-16 and Inception v3 in validation loss and classification accuracy. While VGG-16 has been effective in earlier medical imaging studies, its deeper architecture and high parameter count often lead to overfitting, particularly in small datasets [24]. Inception v3, though efficient in multi-scale feature extraction, did not match EfficientNet B0 in computational efficiency or generalization capability, as evidenced by our validation results [25, 26].
To ensure robustness, the model was validated using stratified 5-fold cross-validation and tested on an external dataset (LIDC-IDRI), demonstrating consistent accuracy (98.3% ± 1.4%) and AUC values exceeding 0.95. These results underscore the model’s ability to generalize across heterogeneous imaging protocols, in alignment with prior studies emphasizing the need for cross-dataset validation in clinical AI tools [27].
Another major challenge in lung nodule classification is class imbalance, especially underrepresentation of malignancy subtypes such as MIA. We addressed this through focused data augmentation for the minority class and by employing class-weighted loss functions. These techniques were effective in reducing bias toward the dominant class and align with recent recommendations for handling medical data imbalance using fairness-aware methods [28].
Importantly, our model also demonstrated superior performance compared to experienced radiologists in a 100-case classification task. The AI model achieved higher consistency in benign and malignant classifications, with 0% of cases marked as “suspicious,” whereas radiologists had an 8.4% suspicious category rate. This reinforces findings by Prashanthi and Angelin Claret (2024) [27] and Yanagawa et al. (2021) [29], who showed that deep CNNs, when integrated with optimized segmentation frameworks, enhance diagnostic certainty and reduce inter-observer variability.
The clinical implications are substantial. Automated lung nodule classification can reduce workload, improve accuracy in early cancer detection, and assist in cases where radiological expertise is limited. These benefits are consistent with those noted by Setio et al. (2017) [30] and Yuan et al. (2006) [23], who showed that CNN-based tools can support radiologists in improving lung cancer detection outcomes.
From a broader AI perspective, our findings align with recent advancements in intelligent diagnostic systems (Table 5). Yang et al. (2024) [31] demonstrated the adaptability of Inception v4 in diabetic retinopathy classification using optimization algorithms, a strategy that could be translated to lung CT analysis. Uddin et al. (2024) [32] proposed multimodal learning with CT, histopathology, and clinical metadata to improve precision in lung cancer detection. Nabeel et al. (2024) [33] emphasized that hyperparameter optimization in CNN architectures significantly enhances classification accuracy. Moreover, Yang et al. (2023) [34] employed Bayesian models and texture-based radiomics to aid in cancer subtype prediction, showcasing the value of integrating handcrafted and deep features.
The comparison of the recent research outcome with the proposed method
| Study/Author | Model/Approach | Domain | Key contributions | Limitations |
|---|---|---|---|---|
| Yang et al. (2024) [31] | Inception v4 | Diabetic retinopathy | High diagnostic accuracy using deep CNN; adaptable to lung imaging. | Not directly tested on lung cancer datasets. |
| Uddin et al. (2024) [32] | Multimodal learning | Lung cancer | Combines imaging with genetic and clinical data for enhanced precision. | Requires access to multiple modalities; complex data integration. |
| Nabeel et al. (2024) [33] | CNN with hyperparameter optimization | Lung cancer classification | Improved accuracy via fine-tuning CNN parameters. | Focuses only on classification; no segmentation or explainability features. |
| Yang et al. (2023) [34] | Bayesian inference + GLCM texture features | MRI-based cancer detection | Combines probabilistic models with handcrafted features for improved prediction. | Primarily based on MRI; lacks deep learning integration. |
| Prashanthi and Angelin Claret (2024) [27] | U-Net + Custom CNN | Lung nodule detection (CT images) | Integrated segmentation and classification with 98.3% accuracy; scalable, efficient for clinical use. | Lack of multi-class subtype differentiation. |
| Proposed Methodology | U-Net + Transfer learning (VGG-16, Inception v3, EfficientNet B0) | Lung nodule classification (CT biopsies) | Enhanced internal features, precise segmentation, and classification using EfficientNet B0 with 99.3% accuracy. Offers computational efficiency and early detection support for clinicians. | Focuses on binary classification; future work needed on subtype differentiation and XAI integration for better clinical practise. |
CNN: convolutional neural network
While the results are promising, limitations must be acknowledged. The current model was trained on a single-center dataset, which may introduce demographic bias. Although external validation partially mitigated this, future studies should include multi-center datasets with metadata such as age, sex, smoking history, and ethnicity to evaluate fairness. The model is also limited to binary classification due to the unavailability of subtype-labeled data.
Future work will focus on expanding the model to support multi-class classification, including subtypes like adenocarcinoma, squamous cell carcinoma, and small cell carcinoma. Additionally, the incorporation of Explainable AI (XAI) techniques such as Grad-CAM and SHAP will improve transparency and build clinician trust. Integration into Picture Archiving and Communication Systems (PACS) and real-time deployment will further enable clinical translation.
In conclusion, this study provides a clinically relevant AI framework for lung nodule classification. The integration of U-Net segmentation with EfficientNet B0 classification yields high diagnostic accuracy and reliability. Supported by robust validation and comparative performance against radiologists, this model sets the foundation for future deployment in precision oncology and AI-assisted lung cancer screening.
AI: artificial intelligence
AIS: adenocarcinoma in situ
BIR: Barnard Institute of Radiology
CNNs: convolutional neural networks
IVA: invasive adenocarcinoma
MIA: minimally invasive adenocarcinoma
WHO: World Health Organization
We acknowledge the support and facilities provided by our institution for conducting this research. We also extend our gratitude to all faculty members and staff who contributed to data collection and analysis. We are especially grateful to the Multidisciplinary Research Unit (MRU—a unit of the Department of Health Research) for their continued support throughout the research process, up to the submission of this manuscript.
AKA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing—original draft, Writing—review & editing. PB: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing—original draft, Writing—review & editing. AR: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing—original draft, Writing—review & editing. KS: Conceptualization, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing—original draft, Writing—review & editing. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Ethical approval for this study was obtained from the Institutional Ethics Committee Review Board of Madras Medical College, Chennai (Ec.No.02122021).
The need for informed consent was waived for this retrospective review of patient records, imaging data, and biomaterials. All CT data and pathological specimens were provided by the host institution.
Not applicable.
The proposed BIR Lung Dataset in the article, version 1 data is available online with the following link https://doi.org/10.34740/KAGGLE/DSV/8288306.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 5809
Download: 62
Times Cited: 0
