Affiliation:
Nanobiointeractions and Nanodiagnostics, Istituto Italiano di Tecnologia, 16163 Genova, Italy
Email: giuseppe.bardi@iit.it
ORCID: https://orcid.org/0000-0001-9662-3007
Explor Immunol. 2022;2:637–647 DOI: https://doi.org/10.37349/ei.2022.00073
Received: May 19, 2022 Accepted: July 05, 2022 Published: August 31, 2022
Academic Editor: Dominique J. Charron, Hospital Saint Louis, France
Chemokines are homeostatic or inflammatory small proteins regulating immune cell migration and are structurally characterized by cysteine disulfide bridges. Around 50 human chemokines binding almost 20 seven-transmembrane G-protein coupled receptors have been discovered. The finding that two of them were the main human immunodeficiency virus (HIV) co-receptors intensified the research on the binding mechanism to block the viral entrance. Blockade of chemokine/chemokine receptor signaling ultimately modulates cell migration, then immune responses. Particular nanotechnologies can be designed to interfere with chemokine signaling or to exploit the ligand-receptor interaction. Surface chemical modification of nanomaterials with chemokines or specific peptides can find several applications in bio-medicine, from tissue-specific drug delivery to reduced cell migration in pathological conditions. Recent highlights on peculiar chemokine-nanoparticle design and their potential to modulate immune responses will be discussed.
The discovery of “Chemo-attractant cytokines” (chemokines) has been nicely reported by Baggiolini and colleagues [1], who first demonstrated the role of interleukin-8 [IL-8, now called Cys-aminoacid chemokine X-Cys ligand (CXCL8)]. Different chemokines and their cognate receptors have been found in several cell types [2], mainly regulating homeostatic and inflammatory leukocyte migration. These small proteins (roughly 10 KDa) have a peculiar structure characterized by disulfide bridges between cysteine residues. The presence or the absence of amino acids placed between the first two cysteines is used to classify the different chemokines, as well as their length, cellular expression, and physiologic or pathologic function [3]. Chemokine receptors belong to the 7-helix trans-membrane Gαi protein-coupled receptor (7TM GPCRs) family. The N-loop of the chemokine allows the interaction with the receptor extracellular domains [4, 5]. This contact is important to lay the chemokine onto the receptor allowing the N-terminus to properly switch-on the downstream signal transduction, as firstly demonstrated for CXCL12/Cys-aminoacid X-Cys chemokine receptor 4 (CXCR4) binding [6]. Integrin-dimer structural change into the active integrin-receptor binding arrangement is one of the first signals triggered by chemokines in leukocytes and facilitates their adhesion to the endothelial cell-surface [7, 8]. Chemokine-dependent integrin activation is fundamental to stopping monocyte or T lymphocyte rolling at the sites of inflammation [9, 10]. The role of chemokines in orchestrating the immune system is so crucial that different viruses also encode chemokine-analog peptides, receptor-analogs, and chemokine binding proteins to subvert immune responses [11–13].
In the last decade of the twentieth century, a sequence of discoveries highlighted the role of two chemokine receptors as primary human immunodeficiency virus (HIV) co-receptors required for cell infection, namely Cys-Cys chemokine receptor 5 (CCR5) for monocyte/macrophage-tropic HIV and CXCR4 for the lymphocyte tropic variant [14–19]. Importantly, these studies demonstrated that chemokines can antagonize the virus entry paving the way for the following research of molecules to block HIV binding and its membrane fusion. The mechanism of HIV binding to CCR5 and CXCR4 involves conformational changes of the viral envelope protein gp120 allowed by previous interaction with CD4 also expressed on the target cells: or CD4-independent interaction with CXCR4 by specific HIV-strains’ proteins [20, 21]. The discussion on intracellular signals triggered by CD4-independent HIV-gp120 interaction with chemokine receptors is still open. Some indications come from experiments with gp120 binding CXCR4 and CCR5 expressed on neurons [22, 23] leading to limited receptor signal transduction or pathological outcomes. Other molecules can also bind to chemokine receptors and antagonize chemokine interaction. For instance, peptides like [Tyr5,12, Lys7]-polyphemusin II (T22) [24] or small molecules, which are investigated for precision medicine therapies related to viral infection or cancer [25–27]. Chemokine-to-chemokine receptor binding is exploited in many ways in immunology and the development of nanotechnologies has increased these opportunities.
The present review will be discussed the interaction between chemokines and certain types of nanomaterial (NM), as well as their potential applications to remove the excess of chemokines by nanoparticles (NPs)-selective adsorption. Furthermore, the possibility of precisely targeting immune cell subsets by NP-surface functionalization with chemokines or other chemokine receptor-binding molecules will be considered.
The adsorption of proteins is a common event when NPs are released in biological media [28–30]. It depends on NP size, shape and surface chemistry and protein structure. Intrinsic properties of the materials can show preferential adsorption of certain molecules that may affect NP cellular targeting and uptake [31, 32]. In fact, NP-material ability to adsorb selected proteins with high efficiency can be exploited to functionalize NPs for specific delivery purposes, although the resulting corona may show unexpected implications [33, 34].
Some NMs seem to selectively bind chemokines with subsequent modulatory effects on the immune responses. Batt and colleagues [35] demonstrated by enzyme-linked immunosorbent assay (ELISA) and gel electrophoresis in vitro that TiO2 NPs with < 25 nm diameter have a surface affinity for CXCL8. These authors showed that CXCR8 interaction with TiO2 NPs was dose-dependent and this specificity was retained even if the chemokine was mixed with human serum proteins. Remarkably, TiO2 administration impaired the chemotaxis of neutrophils by efficient sequestration of the neutrophil-attracting CXCL8 from the medium. This information could be very important for the numerous applications of TiO2 NPs in biomedical fields [36]. Actually, various NMs surfaces may have an affinity for CXCL8. In a research paper regarding methods for extracorporeal detoxification from pro-inflammatory mediators, Zheng et al. [37] report graphene nanoplatelets (GNPs) as valid adsorbents to remove several proinflammatory cytokines including CXCL8. The idea of using materials that adsorb chemokines in excess during uncontrolled inflammatory reactions has been patented by Bruce and Lyngstadaas [38]. They designed a filter for extracorporeal fluids exploiting Group 4 and Group 5-metals’ ability to selectively bind CXCL10 [previously known as interferon gamma-induced protein 10 (IP-10)]. Presumably, the synthesis of such metal particles at a nanometric scale could improve the elimination of this chemokine from the fluids, due to an enormous increase in available adsorbent surface area. In principle, the NP surface could be chosen or modified “ad hoc” to remove the excess of inflammatory chemokine and cytokines when needed, for example during cytokine storms in sepsis or coronavirus disease 2019 (COVID-19) disease, as suggested in Pisani et al. [39].
The consequences of cytokine and chemokine binding to metal-oxide and carbon-NPs were already noted more than ten years ago in a study by Kocbach et al. [40] on “ultrafine particles” present in environmental pollution. The researchers showed significant adsorption of CXCL8 (IL-8) and other cytokines by carbonaceous “very small particles” when present at relevant concentrations. They also warned that testing the cellular release of inflammatory cytokines in the presence of these ultrafine particles could be misinterpreted because of the above-described binding properties of these materials. Moreover, the authors observed that chemokine or cytokine binding in vitro was reduced in a serum-supplemented medium, and suggested preferably preform chemokine/cytokines immunologic test in such complete culture medium to collect more precise results.
In line with these data, unpublished observations by our group suggest that as-synthesized SiO2 NP uptake by THP-1 monocytes is increased when particles are previously mixed with CXCL5. However, our results show that cellular internalization of SiO2 NP:CXCL5 is only slightly reduced when performed in serum-supplemented medium. Apparently, chemokines adsorbed onto the negatively charged particles could facilitate NP uptake through its cognate-receptor expressed on the target cell. Likely, NP brings close to the target high amount of exposed chemokine increasing its local concentration and the chances to hit the receptor. This hypothesis is reinforced by the following studies using chemokines covalently linked to NP surface (see below).
Positive-negative charge attraction between chemokine and NM surface may be one of the main events ruling the adsorption mechanism. The isoelectric point of many chemokines ranges between 9 and 11 [41]. At neutral pH, these proteins are positively charged increasing their affinity for negatively charged metal-oxide NPs. As a direct consequence, a key role to modulate the targeting is played by the chemical modification chosen to solubilize or functionalize the core NM.
As previously mentioned, NP-surface chemistry can be modified on purpose to bind specific chemokines with high affinity. A sophisticated functionalization of heparin-coated poly(lactic acid) (PLA)-NPs was performed by Guryanov and colleagues [42]. These investigators successfully exposed two chemokine receptor modified-fragments responsible for the chemokine binding, specifically, the N-terminal domain and second extracellular loop of CCR5. They demonstrated the functionalized-PLA-NP ability to eliminate Cys-Cys chemokine ligand [(CCL5), previously known as regulated upon activation, normal T cell expressed and secreted (RANTES)] from the culture medium abrogating monocyte adhesion to human endothelial cells. This work made two important steps ahead in chemokine sequestration research. Firstly, the use of biodegradable-polymer NPs with obvious biocompatibility and immunologic advantages [43] with concerning inorganic materials, such as metallic NPs with inflammatory consequences often underestimated by limited immunological characterization [44]. Especially, undegradable accumulation of NPs into phagocytic cells induces deleterious effects on the cells and the surrounding tissue [45–49]. In addition, CCR5 is one of the two main HIV co-receptors, which is fundamental for viral entry into monocytes [50]. From the discovery of CCR5-delta 32 mutation, which is a homozygous expression that can prevent human infection [51], this receptor has been the main target for pharmacological or other therapeutic approaches in the battle against HIV infection [52–54].
Although the demonstration of NP-dependent inflammatory chemokine sequestration in vivo is still a few and no protocols for NM designed for this task have been clinically approved, the observations reported above suggest that NMs modulate the action of chemokines and might improve the treatments for chemokine-mediated inflammatory diseases.
Nanotechnologies aimed at drug delivery have always pursued the highest possible precision to reduce the amount of drugs and avoid side effects. The main strategy concerns the modification of NP surface chemistry to facilitate specific targeting of receptors expressed only, or predominantly, in the needed cell subset. Differentiated immune cells express different chemokine receptor patterns and the same cell can differently upregulate or downregulate these receptors during its life cycle [55]. This diverse receptor expression can be used as an unambiguous cell address to deliver functionalized particles (Figure 1).
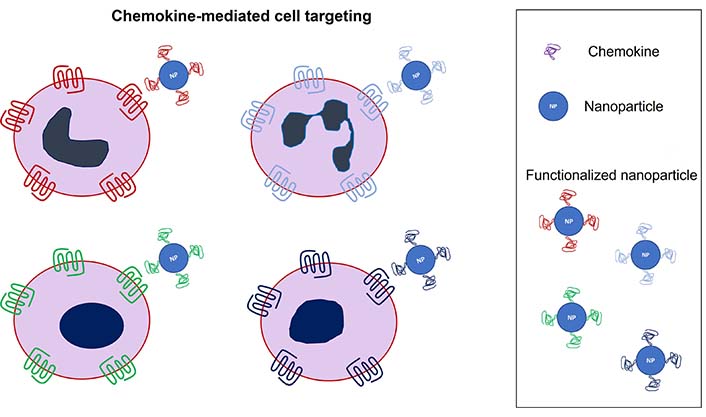
Schematic representation of cellular targeting exploiting selective chemokine-receptor binding
The exposure of receptor binding molecules onto particle surfaces is not so easy as it can be drawn. Different strategies include the functionalization with chemokine-derived peptides, small molecule antagonists, anti-receptor antibodies, entire chemokines, or the NP coating with chemokine receptor-expressing cell membranes. Indeed, the latter method covers the particle with several proteins expressed on the source cell, limiting the selection of the target receptors and will not be discussed here.
So far, the most exploited chemokine/chemokine receptor axis using nanotechnological approaches is CXCL12/CXCR4, which has a key role in several types of cancer and cancer metastasis [56]. Indeed, CXCR4 is overexpressed in different tumor cells. This receptor and its ligand CXCL12 [previously known as stromal cell-derived factor 1 (SDF-1)] have pleiotropic physiological roles in homeostasis and organogenesis, explaining why their dysregulation can facilitate tumorigenesis [57, 58].
Among the non-chemokine CXCR4-binding peptides, T22 is the most successful and it is frequently used to target diverse cancer cells over-expressing the receptor [59–68]. For instance, the release of doxorubicin in B cells from non-Hodgkin’s lymphoma has been enhanced by capping mesoporous SiO2 NPs with a derivative of the T22 peptide [60]. This 18-aminoacid peptide is known to be a potent CXCR4 antagonist, although higher doses induce the receptor internalization allowing intracellular NP delivery. Notably, selective T22-mediated CXCR4+ tumor cell targeting has been demonstrated in vivo in mouse models using protein based nanocarriers [66, 68]. Biochemical modifications of T22 or the mix with other peptides allow self-assembly in protein NPs able to be uptaken through CXCR4 and release toxins or antineoplastic drugs in different types of cancer cells [61, 65, 66, 69, 70]. Shorter peptides have also been used to drive polymer NPs to CXCR4+ cancer cells. The 5-aminoacid peptide LFC131 has been conjugated to poly(lactic-co-glycolic acid) (PLGA) or chitosan NPs efficiently releasing antitumor agents into CXCR4 overexpressing A549 lung cancer cells in vitro [71, 72] or hepatocellular carcinoma [73].
As far as the aminoacid sequence is part of the receptor binding domain (RBD, i.e. N-terminus [6]) of a chemokine, it is likely to induce antagonistic or agonistic effect on its target receptor. Diverse chemokine-derived peptides are currently under investigation and could represent a plethora of potential molecules with high specificity that can be used to decorate NPs for a broad range of biomedical applications. Either human or viral derived chemokine sequences have demonstrated efficient drug delivery properties [74–76].
Modified peptides targeting chemokine receptors are also gaining interest as key targeting moieties for in vivo diagnostic imaging [77], as demonstrated by NP surface linking of d-Ala-peptide T-amide (DAPTA) peptides [78–81]. Detering et al. [78] recently showed that polymer NPs functionalized with the CCR5 targeting peptide DAPTA can precisely detect CCR5 along the progression of atherosclerotic lesions.
Successful results for anticancer drug or small interfering RNA (siRNA) intracellular release delivery have also been obtained with the small-molecule CXCR4-antagonist, 1,1′-[1,4-phenylenebis(methylene)]bis-1,4,8,11-tetraazacyclotetradecane (AMD 3100), attached to PLGA NPs [82, 83]. This HIV-inhibiting molecule can mobilize hematopoietic stem cells with advantages for therapeutic approaches in transplantation [84] but some concerns for unexpected side effects due to its agonistic activity at high doses [85, 86]. Anti-CXCR4 antibodies have also been effectively used to target drug-loaded liposomes for CXCR4+-cancer cell cytotoxicity [87, 88].
From toxicological and immunological points of view, peptides could be preferred to small-molecule-CXCR4 antagonists or anti-CXCR4 antibodies as NP surface moieties for delivery purposes. Unbound molecules dispersed in the solvents or free antibodies released in the bloodstream can induce unexpected complement activation and the creation of immune-complexes with circulating antibodies. Particle synthesis and characterization are very important, especially its surface decoration with receptor ligands.
The creation of chemokine receptor-specific targeting NPs with reduced toxicity could be facilitated by using the entire chemokines to functionalize the NP-surface [89–91]. A chemical binding strategy to covalently link full-length chemokines has been made by our group [89] with CXCL5 onto SiO2 NPs and with CXCL12 on the surface of PLGA NPs by other researchers [91]. Although immunoblot transmission electron microscopy (TEM) images in both papers did not reveal many chemokines on the NPs, cognate receptor selectivity and functional assays clearly demonstrated targeting efficacy in vitro, even in serum enriched medium. In the work by Cagliani et al. [89], CXCL5-SiO2 NPs internalization was facilitated in CXCR2+ THP-1 but not into CXCR2– HeLa cells. Furthermore, pre-treatment with free CXCL5 significantly reduced only CXCL5-SiO2 NP cell uptake, whereas non-functionalized NP internalization was slightly increased, maybe due to the effect of CXCL5 adsorption onto the NP surface, as mentioned in the previous paragraph. Clear advantages of chemical decoration of PLGA NPs with CXCL12 were demonstrated in vitro and in vivo by Xiong and coworkers [91] using a combination of photothermal therapy and targeted chemotherapy to induce cytotoxicity in metastatic lymph nodes in tongue squamous cell carcinoma.
In a different attempt to improve chemokine-mediated specific cell targeting in our lab, we adopted non-covalent streptavidin-biotin bond to cover biodegradable PLGA/Pluronic NPs with CXCL12 biotinylated at the C-terminus [90]. Microfluidics-assisted NP preparation and accurate molarity of biochemical reactions allowed homogeneous size distribution of the particles and higher surface-bound chemokine amount. CXCL12-PLGA/Pluronic NPs confirmed cellular internalization in human THP-1 monocytes through CXCR4 endocytosis in the presence of serum. However, differences between fetal bovine serum (FBS)- and human serum-supplemented media emerged. This could be due to the exposure of cells to different biological milieus stimulating different expressions of CXCR4 on the THP-1 membrane. Crucially, chemokine-decorated NPs did not induce cell chemotaxis as the free CXCL12. Notwithstanding the forgoing, the pre-treatment of THP-1 cells with CXCL12-PLGA/Pluronic NPs abrogated CXCL12-dependent chemotaxis. This phenomenon was seen only in the presence of chemokine-NPs but not with non-decorated ones demonstrating the exclusive interaction with CXCR4 of the functionalized particles. The proved internalization without triggering the CXCR4 pathway to cytoskeleton remodeling for chemotaxis inspires the hypothesis that the complex chemokine-NP could activate biased signaling. On the other hand, the signaling could be delayed by the particle present in the endosomes during the observed time points. Indeed, recent observations with gold nanoparticles (AuNPs) reported that these particles phagocytized by immune cells can retard the sensitivity of some immune responses [92]. This is not true for all NPs, in fact, some NMs have been proved not only to release inflammatory mediators including chemokines, but also to stimulate the up-regulation of particular chemokine receptors involved in many processes of inflammation [93].
NP surface modification with chemokines is emerging as selective-targeting tool for biomedical applications such as drug delivery or imaging. However, immune system modulation can be likewise achieved by the release of chemokines embedded in polymer-NMs with reversible binding [94, 95]. This approach can be useful to keep a drug releasing particle in a defined anatomical place. The release of chemokines creates a local gradient aimed at attracting immune cells. Once the cells are close to the chemokine-releasing material, the receptors will be downregulated by the high chemokine concentration stopping cell migration. The choice of the chemokine to be released in a certain tissue is decisive for the final outcome of the application.
Chemokines and their receptors are fundamental for the regulation of the immune system and for many other cellular events. The possibility to use NM-interactions opens the way to several biomedical approaches. Decreasing overexpression of inflammatory chemokines by NP surface-mediated sequestration and driving selective cell targeting for imaging and delivery purposes represent the main fields of potential applications. Nevertheless, interdisciplinary collaborations among researchers will certainly find new ideas in the future to develop novel therapeutic treatments based on the current results and observations on chemokine-NM interactions.
CCR5: Cys-Cys chemokine receptor 5
CXCL8: Cys-aminoacid chemokine X-Cys ligand 8
CXCR4: Cys-aminoacid X-Cys chemokine receptor 4
HIV: human immunodeficiency virus
NM: nanomaterial
NP: nanoparticle
PLGA: poly(lactic-co-glycolic acid)
T22: [Tyr5,12, Lys7]-polyphemusin II
The author contributed solely to the paper.
The author declares that there are no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2022.
Copyright: © The Author(s) 2022. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
