Affiliation:
Health Sciences program, Federal University of Triângulo Mineiro, Uberaba 38025-440, Brazil
Email: murilo.porfirio@yahoo.com
ORCID: https://orcid.org/0000-0002-7533-0763
Affiliation:
Health Sciences program, Federal University of Triângulo Mineiro, Uberaba 38025-440, Brazil
ORCID: https://orcid.org/0009-0005-4103-8771
Explor Immunol. 2024;4:152–168 DOI: https://doi.org/10.37349/ei.2024.00134
Received: July 16, 2023 Accepted: January 17, 2024 Published: March 05, 2024
Academic Editor: Dominique J. Charron, Hospital Saint Louis, France
This review provides a detailed examination of CD4+ T lymphocyte subsets, crucial components of the immune system originating from the thymus. This study explores the distinct roles and mechanisms of various T helper (Th) cell subsets, including Th1, Th2, Th17, Th22, regulatory T cells (Tregs), Th9, and T follicular helper (Tfh) cells, focusing on their induction by specific cytokines, regulation by transcription factors, and the production of post-induction cytokines. The study traces the historical origins of Th lymphocyte research, emphasizing the unique cytokine profiles and functional implications of each subset in immune regulation and pathology, including autoimmune diseases, allergies, and cancer. Key findings include the delineation of cytokine-mediated induction processes, highlighting factors like interferon-gamma (IFN-γ), interleukin-4 (IL-4), transforming growth factor-beta (TGF-β), and IL-6. The review delves into transcription factors such as T-box transcription factor 21 (T-bet), GATA binding protein 3 (GATA3), and forkhead box P3 (Foxp3), underlying the lineage-specific development of these cells, and discusses the significant roles of post-induction cytokines. The research underscores the clinical relevance of CD4+ T cell subset dysregulation in various diseases, advocating for a nuanced understanding of these subsets for potential therapeutic advancements in immune-related disorders.
The foundational research on T helper (Th) lymphocytes dates back to the 1960s work of Miller [1]. Their studies involved thymectomized mice, a process where the thymus, an upper chest organ crucial for immune system development, is surgically removed. This research revealed that mice lacking a thymus failed to initiate specific immune responses.
Based on the remarkable discoveries, Miller [1] postulated the existence of a distinct subset of immune cells that relied on the thymus for their development and functionality. Subsequently, these cells were identified as T lymphocytes and categorized into two primary types: cytotoxic T lymphocytes (CD8+) and Th lymphocytes (CD4+) [1, 2]. Building upon this classification, in the late 1980s, Mosmann and Coffman [3], both seasoned immunologists, made pivotal advancements in delineating the heterogeneity within lymphocyte populations, previously classified broadly as Th lymphocytes. Their research elucidated a bifurcation in these cells, revealing opposing functionalities and laying the groundwork for the Th1/Th2 paradigm. This breakthrough provided a template for subsequent research, which further identified and characterized additional subpopulations within the Th lymphocyte framework.
The differentiation of CD4+ T cells into various subtypes is intricately directed by specific signals, including the nature and length of antigen exposure and the surrounding cytokine milieu [4]. Recent research indicates that the quantity of T and B lymphocytes present in the microenvironment plays a crucial role in CD4+ T cell differentiation [5]. Significantly, a reduction in lymphocyte count often leads to a Th1-predominant setting, irrespective of the antigenic properties. Prominent subtypes of Th cells encompass Th1, Th2, Th9, Th17, Th22, regulatory T cells (Tregs), and T follicular helper (Tfh) cells. Each of these subtypes manifests unique characteristics and exerts distinct influences on immunological responses (Table 1) [6].
Comprehensive overview of CD4+ T cell subsets: induction, transcription factors, post-induction cytokines, roles in host protection, and associated pathologies
| CD4+ T cell subsets | Inducing cytokines | Transcription factors | Post-induction cytokines | Roles in host protection | Associated pathologies | References |
|---|---|---|---|---|---|---|
| Th1 | IL-12, IFN-γ, IFN-α, IL-2, and IL-4 | T-bet, STAT1, and STAT4 | IFN-γ, IL-2, IFN-α, TNF-α, and TNF-γ | Defense against intracellular pathogens | Autoimmune diseases and pregnancy complications | [24, 27, 32–39] |
| Tumor progression (when decreased) | ||||||
| Th2 | IL-4, IL-2, and IL-6 | GATA3, STAT6, MAF, and NFAT | IL-13, IL-4, IL-5, IL-10, IL-6, and IL-9 | Defense against extracellular pathogens | Allergic diseases such as asthma and eczema | [24, 27, 33, 35, 36, 38–42] |
| Th17 | TGF-β, IL-6, IL-21, IL-23, and IL-1 | RORγt, STAT3, RORα, and RUNX1 | IL-17A, IL-22, IL-17F, IL-21, GM-CSF, TNF-α, IFN-γ, IL-26, IL-10, and IL-9 | Defense against intracellular and extracellular pathogens | Inflammatory bowel disease, rheumatoid arthritis, psoriasis, multiple sclerosis, and bone inflammation | [27, 35, 38, 46–52] |
| Fungal diseases (when decreased) | ||||||
| Treg | TGF-β, IL-2, and IL-35 | Foxp3, STAT5, and RUNX1/3 | IL-10, TGF-β, and IL-35 | Immune homeostasis and preventing autoimmunity | Tumor progression | [27, 35, 38, 48, 57–62] |
| Autoimmune diseases (when decreased) | ||||||
| Th9 | TGF-β, IL-4, IL-1, IL-25, IL-33, IL-2, IL-10, IL-21, TNF-α, IL-6, and IL-7 | PU.1, IRF4, STAT6, GATA3, STAT5, NFκB, STAT1, FoxO1, IRF1/8, and BATF | IL-9, IL-10, IL-5, IL-13, and IL-21 | Skin immunity | Autoimmune and inflammatory diseases including atopic dermatitis and asthma | [35, 38, 71–78] |
| Anti-tumor response | ||||||
| Protection against helminth infections | ||||||
| Th22 | IL-6, TNF-α, IL-1β, IL-21, and IL-26 | AHR, STAT3, STAT1, STAT5, RORγt, and HIF-1A | IL-22, IL-13, IL-26, TNF-α, and IL-10 | Cutaneous, intestinal, and respiratory barrier immunity | Inflammatory bowel diseases, thyroid diseases, immune thrombocytopenia, asthma, multiple sclerosis, rheumatoid arthritis, hepatitis, nephropathy, myasthenia gravis, and certain types of tumors | [35, 38, 79–86] |
| Augmentation of intrinsic immunity | ||||||
| Regeneration of tissues | ||||||
| Tfh | IL-21, IL-12, IL-6, TGF-β, IL-23, IL-27, IL-2, and IFN-γ | Bcl6, STAT1, STAT3, STAT4, STAT5, BATF, IRF4, MAF, FoxO1, and T-bet | IL-21, IL-4, IFN-γ, IL-17, IL-13, IL-10, and IL-5 | Critical role in supporting B cell maturation and differentiation into effector and memory B cells | Autoimmunity and T cell lymphoma | [35, 38, 87–94] |
| Humoral immunodeficiency (when decreased) |
IFN-γ: interferon-gamma; STAT1: signal transducer and activator of transcription 1; TNF-α: tumor necrosis factor-alpha; MAF: v-maf musculoaponeurotic fibrosarcoma oncogene homolog; NFAT: nuclear factor of activated T cell; RORγt: retinoic acid receptor-related orphan receptor gamma-t; RUNX1: runt-related transcription factor 1; GM-CSF: granulocyte-macrophage colony-stimulating factor; PU.1: Spi-1 proto-oncogene; IRF4: IFN regulatory factor 4; NFκB: nuclear factor kappa B; BATF: basic leucine zipper transcription factor; AHR: aryl hydrocarbon receptor; HIF-1A: hypoxia-inducible factor 1-alpha; Bcl6: B cell lymphoma 6; IL-12: interleukin-12; TGF-β: transforming growth factor-beta; T-bet: T-box transcription factor 21; GATA3: GATA binding protein 3; Foxp3: forkhead box P3
Understanding this diversity and differentiation among CD4+ T cell subtypes necessitates exploring beyond surface-level signaling; it involves delving into the realm of epigenetic regulation. This complex layer of control plays a pivotal role in the differentiation and function of these cells. Epigenetic regulation entails changes in gene expression that occur without modifying the DNA sequence itself, primarily achieved through DNA methylation, histone modifications, and non-coding RNA interactions. Such epigenetic landscapes are dynamic, responding to environmental stimuli and thereby influencing the cells’ phenotypic plasticity and diversity. For instance, in Tregs, the transcriptional and epigenetic regulation of the Foxp3 gene is vital for maintaining immune tolerance and preventing autoimmunity [7]. In Th1 and Th2 cells, epigenetic mechanisms selectively orchestrate cytokine gene expressions, like IFN-γ and IL-4, shaping their immune responses [8]. Similarly, Th17 cell development and its role in autoimmune diseases are intricately controlled by epigenetic factors, such as the role of the tripartite motif containing 28 [9]. Likewise, Tfh cells, crucial for B cell assistance and germinal center formation, depend on a coordinated interplay of epigenetic regulations and transcription factors for their differentiation and functionality in autoimmune contexts [10]. This intricate epigenetic regulation is pivotal in understanding the complex functionalities and interactions of Th cell subtypes in various immune responses and diseases.
The differentiation of CD4+ T cells into various subtypes, a crucial aspect of their functionality is the formation and maintenance of memory cells. Immunologic memory, a powerful ability of the adaptive immune system, allows for a rapid and more effective response upon re-exposure to a previously encountered antigen. This is foundational for vaccine-induced immunity and durable protection against reinfections. CD4+ T cell memory encompasses multiple distinct lineages with diverse effector functions, addressing the issues of lineage commitment, plasticity, and the distribution of memory cells within each lineage. Naive CD4+ T cells differentiate into specialized effector subsets that support cell-mediated or humoral responses, and upon infection clearance, a fraction of these cells survive as long-lived memory cells, widely distributed across the body. This memory formation involves critical signaling pathways and cytokines, including IL-2 and IL-12 for Th1 cells, with memory precursors differentiating into effector memory T cells and central memory T cells. Th1 memory CD4+ T cells, upon secondary antigen encounter, are thought to give rise to more Th1-like secondary effector cells, while Tfh memory cells are more plastic and can give rise to both Th1 and Tfh secondary effector cells [11–13].
Delving deeper into the functionality of CD4+ T cells, it is important to consider their adaptability in various physiological states, especially in chronic diseases and cancer scenarios. In such conditions, CD4+ T cells often enter a state of “exhaustion”, characterized by a significant decrease in their effector functions and an upsurge in the expression of inhibitory receptors. This phenomenon is particularly evident in conditions like chronic lymphocytic leukemia, where CD4+ T cells display impaired functionalities and express multiple inhibitory receptors [14]. On the other hand, CD4+ T cells can also exhibit direct cytotoxic capabilities. This role is becoming increasingly recognized, especially in the context of tumor immunity, where CD4+ T cells can directly target tumor cells. This cytotoxic function of CD4+ T cells expands their role in the immune system, making them an active participant in direct immune responses against cancer [14, 15].
Cytokines, small protein molecules secreted by immune cells, are critical in guiding the development and differentiation of CD4+ T cell subtypes. They act as intercellular messengers, modulating cellular activities through signal transduction [16]. Transcription factors, which regulate gene expression, play a vital role in dictating the fate and functionality of CD4+ T cells. Acting as molecular regulators, they direct the development of specific T cell subtypes by selectively activating or repressing gene expression [6, 16, 17].
Distinct cytokine secretion patterns characterize each CD4+ T cell subtype, with these patterns being associated with various pathologies [4, 16]. Th1 cells, for example, secrete cytokines like IFN-γ, IL-2, and TNF-α, essential for cell-mediated immunity and combating intracellular pathogens. Conversely, Th2 cells produce cytokines such as IL-13, IL-4, and IL-5, crucial in allergic responses and defense against helminthic infections [16, 18].
Th17 cells are defined by their secretion of IL-17 and are crucial in defending against extracellular pathogens, such as fungi and bacteria, while also promoting tissue-specific inflammatory responses. Conversely, Th22 cells, characterized by IL-22 production, primarily focus on tissue protection and regeneration amid inflammatory conditions. This subset is notably linked to inflammatory skin disorders, given the significant impact of IL-22 on skin homeostasis and inflammatory response mechanisms [18].
Tregs emit anti-inflammatory cytokines like IL-10 and TGF-β, pivotal in upholding immune tolerance and curbing hyperactive immune responses. Meanwhile, Tfh cells produce IL-21, essential for the maturation of antibody-producing B cells [19, 20].
The Th9 cell lineage, initially associated with diverse pathological conditions including asthma and atopic dermatitis, is now being explored for its potential role in certain cancer types. These cells are characterized by their ability to produce a variety of cytokines, with IL-9 being the most prominent [6, 17].
The induction and differentiation of CD4+ T cell subtypes constitute a highly intricate process, steered by a network of cytokines and transcription factors. This dynamic interplay activates specific genetic pathways, culminating in the formation of distinct T cell subtypes. Furthermore, these differentiated cells are capable of producing a broad-spectrum of cytokines, adding to the complexity of immune system responses [6, 16].
The Th1 subset is pivotal in eliminating intracellular pathogens, including viruses, bacteria, and tumor antigens, characterized by its production of cytokines such as IFN-γ and IL-2 [16]. Th1 cell development is orchestrated by a complex network of cytokines and transcription factors. IL-12 and IFN-γ are crucial for Th1 induction, followed by the production of cytokines like IFN-γ, IL-2, TNF-α, and IFN-α. T-bet, a T-box transcription factor, is vital for Th1 differentiation, activated through IFN-γ signaling and STAT1 (Figure 1) [6, 16, 21]. Emerging studies reveal that leptin can prompt CD4+ T cells to differentiate into the Th1 subset [22].
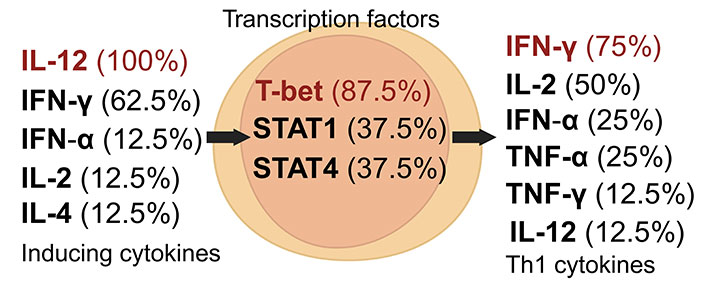
Dynamics of Th1 cell induction and literature citation analysis. This figure presents Th1 pre-induction cytokines [24, 27, 32–38], transcription factors [24, 27, 32, 34–38], and post-induction cytokines [24, 27, 33–38], each accompanied by their literature citation frequency as a percentage, based on ten articles [24, 27, 32–39]. Arrows depict the signaling cascade where inducing cytokines on the left regulate central transcription factors to initiate the synthesis of Th1 cytokines on the right. Created with Biorender.com
The Th1 subset is crucial in fighting microbes that infect the phagosomes of myeloid cells, and any changes in the function or frequency of Th1 cells are linked to various diseases [16]. This is particularly relevant considering that stress-related hormones like glucocorticoids can suppress Th1 CD4+ lymphocytes. Such suppression can lead to a shift towards Th2 cells, potentially weakening the immune system’s ability to effectively combat tumors [23]. Interestingly, a robust Th1 response in the initial stages of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has been associated with better patient outcomes [24].
Moving beyond the pivotal role of Th1 cells in combating intracellular pathogens, it’s important to consider the parallel yet distinct contributions of group 1 innate lymphoid cells (ILC1s). ILC1s, which mirror the functionality of Th1 cells, have emerged as key players in orchestrating type 1 inflammatory responses. Like their Th1 counterparts, ILC1s respond to innate stimuli and produce T cell-associated cytokines such as IFN-γ, aligning with the role of Th1 cells in immune responses. However, ILC1s operate without the need for antigen recognition. The shared transcriptional regulators, including T-bet, further underscore the parallels between ILC1s and Th1 cells. T-bet, crucial for Th1 differentiation, also plays a pivotal role in the development of ILC1s, influencing their cytokine production profile. Despite these functional similarities, ILC1s exhibit distinct phenotypic traits and roles, especially in the context of diseases like systemic sclerosis, highlighting their unique contribution to the immune system [25, 26].
Continuing with the Th1 subset, research has established a connection between these cells and organ-specific autoimmune diseases [21]. A reduction in Th1 cell frequency is linked to an increased susceptibility to bacterial infections, whereas an elevated frequency of Th1 cells is associated with multiple sclerosis. During pregnancy, a Th1 predominance has been correlated with spontaneous abortions [27, 28].
The Th1 subset plays a pivotal role in pathogen-specific immune responses, as exemplified in leprosy. In tuberculoid leprosy, a Th1-mediated response, marked by IFN-γ production, fosters a strong cellular immune reaction, enabling macrophages to effectively eliminate Mycobacterium leprae. In contrast, lepromatous leprosy features a dominant Th2 response, leading to a humoral reaction that is less effective against this intracellular pathogen and results in more severe disease progression [29].
Beyond their involvement in autoimmunity and infectious diseases, Th1 cells are vitally important in the regulation of malaria. CD4+ T cell responses are critical in managing malaria and its associated immunopathology. For example, in malaria-induced anemia and cerebral malaria, the balance between Th1 cells’ protective and pathological roles is crucial. In cerebral malaria, characterized by extreme inflammation, an excessive Th1 response can worsen the disease. Conversely, CD4+ T cells are essential in priming phagocytes and supporting B cell antibody production against Plasmodium parasites, highlighting the need for a delicate equilibrium to effectively combat this complex infection [30].
The interaction between programmed death-ligand 1 (PDL-1) and programmed cell death protein 1 (PD-1), as exhibited by myeloid tumor cells, can transform T-bet+ Th1 cells into Foxp3+ Tregs [31]. Blocking PDL-1 can enhance the differentiation of CD4+ T cells into the Th1 subset, contributing to the development of graft-versus-host disease (GVHD). Furthermore, Th17 cells can transition to Th1 cells under IL-12 influence, which triggers pathways involving STAT4 [32].
Under the “threshold hypothesis” within the Th1 profile, the nature of the immune response, especially in CD4+ T cells, is determined by the interaction intensity with other immune components, such as B cells. This hypothesis suggests that the type of immune response (Th1 or Th2) is modulated by the degree of cellular cooperation in the immune microenvironment, with weak interactions between CD4+ T cells and B cells leading to Th1 differentiation, while stronger interactions result in Th2 differentiation [5].
In summary, the Th1 subset is vital in the immune system’s fight against intracellular pathogens. However, variations in Th1 cell frequency and function are linked to diverse diseases, including autoimmune disorders, tumors, and complications during pregnancy. The intricate network of cytokines and transcription factors regulating the Th1 subset offers potential therapeutic avenues for these conditions.
Th2 cells, a subset of Th cells, predominantly produce IL-4 and IL-13, which stimulate mucus production and activate B cells for antibody synthesis, especially immunoglobulin E (IgE) [16, 33]. The induction of the Th2 subset is facilitated by cytokines including IL-2, IL-4, and IL-6, which promote its differentiation and maintenance (Figure 2) [24, 27, 33–42]. The transcription factor GATA3 is essential in the differentiation and functionality of Th2 cells [43]. Beyond cytokines, Th2 subset induction can also be influenced by factors such as insulin, modulators of peroxisome proliferator-activated receptors (PPAR), and omega-3 polyunsaturated fatty acids (n-3 PUFAs) [33].
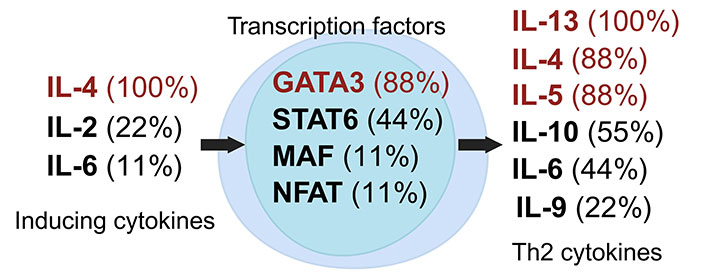
Dynamics of Th2 cell induction and literature citation analysis. This figure illustrates Th2 pre-induction cytokines [24, 27, 33, 35, 36, 38, 40–42], transcription factors [24, 27, 33, 35, 36, 38, 40–42], and post-induction cytokines [24, 27, 33, 35, 36, 38, 40–42], each accompanied by their literature citation frequency as a percentage, based on ten articles [24, 27, 33, 35, 36, 38–42]. Arrows show the signaling cascade where inducing cytokines on the left regulate central transcription factors to start the synthesis of Th2 cytokines on the right. Created with Biorender.com
Stress has a notable impact on the immune system, with studies indicating that stress hormones like glucocorticoids can suppress Th1 cells, favoring a shift towards Th2 cells [39]. This alteration in the Th1/Th2 balance might enhance susceptibility to infections and other diseases linked to chronic stress [16, 39]. The Th2 subset, extensively researched for its role in allergic diseases such as asthma and eczema, contributes both to the immune response against extracellular parasites like helminths and to the pathogenesis of allergic conditions by producing cytokines that are key in recruiting eosinophils and other cells involved in allergic inflammation [16, 40].
Similar to Th2 cells, ILC2s play a critical role in type 2 immune responses. They rapidly respond to environmental cues and produce type 2 cytokines, notably IL-4 and IL-13, which are key in allergic inflammation and responses against extracellular parasites. ILC2s, as tissue-resident sentinels, are instrumental in regulating both innate and adaptive immunity. The development and function of ILC2s are influenced by various environmental and genetic factors, paralleling Th2 cells, particularly with the transcription factor GATA3 playing a pivotal role. While there are similarities in cytokine profiles and regulatory mechanisms between ILC2s and Th2 cells, ILC2s represent a distinct component of the immune system, contributing uniquely to both innate and adaptive responses [25, 44].
The Th2 subset is also implicated in viral infections, including SARS-CoV-2 and influenza A (H1N1). A transition from a Th1 to a Th2 response during SARS-CoV-2 infection correlates with poorer outcomes and a heightened risk of developing acute respiratory distress syndrome (ARDS). In pregnancy, the Th1/Th2 balance is vital for maintaining fetal immune tolerance. Th2 cells are important for maternal tolerance towards fetal antigens, but recent studies emphasize the significance of the Th1/Th2 balance over the absolute levels of Th2 cells [27].
While the Th2 response is crucial in allergy and helminth immunity, it also plays a role in specific pathogen contexts. For instance, in leprosy, a shift towards a Th2 response in the lepromatous form indicates compromised cellular immunity against Mycobacterium leprae, leading to more widespread disease [29]. This highlights the impact of Th2 differentiation on disease outcomes. In malaria, Th2 responses have implications as well; although not directly involved in anemia or cerebral malaria, the Th2 cytokine environment may affect the severity of these conditions, underscoring the need for a balanced Th1/Th2 response for effective immunity and host survival [30, 45]. These insights emphasize the multifaceted nature of Th2 cell functions beyond their known roles in inflammatory and atopic disorders.
Th17 cells, a unique subset of CD4+ Th cells, are crucial in the immune defense against extracellular pathogens [16]. These cells differentiate from naive T cells under the influence of specific cytokines. The generation of Th17 cells is primarily driven by the combined action of IL-6 and TGF-β, or IL-21 and TGF-β [46]. Characteristically, Th17 cells produce various cytokines, including IL-17A, IL-17F, and IL-22, along with other less commonly discussed cytokines like GM-CSF, TNF-α, IFN-γ, IL-9, IL-10, IL-21, and IL-26 (Figure 3) [27, 35, 38, 46–52].
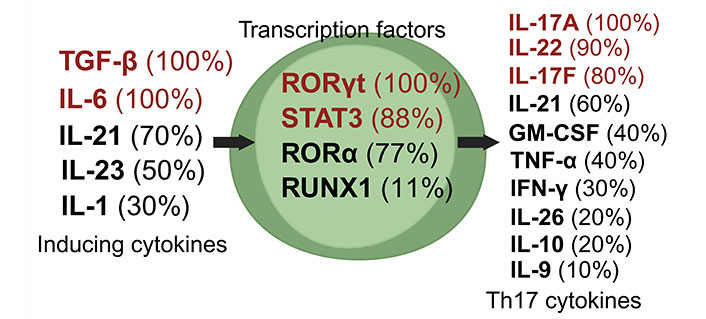
Dynamics of Th17 cell induction and literature citation analysis. This figure displays Th17 pre-induction cytokines [27, 35, 38, 46–52], transcription factors [27, 35, 38, 47–52], and post-induction cytokines [27, 35, 38, 46–52], each with their corresponding literature citation frequency in percentage, based on ten articles [27, 35, 38, 46–52]. Arrows depict the signaling cascade where inducing cytokines on the left guide central transcription factors to trigger the synthesis of Th17 cytokines on the right. Created with Biorender.com
The proliferation of Th17 cells is modulated by factors such as high sodium levels, IL-23, IL-1β, and TNF-α [46, 47]. In contrast, IL-25, IL-27, IL-2, suppressor of cytokine signaling 3 (SOCS3), SOCS1, vitamin D, and retinoic acid are known to negatively regulate Th17 cells [46]. An increase in Th17 cells is linked to inflammatory diseases such as inflammatory bowel disease, rheumatoid arthritis, psoriasis, multiple sclerosis, and bone inflammation [38, 48, 51, 52]. Conversely, a decrease in Th17 cells is associated with susceptibility to fungal infections. Additionally, prostaglandin E2 (PGE2) influences Th17 differentiation and proliferation [27].
Th17 cell generation is triggered by activation of the RORγt, STAT3, and RORα transcription factors [27, 35, 38, 46–52]. IL-23 is vital for maintaining Th17 cell stability and promoting their proliferation. Th17 cells predominantly produce IL-17A, belonging to a cytokine family that includes IL-17A, IL-17B, IL-17C, IL-17D, IL-17E (or IL-25), and IL-17F [47]. These cells play an integral role in mediating immune responses against various extracellular and intracellular bacteria and viruses [27].
Expanding on this, it’s pertinent to explore the role of ILCs, especially ILC3s, which parallel the functions of Th17 cells, a subset of CD4+ Th cells. Both ILCs and Th17 cells are critical in mucosal immunity, producing key cytokines such as IL-17 and IL-22, essential for defending against pathogens, particularly at epithelial barriers. Often described as the innate counterparts of T cells, ILCs share similarities with Th17 cells in their development, transcriptional regulation, and cytokine secretion, underscoring their complementary roles in the immune system [53, 54]. This parallel extends to their involvement in immune homeostasis, response to infection, and tissue repair, emphasizing the interconnected nature of innate and adaptive immunity [53–56].
In the context of coronavirus disease 2019 (COVID-19) infection, Th17 cells contribute significantly to the immune response. IL-17, secreted by Th17 cells, recruits neutrophils and monocytes to infected sites and induces cytokines like granulocyte-colony stimulating factor (G-CSF) and IL-6, which facilitate innate inflammation and chemokinesis [48, 49].
Tregs are a specialized subset of T cells instrumental in maintaining immune homeostasis and preventing autoimmunity [57, 58]. Tregs characteristically express the transcription factor Foxp3, a key regulator of their development and function (Figure 4) [27, 35, 38, 48, 57–62]. However, their role in cancer immunotherapy is debated. While some studies indicate Tregs suppress immune responses against tumors and may aid tumor progression [59, 61], others suggest Tregs can mitigate inflammation and reduce autoimmunity, offering potential benefits in certain pathologies [57].

Dynamics of Treg induction and literature citation analysis. This figure showcases Treg pre-induction cytokines [27, 35, 38, 48, 57, 60–62], transcription factors [27, 35, 38, 48, 57–62], and post-induction cytokines [27, 35, 38, 57, 59–62], each detailed with their literature citation frequency in percentage, based on ten articles [27, 35, 38, 48, 57–62]. Arrows demonstrate the signaling cascade where inducing cytokines on the left influence central transcription factors to commence the synthesis of Treg cytokines on the right. Created with Biorender.com
A defining trait of Tregs is their production of IL-10, a powerful anti-inflammatory cytokine [59]. IL-10 inhibits the activation and proliferation of effector T cells while supporting Treg differentiation and function. This property positions Tregs as a significant target in cancer immunotherapy, where Treg suppression could enhance effector T cell activation and proliferation, aiding in tumor cell eradication [63].
In cancer immunotherapy, inhibiting Tregs necessitates a careful balance between reducing their function to boost anti-tumor immunity and avoiding the onset of autoimmunity. Various factors, including cancer type, disease stage, and patient immune status, are crucial considerations in designing Treg-targeted cancer immunotherapies [64].
Tregs also play a vital role in pregnancy, contributing to immune tolerance towards the fetus and preventing embryo rejection. TGF-β, a cytokine promoting Treg differentiation, is crucial in sustaining this immune tolerance [65–67]. Tregs are capable of inhibiting the proliferation and activation of Th cells, cytotoxic T cells, and antibody production by plasma cells, as well as dampening natural killer (NK) cell cytotoxicity and dendritic cell (DC) maturation [50].
Tregs can be classified into natural Tregs (nTregs), which originate in the thymus, and induced Tregs (iTregs), generated in laboratory conditions [60]. Further subclassification of Tregs is based on their cytokine profiles and functional attributes. For instance, type 1 Tregs (Tr1s) are notable for high IL-10 production, whereas Th3 Tregs primarily secrete TGF-β. Notably, both subgroups typically exhibit low Foxp3 expression levels [61]. The classification of Th3 cells remains a nuanced and debated topic within the immunology community. While some researchers advocate for considering Th3 cells as a distinct subset separate from Tregs, others posit them within the Treg category [68, 69]. This debate stems from their cytokine secretion profile, which includes immunosuppressive factors, yet lacks the primary signaling via the Foxp3 transcription factor typical of Tregs. Regardless of their precise classification, the role of Th3 cells is gaining increasing attention and is considered vital in the study of immunological disorders, particularly those related to autoimmunity and allergies [68–70].
Th9 cells, a subset of CD4+ T cells, are distinguished by their production of IL-9 upon activation [71]. Their differentiation is primarily driven by TGF-β and IL-4, with additional influences from IL-1, IL-2, and IL-33 [71, 72]. Post-induction, Th9 cells continue to secrete IL-9, along with other cytokines like IL-5 and IL-10 [73]. Key transcription factors regulating Th9 cells include PU.1, IRF4, and STAT6 (Figure 5) [35, 72].
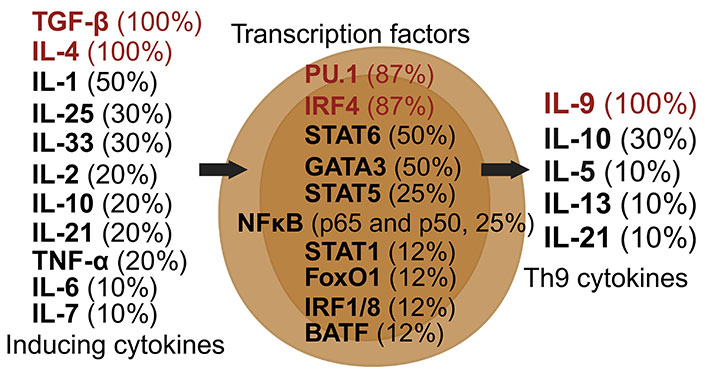
Dynamics of Th9 cell induction and literature citation analysis. This figure illustrates Th9 pre-induction cytokines [35, 38, 71–78], transcription factors [35, 38, 71–73, 75, 76, 78], and post-induction cytokines [35, 38, 71–78], each accompanied by their literature citation frequency in percentage, based on ten articles [35, 38, 71–78]. Arrows depict the signaling cascade where inducing cytokines on the left guide central transcription factors to begin the synthesis of Th9 cytokines on the right. Created with Biorender.com
Th9 differentiation requires both IL-4 and TGF-β, as IL-4 alone is insufficient; conversely, IFN-γ and IL-23 inhibit Th9 differentiation [71, 72]. Although Th17 cells also produce IL-9, Th9 cells are the predominant source [46]. Th9 cells have been identified in the peripheral blood of allergy patients and in the skin of both healthy and allergic individuals, particularly in allergies to dust and cat dander [71].
In the realm of cancer therapy, Th9 cells have shown promise. IL-9 is effective in treating melanoma and lung cancer, with mast cells playing a pivotal role in the anti-tumor response [71, 72]. Furthermore, Th9 cells are linked to certain autoimmune and inflammatory diseases, such as atopic dermatitis and asthma, and play a role in immune responses to parasitic infections [35, 38, 73, 74].
In scientific literature, Th9 cells are regulated by transcription factors like STAT6, IRF4, and PU.1, with occasional mentions of GATA3 and STAT5 [71, 75]. These cells have both protective and pathological roles within the immune system [76]. Beyond their application in cancer therapy, Th9 cells are also considered therapeutic targets for allergies and autoimmune diseases [73]. Chronic allergies, for example, may involve Th9 cells along with Th2 cells [77]. Overall, Th9 cells represent a unique subset of CD4+ T cells with a specific cytokine profile and transcriptional regulation, implicated in various diseases including allergies, cancer, and autoimmune disorders [71, 76].
Th22 cells, a unique subset of CD4+ Th cells known for producing IL-22 (Figure 6), have been identified as key players in a range of autoimmune disorders including inflammatory bowel diseases, thyroid diseases, immune thrombocytopenia, asthma, multiple sclerosis, rheumatoid arthritis, hepatitis, nephropathy, and myasthenia gravis [35, 38, 77, 78]. They also play roles in infectious diseases and tumorigenesis [79]. First described in 2000, IL-22, often acting synergistically with IL-17 and TNF, primarily targets epithelial cells, liver cells, pancreatic cells, and certain fibroblasts [80]. IL-22 receptors are predominantly expressed in the skin, pancreas, intestine, liver, lung, and kidneys, with IL-22 chiefly enhancing innate immune responses and facilitating tissue regeneration [81, 82]. However, IL-22 expression is inhibited by TGF-β [80]. Th22 cells are also integral to the protection of skin, intestines, and airways [35].

Dynamics of Th22 cell induction and literature citation analysis. This figure displays Th22 pre-induction cytokines [35, 38, 79–86], transcription factors [35, 38, 79–86], and post-induction cytokines [35, 38, 79–86], each with their corresponding literature citation frequency in percentage, based on ten articles [35, 38, 79–86]. Arrows show the signaling cascade where inducing cytokines on the left regulate central transcription factors to start the synthesis of Th22 cytokines on the right. Created with Biorender.com
Th22 cells are characterized by the expression of specific chemokine receptors (CCRs), including CCR6, CCR4, and CCR10. While Th17 and NK cells also produce IL-22, they do so in response to different stimuli and at lower concentrations [82]. IL-22 stimulates the expression of antimicrobial molecules such as neutrophil gelatinase-associated lipocalin (NGAL), β-defensins, human peptide LL-37 (cathelicidin antimicrobial peptide), and S100 calcium-binding protein A7 (S100A7) [81]. The induction of Th22 cells is influenced by cytokines like IL-6 and TNF-α, and their transcriptional regulation is mediated by factors such as STAT1, STAT3, STAT5, RORγt, and AHR [35].
Tfh cells are a specialized subset of CD4+ T cells, pivotal in supporting B cell maturation and differentiation into effector and memory cells [35, 38]. Characterized by the C-X-C CCR type 5 (CXCR5) expression and molecules like PD-1, inducible T cell co-stimulator (ICOS), CD200, and B- and T-lymphocyte attenuator (BTLA), Tfh cells are regulated by Bcl6, identified as a master regulator of Tfh cell differentiation and function (Figure 7) [83–87]. Predominantly located in the follicles of lymphoid organs, Tfh cells play a vital role in humoral immunity [35].
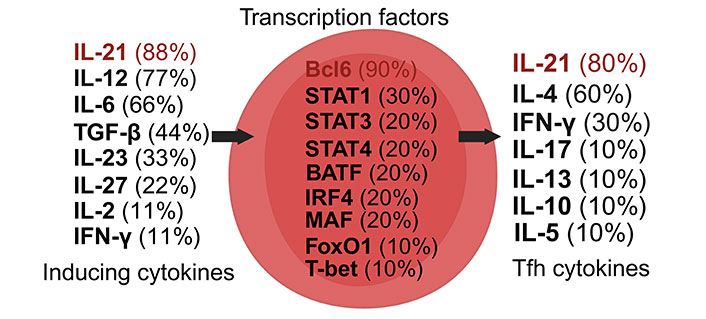
Dynamics of Tfh cell induction and literature citation analysis. This figure presents Tfh cell pre-induction cytokines [35, 38, 88–94], transcription factors [35, 38, 87–94], and post-induction cytokines [35, 38, 87–94], each accompanied by their literature citation frequency in percentage, based on ten articles [35, 38, 87–94]. Arrows illustrate the signaling cascade where inducing cytokines on the left control central transcription factors to initiate the synthesis of Tfh cell cytokines on the right. Created with Biorender.com
The primary cytokine secreted by Tfh cells, IL-21, is crucial for enhancing B cell affinity maturation and class switch recombination. Tfh cells contribute significantly to the formation and maintenance of germinal centers, essential for producing potent and long-lasting antibodies [88, 89]. Without Tfh cells, the immune system’s response to infections and diseases would be significantly compromised [88].
Recent research highlights Tfh cells’ involvement in autoimmune and inflammatory diseases, where they facilitate the production of pathogenic autoantibodies by autoreactive B cells (Figure 8) [88, 90–94]. This process leads to increased inflammation, tissue damage, and potentially irreversible organ dysfunction, underlining the loss of tolerance to self-antigens characteristic of these diseases.
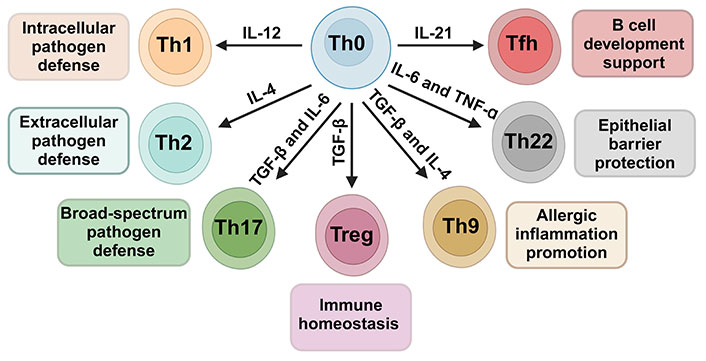
Cytokine-driven differentiation of CD4+ Th cells from the Th0 precursor to specialized subsets. Influenced by cytokines like IL-12, IL-4, IL-6, TNF-α, TGF-β, and IL-21, Th0 cells mature into Th1, Th2, Th17, Treg, Tfh, Th22, and Th9, each fulfilling distinct immunological roles. The arrows indicate the most likely CD4+ Th cell subtype differentiation in response to the corresponding cytokine stimulus. Created with Biorender.com
Elevated frequencies of Tfh-like cells (CXCR5+ and PD-1high and/or ICOShigh) are observed in autoimmune diseases like multiple sclerosis, rheumatoid arthritis, and systemic lupus erythematosus [88]. Understanding Tfh cells’ role in these diseases could unlock new therapeutic strategies for managing autoimmune pathologies.
Induced by cytokines such as IL-6, IL-12, and IL-21, Tfh cells, post-induction, produce cytokines including IL-4 and IL-21 [38, 88]. The transcriptional regulation of Tfh cell differentiation and function involves factors like Bcl6, IRF4, MAF, STAT1, STAT3, and STAT4, highlighting their complex regulatory network [35].
Numerous articles characterize the subsets of Th lymphocytes, associating them with pathologies. Some subsets tend to have a more solid definition regarding the cytokines capable of inducing them, the transcription factors involved, and the cytokines produced after induction. However, a minority of articles tends to broaden the characteristics of these lymphocyte subsets, being more comprehensive in the cited cytokines and consequently expressing details that are not commonly pointed out [27, 32, 37, 38, 41, 61, 85]. Therefore, it is important to be aware that the characterization of these subsets can be more extensive than traditionally acknowledged.
AHR: aryl hydrocarbon receptor
Bcl6: B cell lymphoma 6
CCRs: chemokine receptors
Foxp3: forkhead box P3
GATA3: GATA binding protein 3
GM-CSF: granulocyte-macrophage colony-stimulating factor
IFN-γ: interferon-gamma
IL-4: interleukin-4
ILC1s: group 1 innate lymphoid cells
IRF4: interferon regulatory factor 4
MAF: v-maf musculoaponeurotic fibrosarcoma oncogene homolog
PD-1: programmed cell death protein 1
PU.1: Spi-1 proto-oncogene
RORγt: retinoic acid receptor-related orphan receptor gamma-t
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2
STAT1: signal transducer and activator of transcription 1
T-bet: T-box transcription factor 21
Tfh: T follicular helper
TGF-β: transforming growth factor-beta
Th: T helper
TNF-α: tumor necrosis factor-alpha
Tregs: regulatory T cells
MPdA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing—original draft, Writing—review & editing. JHV: Conceptualization, Data curation, Visualization, Writing—original draft, Writing—review & editing. Both authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
