Affiliation:
1Novosibirsk State University, 630090 Novosibirsk, Russia
2Biolink Ltd, 630090 Novosibirsk, Russia
Affiliation:
3Federal Research and Clinical Center of Specialized Medical Care and Medical Technologies, Federal Medical and Biological Agency of Russia, 115682 Moscow, Russia
Affiliation:
3Federal Research and Clinical Center of Specialized Medical Care and Medical Technologies, Federal Medical and Biological Agency of Russia, 115682 Moscow, Russia
Affiliation:
3Federal Research and Clinical Center of Specialized Medical Care and Medical Technologies, Federal Medical and Biological Agency of Russia, 115682 Moscow, Russia
4Federal Center of Brain Research and Neurotechnologies, Federal Medical-Biological Agency of Russian Federation, 117513 Moscow, Russia
5Pulmonology Research Institute, Federal Medical and Biological Agency of Russia, 115682 Moscow, Russia
6Department of Medical Nanobiotechnology, Medical and Biological Faculty, Pirogov Russian National Research Medical University, 117997 Moscow, Russia
7Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, 119991 Moscow, Russia
Affiliation:
1Novosibirsk State University, 630090 Novosibirsk, Russia
2Biolink Ltd, 630090 Novosibirsk, Russia
Email: petelaktionov@gmail.com
ORCID: https://orcid.org/0000-0003-2174-6496
Affiliation:
1Novosibirsk State University, 630090 Novosibirsk, Russia
2Biolink Ltd, 630090 Novosibirsk, Russia
Email: sp_kovalenko@yahoo.com
ORCID: https://orcid.org/0000-0002-6903-4239
Explor Med. 2025;6:1001322 DOI: https://doi.org/10.37349/emed.2025.1001322
Received: December 25, 2024 Accepted: April 29, 2025 Published: May 21, 2025
Academic Editor: Apostolos Zaravinos, European University Cyprus, Cyprus
The article belongs to the special issue Molecular Diagnostics in Oncology
Aim: Analysis of circulating free DNA (cfDNA) is now broadly used to diagnose, assess treatment response, and recurrence of various tumor types. Detection of aberrant cfDNA methylation in plasma is considered as one of the promising approaches for early-stage cancer detection, giving rise to new diagnostic tools. Colorectal cancer (CRC) has been one of the first malignancies for which relatively reliable diagnostic markers based on methylation analysis have been developed. Here, we aimed to assess the performance of SDC2 and SEPT9 promoter methylation in circulated plasma DNA as potential markers of colorectal precancerous lesions and carcinomas.
Methods: Plasma samples were collected from donors with unknown cancer status and various anamnesis prior to colonoscopy. Methylation of SDC2 and SEPT9 genes promoters was blindly analyzed by multiplex methylation‑specific PCR using Real-time-PCR-Sept9-SDC2-Met test (BioLink, Russia). Sensitivity, specificity, and AUC of SDC2 and SEPT9 tests were calculated for all groups of cancer and precancerous lesions.
Results: Among 253 patients, 18 were diagnosed with CRC, 14 with advanced adenomas, and 17 with sessile serrated lesions according to the results of colonoscopy examination with subsequent biopsy. The plasma cell-free DNA test detected all cases of CRC, 11 out of 14 cases of advanced adenoma, and 10 out of 17 cases of sessile serrated lesions. The specificity of the SDC2 marker was 91.2% and 97.6% of SEPT9 marker.
Conclusions: A minimal-invasive plasma test that detects methylated SDC2 and SEPT9 genes promoters might be considered as a screening method for detecting CRC and pre-cancerous lesions.
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths worldwide, after respiratory system cancers, in both sexes, with around 2 million new cases diagnosed every year globally [1]. The incidence rates of CRC are three to four times higher in high Human Development Index (HDI) countries compared to low HDI ones, whereas CRC-related mortality rates are substantially lower in the former [1]. These effects might be attributed to the availability of appropriate therapy, diagnostics, and population screening programs, with the latter considered a valuable factor in reducing CRC mortality rates [2, 3].
Currently, there are two main CRC screening approaches: instrumental diagnostics, such as colonoscopy, and in vitro laboratory tests, namely the fecal immunochemical test (FIT), fecal occult blood test (FOBT), or liquid biopsy, which involves the detection of circulating free DNA (cfDNA) and other tumor-derived biomarkers in body fluid samples. Colonoscopy is considered the gold standard for CRC screening, demonstrating superior sensitivity and specificity in detecting precancerous and cancerous lesions [3]. However, the drawbacks of the procedure include its invasiveness, uncomfortable preparation, relatively high cost, and logistical difficulties, which lead to moderate acceptance among patients [4, 5]. Fecal laboratory tests are non-invasive and widely available due to their low cost and the relative ease of laboratory analysis. However, the accuracy of these tests is relatively low, with CRC sensitivity of 50% and 73%, and specificity of 77.9% and 89% for FOBT and FIT, respectively [6, 7]. Fecal biochemical tests also demonstrate low sensitivity for precancerous lesions and reduced sensitivity for right-sided colorectal neoplasia [8–10]. Moreover, there is conflicting data regarding the acceptance of fecal tests compared to colonoscopy, suggesting that it may vary greatly depending on socioeconomic and cultural factors [3, 4]. Blood DNA tests might be considered an alternative to fecal tests, as they demonstrate comparable or superior diagnostic accuracy for CRC and precancerous lesions, along with a higher patient acceptance rate [11, 12]. Available DNA tests for CRC diagnostics are based on the analysis of tumor DNA in feces or cfDNA in blood plasma, allowing for the detection of genetic and epigenetic alterations associated with carcinogenesis. A key component of epigenetic regulatory mechanisms—genomic DNA methylation—is dysregulated in cancer, which can be used for diagnostics [13]. For example, aberrant methylation of the SEPT9 gene promoter is one of the first CRC markers to be approved by the FDA in the Epi ProColon kit [14]. The SEPT9 proteins complex interacts with cellular proteins, like microtubules or actin microfilament, and is involved in the cell cycle regulation and autophagy [15]. Methylation of this gene inhibits its expression and promotes CRC development [15]. Another FDA-approved DNA-based test is Cologuard, which analyzes stool DNA for multiple targets, including KRAS mutations and the methylation status of the NDRG4 and BMP3 genes [16]. In a clinical trial involving 9,989 participants, Cologuard demonstrated high sensitivity and specificity in detecting CRC [16]. In 2024, two new options for CRC screening were approved by the FDA: a new version of Cologuard, which includes three methylation marker genes (LASS4, LRRC4, and PPP2R5C) [17] and the Shield next generation sequencing panel, which assesses aberrant methylation status, patterns of DNA fragmentation and cfDNA genomic alterations [18]. Additionally, tests based on digital droplet PCR (ddPCR), such as Trimeth, have emerged as promising tools for CRC screening [19], offering high sensitivity for detecting methylated DNA markers in stool and blood samples. Although these tests surpass traditional FIT in terms of diagnostic accuracy, their availability is limited, and their cost is considerably higher [10, 20].
It is important to note that, all else being equal, real-time PCR-based tests are significantly more affordable than next-generation sequencing (NGS) technologies. Furthermore, the wealth of knowledge about potential methylation markers encourages researchers to develop new diagnostic systems, including those aimed at identifying precancerous lesions. There are many methylation-based biomarkers and their combinations described in the literature [21]. Considering blood-based studies, the promising markers include SFRP1 and SFRP2, which showed CRC sensitivity of 85.1% and 72.3% and adenoma sensitivity of 89.2% and 83.8%, respectively [22]. Other candidates include ALX4 [23, 24], TMEFF2 [24, 25], and SDC2 [26], whose methylation status has been demonstrated in CRC tissue and blood samples. The SDC2 protein is a cell surface proteoglycan that serves as a receptor for extracellular matrix components [27]. Promotor methylation of SDC2 seems to enhance its expression, since it is significantly higher in CRC tissues and is linked to proliferation, migration, invasion, and induces in colon cancer cells [28, 29]. Additionally, it has been shown that the SDC2 methylation marker is sensitive to precancerous lesions [30], which makes it an excellent candidate for an early CRC diagnostic test.
In this study, we evaluated the effectiveness of Real-time-PCR-Sept9-SDC2-Met test for detecting colorectal neoplasms in a single-center trial with 253 participants. Blood samples were obtained prior to colonoscopy, and clinical data, including the results of the colonoscopy, were collected. We evaluated the performance of the test for various types of colorectal lesions, particularly for precancerous lesions, as well as other bowel disorders.
Real-time-PCR-Sept9-SDC2-Met test (Biolink, Russia) consists of three modules: Plasma DNA Extraction module (cat. #31701, Biolink, Russia), DNA Bisulfite Conversion module (cat. #22002, Biolink, Russia), and real-time PCR module (cat. #22011, Biolink, Russia). The kit contains positive and negative samples, ACTB gene was used as a reference for DNA input.
Blood samples (27 mL) were collected prior to colonoscopy, immediately cooled to 4℃, and processed to obtain approximately 15 mL of plasma from each patient by centrifugation at 3,000 g for 15 minutes within four hours after blood collection. The plasma samples were aliquoted and stored at −80℃ for further analysis. All plasma samples were analyzed in a blinded manner. cfDNA was extracted from 5 mL of plasma using the Plasma DNA Extraction Kit (cat. #31701, Biolink, Russia). Bisulfite treatment of all extracted cfDNA was carried out using the DNA Bisulfite Conversion Kit, and cfDNA was eluted in 50 μL of elution buffer (cat. #22002, Biolink, Russia).
The Real-time-PCR-Sept9-SDC2-Met (cat. #22011, Biolink, Russia) kit was used to detect the methylation of the SDC2 and SEPT9 genes in plasma samples. Real-time PCR was performed on a DTprime 5M1 (DNA-Technology, Russia) PCR detection system according to the instructions for the Real-time-PCR-Sept9-SDC2-Met kit. The reaction was performed in three technical replicates for each sample, and positive and negative control samples were processed in the same manner in each experimental setup. All valid replicates must meet the requirement of a quantification cycle (Cq) value of ACTB ≤ 31. According to the kit instruction, mean Cq cut-off values were taken as Cq = 37.1 for SDC2 and Cq = 38 for SEPT9 analysis. When combining two markers in one test, the result was considered positive if mean methylation Cq ≤ cut-off value for at least one gene. All negative samples without Cq values were assigned a value of 43 to enable a quantitative comparison of methylation levels between CRC, advanced adenoma (AA), sessile serrated lesions (SSL), and normal control groups.
The results of the colonoscopy and pathomorphological neoplasm examination were used to classify the participants and stage the CRC. In cases with multiple colorectal lesions, the most advanced was used for classification. Thus, four groups of patients were defined for the statistical analysis: (1) patients with negative findings on colonoscopy; (2) patients with a CRC diagnosis; (3) patients with AAs; (4) patients with SSL. Patients free of neoplasms according to the colonoscopy were considered negative/healthy in the statistical analysis. Receiver operation curve (ROC) analysis was performed to characterize diagnostic performance. In addition, a Mann-Whitney U test was conducted to compare quantitative methylation levels between CRC, AAs, SSL, and normal cases. The Pearson Chi-square test was employed to compare the qualitative methylation levels and the clinicopathological features of patients. Statistical tests were performed in GraphPad Prism version 10.4.0 software (GraphPad, Boston, MA, USA).
A total of 253 participants were enrolled in the current study. Demographic data, along with colonoscopy and biopsy results, are presented in Table 1. Among the participants, 127 had colorectal neoplasms, while 126 were part of the control group. The median age of participants in each group was 65 years (range: 41–87) for the CRC group, 59 years (range: 26–85) for patients with non-malignant lesions, and 55 years (range: 28–78) for the control group. The male-to-female ratio was 1.25, 0.6, and 0.34, respectively. Neoplasms were evenly distributed between the proximal and distal colon, with 15 patients having neoplasms in both segments.
Patients clinicopathological features
| Features | Tumor | Non-malignant lesions | Normal colon |
|---|---|---|---|
| Median age (years) | 65 | 59 | 55 |
| Gender | |||
| Male | 10 | 41 | 32 |
| Female | 8 | 68 | 94 |
| Location | |||
| Proximal colon | 8 | 63 | |
| Distal colon | 10 | 60 | |
| Tumor stage | |||
| I | 3 | ||
| II + III | 14 | ||
| IV | 1 | ||
| Histological features | |||
| Advanced adenoma* | 14 | ||
| Adenoma | 58 | ||
| Serrated lesions | 17 | ||
| Polyps | 20 | ||
* High-grade dysplasia, ≥ 10 mm and or substantial villous component
We tested 253 plasma samples for methylation of the SDC2 and SEPT9 markers, with 69 samples testing positive for SDC2 methylation and 30 samples testing positive for SEPT9 methylation. To evaluate the effectiveness of these markers, we analyzed the test results across four groups: CRC, AAs, SSL, and healthy controls.
The median Cq values for the SDC2 methylation test were 30.85 for CRC, 32.05 for AAs, 32.50 for SSL, and 43.00 for healthy control samples (Figure 1A). For the SEPT9 methylation test, the median Cq values were 34.7, 43.0, and 43.0, respectively, for CRC, non-malignant neoplasms, and healthy control samples (Figure 1B). These findings suggest that the SDC2 methylation marker was able to distinguish CRC, AAs, and SSL from healthy controls, while the SEPT9 marker was more specific for distinguishing CRC from normal samples. We also analyzed the difference between the Cq values of the reference gene (ACTB) and the target methylation markers (Figure S1). The comparison of ΔCq values confirms the trends observed in the absolute Cq analysis described above.
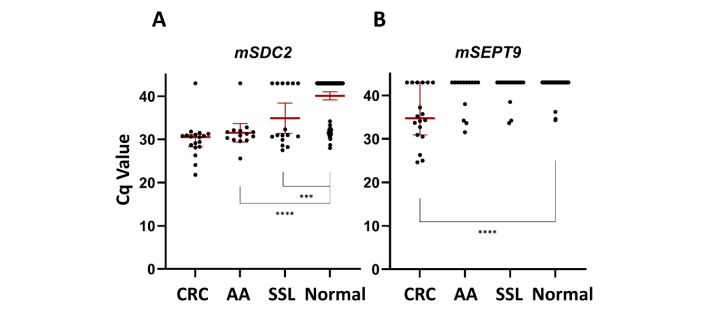
Cq values of CRC samples, advanced adenomas samples, serrated sessile lesion samples, and 126 normal plasma samples. (A) The median Cq values for the mSDC2 test; (B) the median Cq values for the mSEPT9 test. *** indicates P-value < 1 × 10–3 (Mann-Whitney U test); **** indicates P-value < 1 × 10–4 (Mann-Whitney U test). CRC: colorectal cancer; AA: advanced adenoma; SSL: sessile serrated lesions
We then examined whether there was any correlation between positive test results for each marker and other clinical characteristics. Additionally, we assessed the combined use of both markers. Further analysis revealed no significant correlation between positive test results and lesion location (P-value = 0.394), sex (P-value = 0.485), or age of the patients (P-value > 0.05, Chi-square test) (Table 2).
Clinicopathologic characteristics of patients with positive cfDNA plasma test results by methylation status of SDC2 and SEPT9 markers
| Variable | All | Combined test positive (n = 78) | No. of mSDC2 positive (n = 69) | No. of mSEPT9 positive (n = 30) |
|---|---|---|---|---|
| Age (years) | ||||
| ≤ 49 | 108 | 19 (24.4%) | 18 (26.1%) | 9 (30.0%) |
| 50–59 | 60 | 20 (25.6%) | 16 (23.2%) | 7 (23.3%) |
| 60–69 | 60 | 25 (32.1%) | 23 (33.3%) | 7 (23.3%) |
| ≥ 70 | 25 | 14 (17.9%) | 12 (17.4%) | 7 (23.3%) |
| Gender | ||||
| Male | 83 | 28 (35.9%) | 25 (36.2%) | 13 (43.3%) |
| Female | 170 | 50 (64.1%) | 44 (63.8%) | 17 (56.7%) |
| Location | ||||
| Proximal colon | 70 | 33 (47.1%) | 28 (45.2%) | 18 (66.7%) |
| Distal colon | 68 | 37 (52.9%) | 34 (54.8%) | 9 (33.3%) |
| CRC stage | ||||
| I + II | 8 | 8 | 8 | 6 |
| III + IV | 10 | 10 | 9 | 6 |
| Histological classification | ||||
| Advanced adenoma | 14 | 11 | 9 | 5 |
| Serrated lesions | 17 | 10 | 10 | 4 |
| Non-advanced adenoma | 57 | 19 | 17 | 4 |
| Hyperplastic polyps | 19 | 8 | 5 | 3 |
To evaluate the diagnostic performance of the SDC2/SEPT9 methylation test in circulating tumor DNA in plasma, an ROC curve was constructed using all Cq values for single markers (Figures 2A and 2B) and cut-off values for the combined biomarkers test (Figure 2C). The methylated SDC2 test in plasma DNA had a CRC sensitivity of 94.4% with an AUC of 0.94 (Table S1). When CRC and AAs were consolidated into one group, the mSDC2 test detected 81.12% of advanced colorectal lesions. The sensitivity of AA and SSL detection was 64.3% and 58.8%, respectively. Specificity of SDC2 methylation test alone was 91.2%.
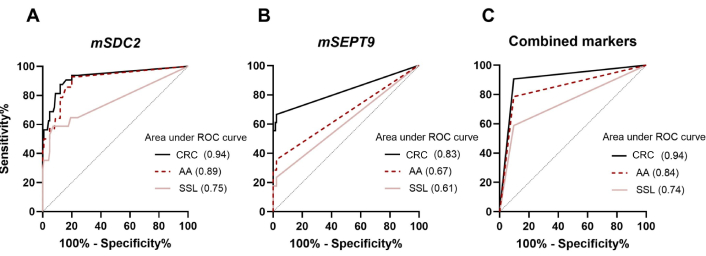
ROC curves for CRC, AA, and SSL detection. (A) ROC of SDC2 methylation status biomarker; (B) ROC of SEPT9 methylation status biomarker; (C) ROC of combination of two biomarkers for colorectal neoplasia detection. ROC: receiver operation curve; CRC: colorectal cancer; AA: advanced adenoma; SSL: sessile serrated lesions
The mSEPT9 marker alone showed higher specificity—97.6% for CRC, but sensitivities were 66.6%, 35.7%, and 23.3% for advanced neoplasia, AA, and SSL, respectively. Combining the two markers improved the diagnostic characteristics for colorectal neoplasms detection. Thus, the Real-time-PCR-Sept9-SDC2-Met test detected 18/18 CRC samples, 11/14 AA samples, and 10/17 SSL samples with a specificity of 90.5%.
Based on the results of colonoscopies, some patients were diagnosed with gastrointestinal diseases (GIDs), including inflammatory conditions such as colitis, ileitis, diverticulitis, and bowel adhesions (Table 3).
Results of the cfDNA plasma test on gastrointestinal diseases
| Gastro-intestinal disease | All | SDC2 positive | SEPT9 positive | Combined test positive |
|---|---|---|---|---|
| Colitis | 21 | 3 | 1 | 2 |
| Ileitis | 16 | 2 | 0 | 0 |
| Diverticulitis | 6 | 2 | 1 | 3 |
| Bowel adhesions | 2 | 1 | 0 | 0 |
To explore whether intestinal conditions influenced the test results, we divided the group of patients without neoplasia into two subgroups based on their GIDs status. Out of 82 samples from the first subgroup, 7 (9%) were positive for the combined methylation markers test. In the second subgroup, consisting of patients diagnosed with GIDs, 5 (11%) samples showed a positive result for the test. Thus, no statistically significant difference was found in methylation status between the two groups (P-value = 0.28, Chi-square test).
Interestingly, our data suggest that diverticulitis may impact methylation test results, as half of the 6 cases in this group tested positive (P-value = 0.001, Chi-square test). However, given the small sample size, this preliminary finding warrants further investigation with a larger cohort of patients with diverticulitis before drawing any definitive conclusions.
Early detection of CRC through screening is associated with a more favorable prognosis, with a 5-year survival rate as high as 90% when the malignancy is detected at the localized stage [31]. Therefore, it is essential to develop a test that can accurately detect early-stage CRC and precancerous lesions. In this study, we investigated the performance of the Real-time-PCR-Sept9-SDC2-Met test based on two methylation markers—SEPT9 and SDC2 on 253 plasma samples. We focused on precancerous lesions and, therefore, included a small group of CRC samples as a positive control group.
SEPT9 has long been considered a potential marker for CRC, and numerous studies have assessed its diagnostic potential [14, 23, 32, 33]. In general, the sensitivity of methylated SEPT9 to CRC ranged from 48.2% to 95.6%, and the specificity ranged from 79.1% to 99.1% [14, 34–37]. The sensitivity and specificity of mSEPT9 may be influenced by the algorithm of the test [37], biospecimen [38], the number of recruited patients, and interfering diseases, such as diabetes, arthritis, and arteriosclerosis [35]. Furthermore, the sensitivity of the assay may also depend on the design of oligonucleotides, as is the case in our study, where we prioritized high specificity at the expense of the sensitivity of mSEPT9 test.
SDC2, though a later addition as a methylation marker, has already been used in commercial tests, such as Colosafe (Creative Biosciences CO., Ltd) [39]. The ColoSafe trial showed a sensitivity of 83.8% and a specificity of 98% in a sample of 1,110 stool samples [39]. One of the first studies on SDC2 as a marker for CRC was conducted by Oh et al. [26], researchers performed a search and validation of CRC markers using genome-wide sequencing of tumors and healthy tissues. When tested on blood samples, SDC2 demonstrated sensitivity and specificity of 87% and 95.2%, respectively [26]. These findings were then supported by the study by Barták et al. [22], which showed SDC2 sensitivity of 89.4% for CRC and 81.1% for adenomas, with a specificity of 97.3%. In the current study, overall sensitivity and specificity to CRC were 100% and 90.5%, respectively. The sensitivity of SEPT9 and SDC2 individually was 66.7% and 94.4%, while the specificity of SEPT9 (97.6%) was higher than that of SDC2 (91.2%).
In CRC diagnostics, it is important to identify patients with precancerous lesions as well. AAs have a high risk of becoming malignant [40], so detecting and removing them can reduce the incidence of CRC [41]. The inclusion of AA in the category of true positives when calculating diagnostic accuracy, along with cancer, has been discussed [41]. In our study, the sensitivity of the test for AA was 78.6%. The SDC2 methylation test, which plays a major role in detecting AA, had a noticeably higher sensitivity (64%) than methylated SEPT9 test (35%), consistent with other studies on the methylation status in AA [22, 39, 42]. When combining AA cases with CRC into progressive colorectal neoplastic group, sensitivity and specificity were 90.6% and 90.4%, respectively, with an AUC of 0.9. SDC2 has high diagnostic potential for detecting early stages of CRC and AAs, as demonstrated in the study of Oh et al. [26] and also in the study of Barták et al. [22]. Our results are in agreement with the SDC2 study of Han et al. [43] on stool samples, in which CRC was detected in 90.2% of cases, AAs in 66.7%, and adenomas in 24.4%, although the sample size for adenomas was relatively small. The SpecColon test showed relatively low sensitivity of SDC2 alone—33.3%, 56.6% for AA and CRC detection, respectively [44]. However, this may be attributed to a small amount of cfDNA (from 1 mL of plasma), as concentrations of cfDNA in plasma have been reported to correlate with tumor burden [45]. So, for detection of early stages of cancer and adenomas, a larger volume of plasma might be needed. According to the investigation, SDC2 methylation seems to be a promising marker for detection of advanced colorectal lesions.
SSL are colorectal neoplasms, which we have also highlighted in our research. Several studies have shown that these lesions differ in their morphology, as well as in their molecular pathway leading to CRC [46]. They also have a less favorable prognosis compared to non-serrated adenocarcinomas [47, 48]. Sensitivity of SSL detection in our study was lower, accounting for 58.8%. SEPT9 methylation marker did not affect the sensitivity to SSL. Additionally, our test identified 35.5% of non-progressive adenomas and hyperplastic polyps. The localization of neoplasms did not affect the detection frequency, either in the case of SDC2 or in the case of SEPT9, which is consistent with other studies [34, 39].
We also investigated the effects of other intestinal diseases detected during colonoscopy. We found no noticeable effect of the test on inflammatory diseases such as colitis and ileitis. Further research is required to establish a precise link between the methylation of the SDC2 and SEPT9 genes and inflammatory bowel diseases. It is difficult to thoroughly assess the potential association of GIDs with methylation status according to the literature, as they are often used as a criterion for excluding samples from analysis [32, 42, 44, 49, 50]. However, some previous studies suggest that GIDs do not affect the results of SEPT9 methylation test: in the study by Wu et al. [36] 108 patients with GIDs were recruited for mSEPT9 analyses and only 3.7% of samples showed positive results, Fu et al. [51] also showed no significant differences between normal group and 30 GID samples regarding SEPT9 methylation. The study by Wang et al. [39] regarding methylated SDC2 includes 20 GID samples of which only 1 had a positive result. Thus, additional studies are needed to establish an accurate link between the methylation of promoters of the SDC2 and SEPT9 genes and GIDs.
This combination of biomarkers in one real-time PCR test is not novel. The ColoDefence assay is based on this approach and has demonstrated similar overall performance in trials—66.7% sensitivity for AAs, 92.3% for CRC, and 93.2% specificity [42]. However, the contribution of SDC2 to sensitivity in this case is less than that of SEPT9. This can be due to the fact that the test material used is stool rather than plasma samples. Here, we focused on the development of plasma-based test, because another main objective of our study was to ensure a high acceptance rate [12].
In conclusion, our clinical trial demonstrated the promising potential of the dual-targeted cfDNA test for CRC screening. However, our study has certain limitations. First, only a small percentage (12.6%) of patients had advanced colorectal lesions. Second, the cfDNA test was not directly compared with FIT, another routine CRC screening method based on stool analysis, which limits our ability to draw conclusions about its superiority. Thirdly, although we mentioned the impact of various interfering diseases on the cfDNA test results, none of these diseases were tested in detail to draw a statistically significant conclusion.
AA: advanced adenoma
cfDNA: circulating free DNA
Cq: quantification cycle
CRC: colorectal cancer
FIT: fecal immunochemical test
FOBT: fecal occult blood test
GIDs: gastrointestinal diseases
HDI: Human Development Index
ROC: receiver operation curve
SSL: sessile serrated lesions
The supplementary figure for this article is available at: https://www.explorationpub.com/uploads/Article/file/1001322_sup_1.pdf. The supplementary table for this article is available at: https://www.explorationpub.com/uploads/Article/file/1001322_sup_2.xlsx.
V Borobova: Formal analysis, Investigation, Software, Visualization, Writing—original draft. AA: Data curation, Resources. DS: Data curation, Methodology. V Baklaushev: Resourses. DI: Resources, Investigation. EV: Investigation, Data curation. AV: Methodology. NO: Data curation. PL: Methodology, Project administration, Supervision, Writing—review & editing. SK: Conceptualization, Validation. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
This study was approved by the ethics committee of the Federal Research and Clinical Center of Specialized Medical Care and Medical Technologies, Federal Medical and Biological Agency of Russia, Moscow (Protocol No. 5_2024, 04.06.2024). Biopsy material was obtained in compliance with the legislation of the Russian Federation, and written informed consent was provided by all the patients.
Informed consent to participate in the study was obtained from all participants.
Informed consent to publication was obtained from relevant participants.
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Tatiana L. Nekhaeva ... Irina A. Baldueva
Noor Mey Wardhani ... Bulkis Natsir
Evgeny Imyanitov, Anna Sokolenko
