Affiliation:
1Faculty of Medical Sciences, University of Oxford, OX3 9DU Oxford, UK
ORCID: https://orcid.org/0009-0002-4956-3050
Affiliation:
2Saw Swee Hock School of Public Health, National University of Singapore, Singapore 117549, Singapore
ORCID: https://orcid.org/0000-0002-5484-241X
Affiliation:
3Centre for Human Genetics, Pandemic Sciences Inst, University of Oxford, OX3 7BN Oxford, UK
Email: miles.carroll@ndm.ox.ac.uk
ORCID: https://orcid.org/0000-0002-7026-7187
Explor Med. 2025;6:1001333 DOI: https://doi.org/10.37349/emed.2025.1001333
Received: December 24, 2024 Accepted: May 13, 2025 Published: June 18, 2025
Academic Editor: Marcos Roberto Tovani-Palone, Saveetha Institute of Medical and Technical Sciences (SIMATS), India
The article belongs to the special issue Emerging Infectious Diseases
Ebola virus (EBOV) infection usually leads to highly lethal EBOV disease (EVD) with associated viraemia. Viraemia is cleared in those that do survive, however, EBOV may remain hidden in the testes and other immune privileged niches (IPNs) where it can persist for years during asymptomatic convalescence. Viral shedding into seminal fluid may result in sexual transmission to naive contacts years after EBOV outbreaks have been declared over. This leads to flare-ups of cases, redefining our understanding of the shaping and origin of EBOV outbreaks. Such delayed sexual transmission eliminates the geographical boundaries which typically constrain EBOV outbreaks, thus posing a significant global health security threat. Despite hints of EBOV persistence dating over half a century, it was only until the unprecedented scale of the 2013–2016 Western Africa EBOV epidemic that the true importance of this phenomenon was revealed to scientists, public health officials and policy makers alike. This review summarises the evidence for EBOV persistence, suggests the possible underlying molecular mechanisms and proposes future directions for research in the field. A meta-analysis is presented to further investigate the duration of EBOV shedding in seminal fluid. The ultimate aim is to develop therapeutics that clear sites of persistence. Such therapeutics could prevent the re-emergence of the persistent virus, eliminating the chance of new outbreaks whilst alleviating the severe stigmatisation facing the EBOV survivor population.
The first reported case of filovirus persistence was in 1967 when viral RNA was detected in the semen of a Marburg virus (MARV) survivor and resulted in the infection of a sexual partner [1]. Similar small, sporadic outbreaks of filoviruses were recorded over the following decades, however, little insight was gained into the seemingly rare phenomenon of filovirus persistence. This was changed by the unprecedented scale of the 2013–2016 West Africa Ebola virus (EBOV) epidemic; > 28,000 cases and > 11,000 deaths were recorded [2]. The large number of EBOV survivors provided the unprecedented opportunity for studying filovirus persistence, sparking momentum in the field.
Here, we define persistence as the presence of viral RNA within an isolated tissue but not peripheral blood of the host. Persistence in immune privileged niches (IPNs), such as the brain, eyes, and testes, can lead to disease recrudescence and transmission months-to-years after clearance of initial infection which pose risks to both the individual and to public health. Additionally, EBOV persistence contributes to stigmatisation facing survivors within their communities which in turn contributes to transmission due to individuals hiding their infection [3]. Immune responses are dampened in IPNs by active immunosuppression and the presence of tight blood-tissue barriers [4], slowing the clearance of EBOV. Persistent EBOV may be shed into bodily fluids from IPNs, potentially resulting in sexual or vertical viral transmission. Additionally, the persistent virus may reactivate into its pathogenic life cycle leading to recrudescence with associated transmission risks like those in acute EBOV disease (EVD). EBOV pathogenesis, lifecycle and host-pathogen interactions are beyond the scope of this review, however, understanding of these core principles is incomplete, hampering the study of EBOV persistence which builds on this foundation.
This review will explore the evidence-base for the different sites of EBOV persistence in the body and the associated epidemiological implications, with a particular focus on the testes. We additionally present a meta-analysis of longitudinal semen sampling studies from EVD survivors to further evaluate the threat posed by viral shedding. Understanding of the molecular mechanisms underpinning the establishment and maintenance of the persistent reservoir is lacking and so we outline the evidence for three possible theories. Lastly, we examine the solutions to tackling EBOV persistence and propose future research priorities in the field to inform researchers and policymakers.
EBOV (aka Zaire) is one of five species in the Orthoebolavirus genus and, like MARV, belongs to the Filoviridae family. It is assumed filoviruses share the same sites and mechanism of persistence. Filoviruses have a non-segmented negative sense single-stranded RNA genome [5]. Their infection in humans is usually highly lethal; EBOV is no exception with a case-fatality rate up to 90% [6]. Zoonotic spillover from the EBOV animal reservoir, likely bats, has led to numerous outbreaks in Africa with the largest two in the past decade; never in recent history have there been more survivors of EBOV [7]. EBOV incubation period can last 2–21 days, often resulting in non-specific febrile illness followed by more severe symptoms that may include haemorrhagic fever [8]. EBOV is usually spread by the mucosal route and initially infects dendritic cells and macrophages which migrate to lymph nodes, facilitating the dissemination of EBOV throughout the body. The pathogenesis underlying acute EVD is a combination of direct cytopathic effects of viral replication plus immune dysregulation that can lead to fatal hypovolaemia [9].
Figure 1 summarises the immune privileged sites that EBOV has been reported to reside and Table 1 provides the supporting literature. The strongest clinical evidence of persistence in the eye and brain come from just two, but well-documented, cases of recrudescence in foreign healthcare workers volunteering during the 2013–2016 EBOV epidemic. Both received aggressive supportive care and experimental EBOV therapies. The first report of late severe EBOV recrudescence involved a Scottish nurse with meningoencephalitis 10 months after initial EBOV diagnosis, despite prior clearance of viraemia. EBOV RNA was detected in cerebrospinal fluid, strongly indicating persistence within the brain [10]. Recently two further case reports have been published regarding fatal meningoencephalitis at day 137 following initial EBOV infection in an older man and day 342 in a young woman. However, the young woman became pregnant during convalescence which questions whether the brain was the true site of the persistent reservoir [11].
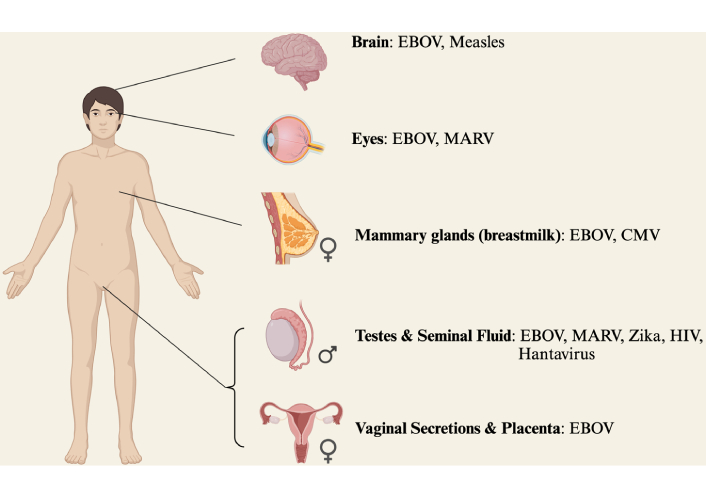
Sites of viral persistence. The phenomenon of persistence is not unique to filoviruses. 22 viruses have been detected in seminal fluid alone following acute infection [12], with evidence of persistence for Hantavirus [13], Zika [14], and HIV [15]. Measles can persist in the brain [16] and CMV in mammary glands [17]. List not exhaustive. EBOV: Ebola virus; MARV: Marburg virus; CMV: cytomegalovirus; HIV: human immunodeficiency virus. Created in BioRender. Tipton, T. (2025) https://BioRender.com/d76j665
Summary of key studies evidencing sites of EBOV persistence
| Study | Experimental design | Key finding | Site |
|---|---|---|---|
| Human clinical studies | |||
| Varkey et al. [18] (2015) | US doctor—recrudescence case study | 1st case of ocular-related recrudescence. | Eye |
| Jacobs et al. [10] (2016) & Bosworth et al. [19] (2021) | Scottish nurse—recrudescence case study | 1st case of severe EBOV recrudescence—meningitis. | Brain |
| Bower et al. [20] (2016) | Case study of pregnancy | Delivery of EBOV-positive stillborn. | Placenta |
| Sissoko et al. [21] (2017) | Longitudinal 26 male survivors | 50% clear semen RNA by day 115, 90% clear by day 394. | Semen |
| Deen et al. [22] (2017) | Cohort convenience sample study 220 male survivors | 27% positive for EBOV RNA at any given timepoint, 11% positive at 10–12 months. | Semen |
| Barnes et al. [23] (2017) | Case study of male survivor | Detection of viable EBOV in semen. | Semen |
| Fischer et al. [24] (2017) | Longitudinal 149 male survivors | 8% men EBOV RNA semen positive > 2 years after initial infection. | Semen |
| Sissoko et al. [25] (2017) | Case study of infant death | Detection of EBOV in breast milk likely causing vertical transmission. | Breast milk |
| Whitmer et al. [26] (2018) | Sequencing of clinical isolates from survivors | Ongoing EBOV replication and heterogeneous evolutionary rates in persistence. | Eye; Semen |
| Sneller et al. [27] (2019) | PREVAIL III: longitudinal study 966 survivors—sequelae | 26% uveitis. EBOV RNA shedding in semen is intermittent. Max 40-month semen persistence. | Semen |
| Liu et al. [28] (2019) | Longitudinal case study of female survivor | EBOV RNA in vaginal fluid 36 days after EVD. | Vaginal secretions |
| Keita et al. [29] (2019) | PostEboGui: 40-month longitudinal study of survivors | 2/168 breast milk positive for EBOV RNA—210 and 500 days after EVD discharge. | Semen; breast milk |
| Luo et al. [30] (2019) | Modelling EBOV transmission dynamics | Sexual transmission important contribution to outbreaks. | Semen |
| Khurana et al. [31] (2020) | Longitudinal clinical antibody measurements | Immunological evidence persistence may still occur even if not detected in semen. | Semen |
| Kofman et al. [32] (2021) | Longitudinal study of 131 male survivors | Semen sample EBOV RNA positive 988 days following EVD hospital discharge. | Semen |
| Medina-Rivera et al. [33] (2021) | Systematic review of clinical studies | Detection of EBOV in breast milk likely resulting in transmission. | Breast milk |
| Thorson et al. [34] (2021) | Longitudinal 220 male survivors | Max duration 696 days. Risk factors: older age, increased EVD severity. | Semen |
| Dyal et al. [35] (2023) | Longitudinal 131 male survivors | Risk factors: older age, decreased EVD severity, high IgG3. | Semen |
| Fallah et al. [36] (2023) | Observational cohort study pregnant women | Largest pregnancy study of EVD survivors. 2/354 breastmilk samples positive. | Breast milk; vaginal secretions; placenta |
| Mukadi-Bamuleka et al. [11] (2024) | Case report of 2 fatal meningoencephalitis | 2nd evidence of brain recrudescence in humans. Persistent EBOV very slow mutation rate. | Brain |
| Animal studies | |||
| Zeng et al. [37] (2017) | NHP model—112 | 1st animal model studying EBOV persistence in IPNs. | Testes; eye; brain |
| Coffin et al. [38] (2018) | MARV NHP model—73 | Sertoli cells as reservoir, breakdown of blood-testes barrier leads to infiltration of immunosuppressive Tregs. | Testes |
| Cooper et al. [39] (2018) | EBOV guinea pig model | Identifies many tropisms in EVD previously overlooked. | Female repro tract |
| Watson et al. [40] (2021) | Ferret model | EBOV RNA in eye with polymerase stop codon mutations. | Eye |
| Johnson et al. [41] (2021) | NHP model | Rare in vivo identification of low levels of defective interfering genomes (DIGs) in testes. | Testes |
| Worwa et al. [42] (2022) | NHP case study | Association between persistent intraocular EBOV RNA with severe uveitis. | Eye |
| Liu et al. [43] (2022) | NHP model—24 | Discovery of new tropisms in male & female repro tracts and fluid. | Male & female repro tracts |
| Liu et al. [44] (2022) | NHP model—36 | 7/36 persistence within the brain, of which 2/7 associated with recrudescence. | Brain |
| Gao et al. [45] (2023) | In vitro & rat model | EBOV stimulated vascular endothelial growth factor production by pericytes leading to blood-brain barrier breakdown. | Brain |
| Clancy et al. [46] (2023) | Mouse-adapted EBOV sexual transmission model | 1st animal model of EBOV sexual transmission. Epididymal epithelium as EBOV reservoir. | Semen |
EBOV: Ebola virus; EVD: EBOV disease; IPN: immune privileged niche; MARV: Marburg virus; NHP: non-human primate
The second healthcare worker involved a US doctor who developed vision-threatening unilateral uveitis nine weeks after viraemia clearance. qRT-PCR showed presence of EBOV RNA in an aqueous humour sample with viral culture confirming presence of viable EBOV [18]. Additional supporting evidence of the eye as a possible site of persistence include a case report of MARV ocular-related recrudescence and non-human primate (NHP) EBOV studies [37, 42, 47]. However, numerous prospective studies of EBOV survivors did not detect EBOV RNA in the aqueous humour [48–50]. One such prospective study investigated 22 EVD survivors undergoing cataract surgery and did not detect EBOV RNA in any aqueous humour samples. The study suggests cataract surgery is both effective and safe in EVD survivors who are many months past their initial infection and who show no signs of active inflammation at the time of surgery [48].
The sites of EBOV persistence identified in clinical studies during the 2013–2016 epidemic were later validated in the NHP model, the gold standard for filovirus research. The landmark study by Zeng et al. [37] was the first to investigate EBOV persistence in IPNs in an animal model. The retrospective nature of using archived tissue facilitated a large sample of 112 rhesus macaques. 11.5% of animals were EBOV RT-PCR-positive in the eye, 1.3% in the epididymides and 1.25% in the brain 28–43 days after viral challenge. Consistent with clinical studies, these findings validate NHPs as a suitable model for studying EBOV persistence although the timeframe is much shorter than possible with human studies.
It is even more unclear whether the female reproductive tract and mammary glands are sites of persistence. Limited evidence suggests viable EBOV is shed into vaginal secretions up to several weeks following acute EVD [28]. However, the largest cohort study of pregnancy in EBOV survivors revealed all 367 vaginal swabs from 79 women were EBOV negative [36]. Although not an IPN, EBOV RNA has been detected in breastmilk of lactating mothers who have recovered from EVD [29, 33, 36]. Whether this means the mammary gland acts as a dormant EBOV reservoir is uncertain. Recently, adipose tissue has been identified as an additional possible site of EBOV persistence. Adipose tissue supported high levels of viral infection for 28 days without cytopathic effects during in vitro EBOV infection of human and bat-derived cell lines [51]. The study illustrates the ongoing evolution of knowledge about the sites of EBOV persistence.
Animal models have been valuable to study EBOV persistence but have also proved challenging. EVD in NHPs is almost uniformly lethal, enabling the modelling of acute disease but less so long-term persistence. Exploration of mucosal routes of inoculation and treatment with mAbs may prove useful routes to improve the NHP persistence model [44, 52]. In contrast, mice are resistant to EBOV, but mouse-adapted EBOV (MA-EBOV) strains have been developed to enhance susceptibility to infection. Ferrets have been explored more recently as an intermediary between the high lethality of EBOV in NHPs and the resistance in mice. However, given the timeframe required to study persistence, there are significant ethical and financial challenges facing animal research in this field.
Clinical studies of EBOV persistence have particularly focused on the testes, largely due to the ease and relative non-invasiveness of seminal fluid sampling and the consequential risk of sexual transmission. Longitudinal cohort studies of male survivors from the 2013–2016 EBOV epidemic used qRT-PCR to detect EBOV RNA in semen samples, the results of which can be used to generate mathematical models for survival analyses of persistence [22]. Estimates of duration and frequency of persistence vary, but overall show an initial rapid drop in number of persistent survivors with 50% survivors persisting at ~ 4 months followed by a levelling off such that a small number of individuals experience persistence for 12+ months (< 10%) [21, 29]. The longest EBOV RNA detection in semen is 988 days after EVD discharge [32]. However, the longest detection time of any virus in semen is Hantavirus with six years [13]. The conflicting estimates between longitudinal studies could be addressed using meta-analyses, however, the variation in experimental designs makes comparison of studies challenging. Similar future investigations of filovirus outbreaks should therefore aim to adhere to standardised protocols, incorporating an international standard, so all studies can be collated.
Though longitudinal studies on the detection of EBOV RNA in the semen of male survivors provide insight into the potential of virus shedding, they are all limited by the number of individuals in each study. Published studies have captured longitudinal shedding in approximately 500 male survivors, however, it is estimated that there are > 5,000 men in West Africa that survived EVD. Added to the issue of a < 10% sampling size is the paucity of repeat sampling timepoints of individuals which may miss the relatively infrequent shedding of EBOV in semen. Furthermore, we know from the 2016 and 2021 sexual transmission flare ups [21, 53], that are linked to virus testicular persistence for ~ 500 days and ~ 5 years respectively, longitudinal EBOV RNA shedding studies may underestimate the true threat of virus persistence and humans as a source of future EBOV outbreaks.
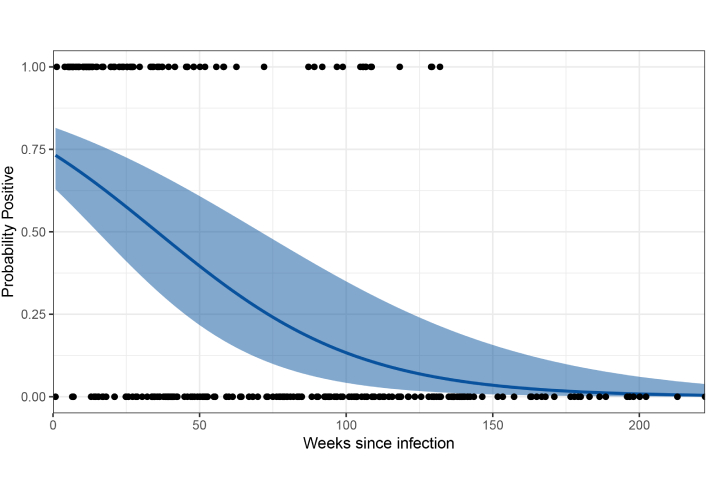
Using prior published studies on EBOV persistence in male survivors, we conducted a meta-analysis of the probability of detecting EBOV in seminal fluid [21, 24, 29]. We included all cohort studies reporting longitudinal data on Ebola RNA carriage in seminal fluids from male survivors with dates from EBOV diagnosis and used a similar cycle threshold (Ct) criteria for definition of positive, in addition to providing full access to meta data on individuals within study. We excluded one study [32] as this was a case control study preferentially sampling from Ebola survivors with known EBOV persistence. For all included studies, we classified EBOV RNA presence as a Ct value of less than 41 from at least one RT-PCR assay. From these binary classifications, we fitted a binomial model for the probability of detecting EBOV RNA in seminal fluid by days since diagnosis. To adjust for repeat measurements on individuals, we included a random intercept for each participant. Models were implemented in a Bayesian framework using Integrated Nested Laplace Approximation in R statistical software (version 4.4.0) [54]. Predictions were made using 1,000 posterior samples.
Our final analysis included 287 observations from 66 unique individuals surveyed by three studies [21, 24, 29]. The number of observations for each individual ranged from 1 to 12 (mean 4.3) from 6 to 1,557 days post diagnosis (mean 504 days). No positive individuals were detected after 925 days since diagnosis. Results showed a strong decrease over time, however, with considerable uncertainty (Figure 2). For 5,000 male EBOV survivors, this estimates a median of 7 individuals [95% confidence interval (CI): 0–43] would have detectable EBOV RNA in semen at 5 years post infection.
The cellular reservoir at sites of persistence is currently unknown. Direct tissue sampling in NHPs identified CD68+ macrophages as a cellular reservoir of persistence in the eye [37]. Subsequent NHP studies by the same group confirmed the same cellular EBOV reservoir was found in the brain [44] and Sertoli cells as a site of MARV persistence in the testes [38]. Furthermore, epididymal epithelial cells have been identified as a reservoir for MA-EBOV in an immunocompetent mouse model [46]. Whether the same cellular reservoir exists in humans remains to be determined.
Figure 3 shows the Sertoli cells forming the walls of seminiferous tubules that make up the testis. This creates the blood-testis barrier which prevents developing auto-immunogenic spermatocytes from being recognised as “foreign”. Sertoli cells are susceptible to Zika [55] and can sustain viral replication in vitro, suggesting these cells as a possible persistent reservoir [56]. Theoretically virus may be shed from the reservoir in the Sertoli cells and infect the developing spermatocytes as they migrate towards the lumen of the seminiferous tubule. Interestingly, the shedding of Zika virus in semen coincides with the 74 days required for renewal of spermatogonia, the stem cell population responsible for spermatocyte production, suggesting these cells may also be a contender for the site of persistence [47]. Other potential contenders include Leydig cells which produce testosterone and peritubular myoid cells which contract the tubules, although both these cells lie outside the blood-testis barrier and therefore outside the IPN. Investigations of the EBOV persistence reservoir in human testicular tissue is absent in the literature and warrants further study.
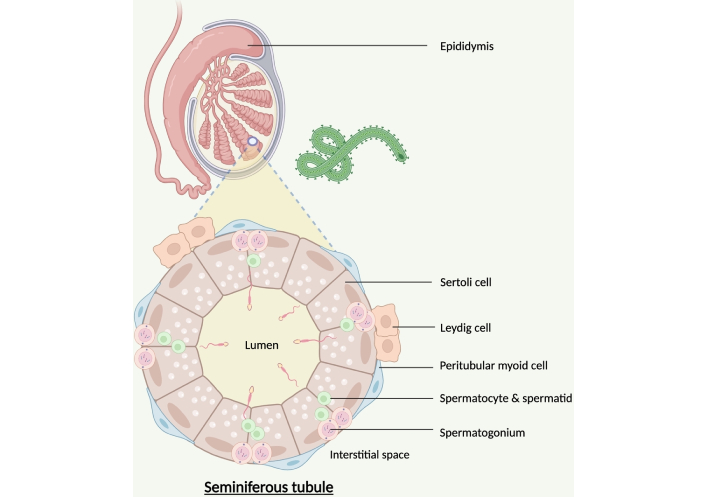
Possible testicular cellular reservoirs of persistent EBOV. EBOV: Ebola virus. Created in BioRender. Tipton, T. (2025) https://BioRender.com/g37g085
Most longitudinal semen studies did not perform viral culture from the sample, therefore understanding of the viability and therefore risk of transmission from EBOV RNA positive semen is unclear. However, numerous studies have now shown this theoretical risk of sexual transmission has already resulted in real-world transmission events [58–62]. EBOV sexual transmission sparks later flare-ups whilst also contributing to the viral basic reproduction rate during outbreaks [30]. The most striking evidence comes from phylogenetic analysis by Keita et al. [53] which showed the likely source of a 2021 EBOV outbreak in Guinea as the persistence in a male survivor infected five years prior in the 2013–2016 epidemic. This is especially alarming given the limited number (likely < 10) male survivors from the 2016 sexual transmission flare up, which is the proposed source of the 2021 outbreak, suggesting long-lasting persistence may be more common than initially indicated in some individuals [61]. The 2021 viral sequences shared mutations unique to the 2013–2016 virus, including the A82V glycoprotein mutation which is indicative of human adaptation [63]. This proves the 2021 flare-up was human in origin, representing a paradigm shift in the field that the initiation of EBOV outbreaks is not always zoonotic in nature. The study also highlights the synergy of molecular and classical epidemiology to trace the origin of outbreaks. Recently, molecular epidemiology has been facilitated by the advent of highly portable technologies such as MinION [64]. It is important to note it is likely several factors must align for transmission to occur; the contact must be EBOV-naive, and the intermittent nature of viral shedding (hence the need for consecutive negative results in longitudinal studies) means timing must be right.
The literature has solely focused on male-to-female transmission with complete absence of discussion regarding male-to-male sexual transmission. Contact tracing of male-to-male transmission is challenging given the severe discrimination facing men-who-have-sex-with-men (MSM) groups in Africa. However, if EBOV behaves in a similar manner to HIV in that transmission by anal sex is 18 times more likely than vaginal sex [65], then EBOV male-to-male sexual transmission may have a significant hidden role in shaping EBOV outbreaks. NHP challenge studies comparing susceptibility to infection between rectal vs. vaginal mucosal EBOV exposure may prove a valuable initial line of enquiry.
EBOV persistence poses a risk of vertical transmission from mother to baby. Live virus has been isolated from breast milk over 500 days after EVD discharge of a seronegative mother [29] whose initial infection occurred before pregnancy began. Viral sequencing has evidenced events of EBOV transmission by breastfeeding resulting in infant death [25]. These events are exceptionally rare, and evidence suggests breastfeeding is safe when conception is greater than one year after maternal EVD recovery [36]. Testing of breastmilk for EBOV should be expanded with further investigations of other maternal fluids to rule out alternative routes of transmission to infants [33]. Acute maternal EVD has a ~ 80% fatality for the foetus despite the immune privilege status of the placenta [66]. High viral loads in a stillborn suggests EBOV can persist in the placenta following maternal recovery which may expose healthcare workers to the virus during delivery [20]. Two studies in Liberia suggest EVD worsens outcomes of future pregnancies, although both studies had no control groups which is concerning given the lack of population-level data in the region [67, 68]. However, a recent large study revealed in women who have recovered from EVD before conception, there is no increased risk of transmission to the baby or healthcare workers during delivery [36].
Case studies of organ-specific inflammation show EVD recrudescence results in poor clinical outcomes [10, 18] which is consistent with NHP models [42, 43]. Uveitis in the recrudescent US doctor [18] hints towards the unusually high incidence of ophthalmic complications seen in EBOV survivors [27, 50]. This suggests a role for persistence not only in recrudescence [42] but within post-Ebola sequelae. This is the chronic disease burden in survivor populations, involving symptoms such as fatigue, myalgia and arthralgia [27].
Recrudescence can also lead to onward transmission; Mbala-Kingebeni et al. [69] report a case of recrudescence with viraemia six months after initial EVD recovery which sparked a transmission chain of 91 cases, extending the 2018–2020 EBOV outbreak an additional six months [69]. The overall incidence of recrudescence is low [70], but it only takes one case to start a new outbreak. It is plausible the incidence of recrudescence may rise in future outbreaks with increased use of vaccination and monoclonal antibody therapies. These medical countermeasures will save lives, including keeping the sickest EVD patients alive but these patients are possibly at the greatest risk of recrudescence [34]. Without extraordinary supportive care and experimental therapies to treat acute EVD, the two recrudescent healthcare workers likely would have died [19]. Additionally, the two other documented cases of recrudescent meningoencephalitis both received monoclonal antibody treatment at initial infection, further suggesting a link between the use of monoclonals and later EBOV persistence.
Overall, there is a reasonable evidence base for the risk and duration of persistence especially in seminal fluid. However, study of the mechanisms resulting in the establishment and maintenance of the persistent reservoir are absent. The lack of immune response in IPNs does not explain why the cytopathic effects of EBOV replication are not observed during asymptomatic persistence. The detection of antigenomic EBOV RNA using fluorescence in situ hybridisation in semen from human survivors and in tissue in NHPs indicates ongoing viral replication during persistence [23, 37]. Phylogenetic analyses of EBOV spanning years over the course of transmission events suggest a low viral evolutionary (mutation) rate [11, 53, 71]. For instance, just two new nucleotide substitutions were identified in the viral RNA sequence from a case of CSF recrudescence compared to the sequence 10 months prior during initial infection [10]. Given the infidelity of the RNA-dependent RNA polymerase of EBOV, 22 substitutions would be expected in the same period [19]. Overall, the evidence strongly suggests a very low rate of EBOV replication during persistence. The underlying mechanism for this low viral replication rate has received little attention in the literature, but there are three theories supported by conflicting bodies of evidence.
A favoured theory is EBOV quasispecies found in acute EVD may contain a minor variant that possesses mutations impairing the function of the RNA polymerase. This minor variant may seed itself within an IPN with low rates of viral replication and lead to persistence [19]. Spontaneous reversion of the mutation during persistence may then lead to lytic replication and recrudescence. This theory is inspired by paramyxoviruses where a single amino acid substitution in the RNA polymerase switches between acute and persistent infection [72]. Similar substitutions in EBOV RNA polymerase have been identified in minor variants found in plasma samples from acute EVD patients and shown in an in vitro replicon system to reduce RNA replication [73]. An important source of evidence comes from a recent EBOV natural history transcriptomics study of 21 NHPs that included isolates from IPNs. Normandin et al. [74] showed presence of specific variant compartmentalisation, with tissue-specific populations acting as isolated infections. Use of a minigenome system revealed emergence of variants reducing viral fitness. However, samples were obtained over just eight days post inoculation, highlighting the uniformly lethal nature of EBOV in the NHP model which makes studying longer-term persistence challenging [74]. In an alternative animal model, deep sequencing of tissues in an EBOV ferret model shows variants within the viral population contain polymerase mutations that contribute to organ-specific phenotypes [75]. Interestingly, a nonsense mutation has been identified in 16% of EBOV genomes within an EBOV ferret eye persistence model that results in a truncated (likely defective) polymerase [40]. Despite this growing evidence supporting the polymerase mutation theory, such minor variants are yet to be found in humans at sites of IPNs or in cases of recrudescence or persistence-related transmission. In vitro testicular models should be explored in the future to further investigate the molecular mechanisms of persistence (Figure 3). Such studies would be well placed given the advancing insights into EBOV replication regulation and polymerase structure [76, 77].
Defective interfering genomes (DIGs) are replication-incompetent sequences of viral RNA genome which can outcompete the intact viral genome for replication machinery, hence reducing the replication of the wild-type virus [78]. DIGs have been shown as a mechanism of measles persistence; high levels of DIGs have been detected in brain tissue of subacute sclerosing panencephalitis, a complication of measles recrudescence [16]. The first evidence for DIGs in EBOV infection involved in vitro serial passaging [79]. However, given 10 viral passages were carried out at high multiplicity of infection it is perhaps unsurprising DIGs were detected [80]. Furthermore, viral RNA sequencing in vivo and from clinical isolates have either not detected DIGs or they were at such low levels that they are unlikely to have a meaningful biological significance [19, 26, 78]. The fact polymerase mutations have been identified in vivo and that these have shown to significantly reduce EBOV replication weighs currently in favour of the mutation theory rather than a central role for DIGs.
The previous two theories for the low EBOV replication are virus centric. Barnes et al. [23] suggests the low evolutionary divergence seen in EBOV persistence is rather due to the absence of immune selection within IPNs and argues high viral loads would be expected even with a low rate of viral replication [23]. Although plausible, immune attenuation of EBOV to facilitate persistence is supported by limited evidence, such as the reactivation of EBOV in vitro when the type 1 response is antagonised [81]. This theory conflicts with the potent inhibition of innate immunity seen by EBOV, although perhaps the virus and the host form a mutualistic relationship as seen in the bat EBOV reservoir. In conclusion, host immune responses may play a role in permitting EBOV persistence but given the current evidence it is difficult to see this as the core underlying mechanism.
In summary, the mechanism of EBOV persistence in survivors’ testes is likely a result of the performance of the viral polymerase in this unique immune privileged site. As there is no obvious evidence of polymerase specific mutations in persistently shed virus, the mechanism it is most likely associated with unique environmental factors found within the testes and other immune privileged sites.
Lack of an evidence-base makes it challenging to target the underlying mechanisms causing persistence. Therefore, semen testing programs and guidelines regarding safer sex and breastfeeding by the World Health Organisation (WHO) are important to reduce the risk of EBOV transmission. However, socioeconomic factors contribute to low adherence with the advice [34].
A compartmental population model predicted antibody levels cyclically rise and fall in survivors, leading to the suggestion of vaccinating survivors to prevent recrudescence [82]. This antibody decay-stimulation may reflect de novo EBOV antigen restimulation from IPNs to boost the residual primary antibody response, paralleling events seen in measles persistence [83]. However, this theoretical model is contradicted by recent experimental findings; ELISA and ELISpot reveal sustained high levels of antibodies and T cells over a 3-year follow-up [84]. It was also shown antibody levels in survivors are approximately ten-times higher than those in EBOV vaccinated individuals [84], with speculation continuous antigen stimulation from IPNs maintains this potent, long-term adaptive immunity in survivors [85, 86]. However, CD8+ T cell phenotyping revealed the presence of memory T cells and absence of effector T cells, indicating these cells have not recently been exposed to antigen. This strongly suggests antigen does not leak out of IPNs into the systemic circulation to boost adaptive immunity; instead, EBOV and its antigens remain restricted inside IPNs during convalescence [84]. In conclusion, EBOV survivors maintain long-term potent adaptive immunity to EBOV and therefore vaccinating these individuals during convalescence is unlikely to have any protection against recrudescence. Instead, ring vaccination may prove a better vaccination strategy, a regime already showing success [87]. Vaccinating the community around male survivors, especially current and potential future sexual partners, may reduce the risk of transmission into the population, hence avoiding flare-ups of EBOV.
There are emerging innovative solutions to mitigate the risk of transmission. For example, seminal amyloid fibrils are a novel target that facilitate EBOV sexual transmission [88]. The repurposed Parkinson’s drug tolcapone has shown to potently inhibit fibril formation and block EBOV-pseudovirus entry in vitro, therefore could be used for topical vaginal administration to prevent sexual transmission [89].
Understanding the long term viral shedding potential of male EVD survivors has already impacted on public health response and policy. When the 2016 EVD flare up in Gueckedou was linked to a male survivor who had recovered 500 days previously, WHO and Guinean authorities initiated a program to immunise potential future contacts surrounding registered EVD survivors [90]. This action may have been linked to a lack of diagnosed sexual transmission flare ups over the next five years. It is accepted that there is a clear need to protect the future sexual partners of EVD survivors. Clinical research has also been actioned to assess the efficacy of antiviral treatment of infected men and the impact on subsequent EBOV RNA shedding in semen. An obvious policy implication for male survivors is the use of condoms to protect sexual partners but this is not always practical to implement in the regions that EVD is prevalent. Furthermore, such policies need to be mindful of exacerbating stigmatism of EVD survivors.
Guidelines, vaccinations and topical prophylactics do not however address the root cause of the persistence, the underlying replication mechanism. Instead, broad-spectrum small molecule antivirals hold promise. The nucleoside analogue remdesivir has shown to penetrate IPNs in NHPs [91] and a small phase 2 clinical trial suggests it reduces the incidence of EBOV RNA detection in the semen within 6 months [92]. However, the small sample size combined with high baseline negativity rates due to delayed recruiting resulted in low statistical power of the investigation. Favipiravir, another nucleoside analogue, is yet to show for efficacy treating acute EVD or for clearing persistence [21, 93, 94]. Higher doses were explored in a subsequent dose-escalation phase 2 trial in two male survivors, showing safety but inconclusive for efficacy due to lack of recruitment [95].
Larger scale trials are required in future filovirus outbreaks, especially a phase 3 trial for remdesivir. Higher dosage may be required than previously trialled to overcome the low levels of viral replication and poor biodistribution to IPNs. For instance, the phase 2 remdesivir trial regime involved 100 mg once daily for five days whereas when used to treat COVID-19 an additional 200 mg loading dose is given and in immunocompromised patients treatment can last up to 10 days [92, 96]. The renal and hepatic damage caused by EVD adds to the complexity of dosing and so further study of the poorly understood pharmacokinetics of these drugs in relation to IPNs is key. Overall, the challenges of developing these antivirals highlight the importance of understanding viral replication during persistence to inform repurposing of broad-spectrum antivirals and to develop specific persistence-targeting therapeutics.
During this EBOV-free period, research is shifting away from clinical studies towards use of animal studies and in vitro models. Human tissue studies of persistent reservoirs combined with modern omics technologies could provide a valuable approach to uncover the molecular mechanisms underpinning testicular persistence. Such models could enable the screening of small molecule antivirals at a scale currently not possible and would be particularly useful during this inter-epidemic period while there are too few survivors to perform clinical trials.
The absence of reports of male-to-male EBOV sexual transmission is worrying. Emphasis on strict confidentiality and impartiality is vital in contract tracing. Animal challenge studies comparing rectal vs. vaginal EBOV exposure could prove useful in determining if anal sex has a higher risk of transmission in the same way HIV behaves. Although the limited evidence has thus far indicated the low risk of persistence and shedding from the female reproductive tract, further longitudinal studies of EBOV shedding into vaginal secretions are important. Additionally, further molecular epidemiological investigations into the origin of transmission events will help uncover the likelihood of female-to-male sexual transmission.
Develop an in vitro testicular EBOV persistence model to:
investigate the molecular persistence mechanism;
screen small molecule antivirals.
Standardise protocols and expand future longitudinal clinical sampling studies including:
four consecutive negative samples before patient dropout;
a standardised PCR Ct threshold;
investigate association of persistence with post-Ebola sequelae.
Phase 3 remdesivir trial (and repeat phase 2 favipiravir) in future EBOV outbreaks to clear seminal fluid EBOV shedding.
Investigate male-to-male sexual transmission:
animal EBOV challenge model to assess susceptibility of anal vs. vaginal mucosa;
ensure utmost confidentiality and impartiality during contact tracing.
Longitudinal clinical studies in EVD survivors who receive vaccination or monoclonal antibodies to assess efficacy of these interventions for preventing persistence.
Continued evidence of the efficacy of ring vaccination around male survivors [90].
The testicular EBOV reservoir holds the greatest threat for human-to-human transmission and the sparking of new outbreaks. Indeed, this reservoir is already responsible for a flare-up 5 years after an initial outbreak was declared over. Maternal transmission via breastfeeding is additionally concerning. Particularly the brain, but also the eyes, are likely further sites of persistence. Although these IPNs pose less of a risk of EBOV transmission due to the contained nature of the organs, they are associated with EVD recrudescence and post-Ebola sequelae. The incidence of recrudescence may increase in future outbreaks with the distribution of life-saving EBOV vaccinations and monoclonals. To address this concern long term follow-up of survivors is key and will also help elude the connection between persistence and post-Ebola sequalae, revealing the complete long-term burden of EBOV persistence in the survivor population.
Longitudinal semen studies have provided valuable indications for the length of persistence; however, they do likely provide an underestimate. The meta-analysis presented in this review aims to address some of the variability between studies but highlights small sample sizes increase the uncertainty of predictions. Based on our estimates, a median of 7 (95% CI 0–43) male survivors from a cohort of 5,000 will be shedding EBOV in semen 5 years following infection. Although long-term persistence is uncommon, it only takes a single case to start an entirely new outbreak, emphasising the importance of long-term follow-up of survivors. Identification of the molecular mechanisms sustaining the persistent reservoirs, facilitated by in vitro models, will help inform the use of broad-spectrum antivirals to clear these sites whilst opening the door to create novel reservoir-specific antivirals. Success of such therapeutics is the ultimate goal, reducing both the risk of future outbreaks and reducing stigmatisation facing survivors in communities.
EBOV and MARV are just two of a multitude of viruses that can establish persistence, therefore benefit from understanding of this phenomenon is not limited to the filovirus field. Filovirus persistence represents a paradigm shift regarding the origin of outbreaks. However, the recent 2023 and 2024 MARV outbreaks [97, 98] and 2025 Sudan virus outbreak (belonging to the same family as EBOV) [99] illustrate this human reservoir should not overshadow the threat imposed by the vast zoonotic reservoir. In a world of increasing human-animal conflicts a One Health approach must be taken. Mitigation of spillover events and initial containment of filovirus outbreaks must maintain priorities and in fact would prevent the establishment of human persistence in the first place.
CI: confidence interval
CMV: cytomegalovirus
Ct: cycle threshold
DIGs: defective interfering genomes
EBOV: Ebola virus
EVD: Ebola virus disease
IPNs: immune privileged niches
MA-EBOV: mouse-adapted EBOV
MARV: Marburg virus
MSM: men-who-have-sex-with-men
NHP: non-human primate
WHO: World Health Organisation
OM: Conceptualization, Investigation, Writing—original draft. KF: Investigation, Writing—review & editing. MWC: Conceptualization, Investigation, Writing—review & editing.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Data used to create Figure 2 can be accessed in within references 21, 24, and 29.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Phey Liana ... Tungki Pratama Umar
Kamran Zaman ... Ranjit Sah
Wael Abu Ruqa ... Antonio Minni
Anna Antipov, Nikolai Petrovsky
Hend Radwan ... Mohamed El-Kassas
