Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0009-0002-8809-9203
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0009-0007-4171-1334
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0009-0006-8524-0127
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0009-0000-5103-5500
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0009-0009-9480-7879
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0000-0002-8041-7935
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0000-0002-5848-3020
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0000-0002-2886-6450
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0000-0001-9396-659X
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0009-0008-3254-0729
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0000-0002-3809-0377
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
ORCID: https://orcid.org/0000-0003-3109-9683
Affiliation:
Department of Clinical and Toxicological Analysis, Federal University of Ceará, Fortaleza 60430-370, Brazil
Email: tiagosampaio@ufc.br
ORCID: https://orcid.org/0000-0002-3962-6508
Explor Endocr Metab Dis. 2026;3:101455 DOI: https://doi.org/10.37349/eemd.2026.101455
Received: September 11, 2025 Accepted: December 01, 2025 Published: January 09, 2026
Academic Editor: Dawood Khan, Ulster University, United Kingdom; Karel Pacak, AKESO Holding, Czech Republic
Background: Metabolic syndrome and dyslipidaemia increase the risk of death by two or three times. In this context, the role of apolipoprotein A-I (Apo A-I), the main structural protein of high-density lipoprotein (HDL), stands out, since its anti-inflammatory potential reduces cardiovascular risk. Further, genetic modifications, such as the rs670 single-nucleotide polymorphism (SNP), in the promoter region of the APOA1 gene are associated with the development of cardiovascular events, dyslipidemia, and diabetes, as well as metabolic syndrome. Thus, this study aims to investigate the relation between the occurrence of dyslipidemia and the rs670 SNP genotypes.
Methods: An integrative and systematic review was performed with the LitVar2 database according to the PRISMA protocol standards. Studies were researched up to August 2025. Then, a meta-analysis was performed using the fixed-effects model, since the study was considered homogeneous based on the I2 value (< 50%).
Results: Of the 99 found articles in the database, 5 referred to metabolic disorders (n = 7,705—4 Chinese studies and 1 Iranian study) and were published between 2015 and 2018. Three (n = 2,784 patients or 36.13%) of the articles indicated an association between the polymorphic allele and a higher risk of developing dyslipidemia with a relative risk of 1.16 (IC 95% 1.09–1.23, p < 0.01, I2 = 0%). Relative risk (IC 95%) was presented, and p < 0.05 was defined as the significance criterion.
Discussion: This study reinforces a possible association between the influence of SNP rs670 and dyslipidemia. This emphasizes the importance of conducting further research incorporating a larger and more diverse study group, as well as investigating the genetic and environmental influence on the phenotypic expression of the rs670 SNP.
Apolipoproteins (Apo) are crucial modulators in the transport and metabolism of lipoproteins. Apo A-I, the main structural protein of high-density lipoprotein (HDL), is synthesized in the liver and small intestine and is an activator of lecithin-cholesterol acyltransferase (LCAT), an enzyme responsible for converting free cholesterol into cholesterol ester. Therefore, high levels of Apo A-I are associated with a reduced risk of atherosclerosis, mainly due to its ability to decrease the synthesis of inflammatory cytokines in endothelial cells and macrophages, indicating its anti-inflammatory potential [1, 2].
Apo A-I has a protective and inverse association with cardiovascular risk, with known atheroprotective effects, showing a negative correlation with triglyceride (TG) levels and body mass index (BMI) [3]. Environmental determinants, such as medications and diet, influence the synthesis and secretion of Apo A-I, especially with regard to the modulation of the peroxisome proliferator-activated receptor alpha (PPAR-α), which induces the transcription of genes related to lipid and carbohydrate metabolism, such as APOA1 [4]. However, there are reports of a relationship between cardiovascular events, dyslipidemia, and diabetes with expression of modified Apo A-I [5].
Polymorphisms in the APOA1 gene are associated with changes in fasting and postprandial plasma lipid levels [6, 7]. The single-nucleotide polymorphism (SNP) rs670, located in the promoter region of the APOA1 gene, has been described as a determinant of HDL levels, as well as the occurrence of metabolic syndrome (MS) and type 2 diabetes (T2D) [8].
The APOA1 gene, located on chromosome 11 (11q23.3), has approximately 8,870 base pairs. The exons generated by the transcription are processed, encoding pre-pro-Apo A-I. Pro-Apo A-I is formed in the intracellular environment and is subsequently released into the plasma. After this, mature Apo A-I is produced after the final cleavage of the N-terminal chain, consisting of 243 amino acid residues. In addition, the APOA1 gene comprises the APO cluster on chromosome 11 (APOA1/C3/A4/A5 gene cluster), responsible for encoding Apo A-I, Apo C-III, Apo A-IV, and Apo A-V [5].
A cytosine-phosphate-guanine (CpG) site in the APOA1 promoter region was discovered in 1984 as an indicator for polymorphisms using the restriction enzyme MspI. Due to the high methylation rate of CpG sites in promoter regions, a transition from cytosine (C) to thymine (T) may occur, resulting in SNP rs670 at position 75 upstream of the APOA1 gene [9]. The occurrence of SNP rs670 in the promoter region of the APOA1 gene is considered the most relevant in relation to disorders in human lipid metabolism. Considering the importance of understanding the true impact of polymorphism on population health, there is a need for studies capable of clarifying this fact [10, 11].
Thus, the objective of this study is to investigate the association between the occurrence of dyslipidemia and the occurrence of the rs670 SNP genotypes. A systematic review and meta-analysis were conducted, considering the discrepancies between the studies conducted to date. Thus, this study aims to clarify and provide, statistically, a more reliable and trustworthy result.
An integrative and systematic review was made, considering studies addressing the rs670 polymorphism of the APOA1 gene. Studies available until August 2025 were analyzed. The search was performed by applying the variant identifier (rs670) in the LitVar2 database (www.ncbi.nlm.nih.gov/research/litvar2/). This review was conducted in accordance with the PRISMA protocol recommendations [12].
The inclusion criteria applied for the research were: (1) articles available in full text, thus allowing for complete critical analysis, and (2) articles including the minor allele frequency (MAF) or a contingency table including genotypic distributions. The following criteria were used for exclusion: (1) being a review article, (2) being a meta-analysis, (3) being a genome-wide association study (GWAS), and (4) not evaluating metabolic disorders. Studies were selected regardless of their publication, language, or year of publication. The studies were independently evaluated by separate reviewers in order to reinforce the methodological selection of the study.
To organize the data, a table was created containing the author, title, study type, year of publication, country, sample size, MAF, risk ratio, and conclusions regarding the presence of the polymorphic allele.
For studies that did not report the odds ratio (OR) or the corresponding p-value, both measures were derived from the 2 × 2 contingency tables provided in the original articles. The p-value was obtained using either the chi-square test or Fisher’s exact test, according to cell frequencies. This procedure permits the computation of effect estimates when sufficient raw data are available, ensuring consistency and comparability across included studies.
The meta-analysis was performed using the fixed-effect model, given that, when compared, heterogeneity did not exceed 50%. The results presented the relative risk (RR) with a 95% confidence interval (CI) and statistical significance with p < 0.05. Statistical analysis was conducted using R software, version 4.5.1 for Windows. Heterogeneity was assessed using the I² value, where values above 50% indicate the study is heterogeneous and lower values indicate the study is homogeneous. To present the RR, the following analyses were performed: the Mantel-Haenszel method and DerSimonian-Laird estimator were used for τ², and the Mantel-Haenszel estimator was used to calculate Q and τ² (such as RevMan 5), and the continuity correction of 0.5 in studies with zero cell frequencies.
A total of 99 articles were found in the LitVar2 database. After applying the inclusion and exclusion criteria, except for the theme criterion, 33 articles were eligible for critical analysis. Among the excluded works were 4 articles not available in full text, 32 review or meta-analysis articles, 12 GWAS, and 18 articles without MAF. Finally, after applying the theme criterion, 5 articles were selected because they addressed metabolic disorders (Figure 1).
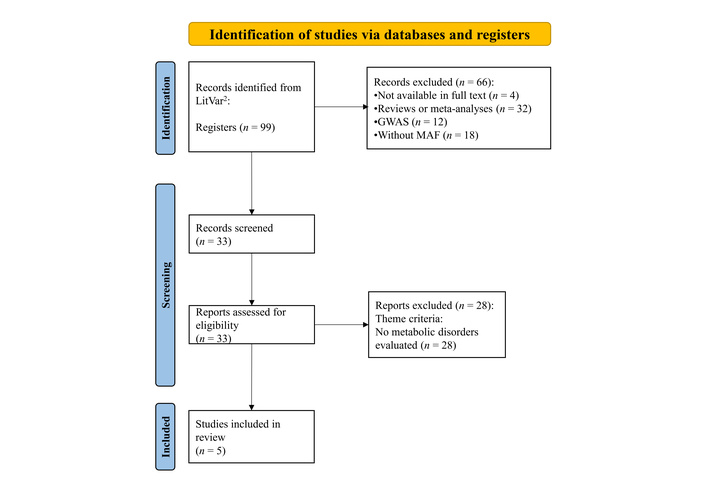
Graphical representation of the screening flow used for article selection. GWAS: genome-wide association study; MAF: minor allele frequency. Adapted from [12]. © 2019 The Authors. Licensed under a CC BY 4.0.
The selected articles were published between 2015 and 2018, comprising four Chinese articles and one Iranian. All studies were classified as case-control. Furthermore, three articles indicated that the presence of the polymorphic allele was associated with a high risk of developing metabolic disorders, and two showed no association between them. Three studies are significant, with a p-value < 0.05, and these are the same ones that presented a high-risk ratio, as indicated in Table 1.
Summary of studies evaluating the association between APOA1 gene polymorphisms and the risk of dyslipidemia.
| Authors | Title | Study type | Year | Country | n | MAF | Odds ratio | Conclusions |
|---|---|---|---|---|---|---|---|---|
| Wu et al. [6] | Interactions of Environmental Factors and APOA1-APOC3-APOA4-APOA5 Gene Cluster Gene Polymorphisms with Metabolic Syndrome | Case-control | 2016 | China | 3,441 | 26% | 1.31 (1.14–1.50);p < 0.001 | The polymorphic allele was associated with a higher risk of metabolic syndromes |
| Wang et al. [13] | Interactions of six SNPs in APOA1 gene and types of obesity on low HDL-C disease in Xinjiang pastoral area of China | Case-control | 2017 | China | 1,267 | 17% | 1.34 (1.03–1.74);p = 0.0262 | The polymorphic allele was associated with a higher risk of low HDL-C |
| Feng et al. [14] | Association of APOA1 gene polymorphisms (rs670, rs5069, and rs2070665) with dyslipidemia in the Kazakhs of Xinjiang | Case-control | 2016 | China | 736 | 14% | 1.17 (0.84–1.64); p = 0.3406 | The polymorphic allele was not associated with the risk of dyslipidemia |
| Wang et al. [15] | Interactions among genes involved in reverse cholesterol transport and in the response to environmental factors in dyslipidemia in subjects from the Xinjiang rural area | Case-control | 2018 | China | 1,433 | 16% | 1.34 (1.06–1.69);p = 0.0141 | The polymorphic allele was associated with a higher risk of dyslipidemia |
| Hosseini-Esfahani et al. [16] | Dietary patterns interact with APOA1/APOC3 polymorphisms to alter the risk of the metabolic syndrome: the Tehran Lipid and Glucose Study | Case-control | 2015 | Iran | 828 | 29% | 1.11 (0.85–1.46);p = 0.4433 | The polymorphic allele was not associated with the risk of metabolic syndromes |
HDL-C: high-density lipoprotein cholesterol; MAF: minor allele frequency.
The development of dyslipidemia associated with the presence of the polymorphic allele was evaluated in the five selected articles (including their references), with 3,539 individuals manifesting dyslipidemia associated with the presence of the polymorphic allele, compared to 4,166 individuals who did not. The presence of the polymorphic allele was associated with the development of dyslipidemia in relation to non-carriers (RR = 1.16; 95% CI 1.09–1.23; p < 0.01). No significant heterogeneity was observed between the selected studies (I² = 0%; τ² = 0; p = 0.57) (Figure 2).
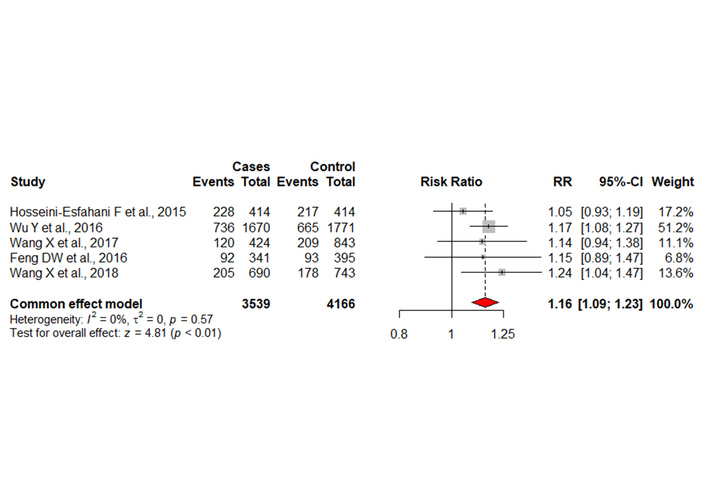
Table and graph representing the meta-analysis. Cases: individuals with dyslipidaemia; Control: healthy individuals; Events: individuals with APOA1 rs670 polymorphisms. RR: relative risk.
This meta-analysis showed that the presence of the rs670 polymorphic allele in the APOA1 gene is associated with a significant increase in the risk of developing metabolic disorders, with a RR of 1.16 (95% CI: 1.09–1.23; p < 0.01). This finding, obtained from the joint analysis of five case-control studies, demonstrates a consistent association, reinforced by the absence of significant heterogeneity (I² = 0%; τ² = 0; p = 0.57). Such consistency strengthens the robustness of the effect of the rs670 polymorphism on predisposition to lipid disorders.
The results presented corroborate previous evidence pointing to the functional role of rs670 in regulating APOA1 gene expression. In addition, reduced Apo A-I has been associated with unfavorable lipid profiles and increased cardiovascular risk [6, 16–19]. Furthermore, the interaction between genetic and environmental factors, such as the dietary patterns of each population, has also been highlighted in previous studies. It has been found, for example, that Western diets, which are generally rich in saturated fats, increase the risk of MS in carriers of the polymorphic allele of SNP rs670 [17].
Another relevant aspect is the influence of this polymorphism on the modulation of the inflammatory response, given that evidence suggests that, through the regulation of the ATP-binding cassette transporter A1-Janus kinase 2-Signal transducer and activator of transcription 3 pathway—tristetraprolin (TTP) [ATP-binding cassette transporter A1 (ABCA1)–Janus kinase 2 (JAK2)–signal transducer and activator of transcription 3 (STAT3)–TTP pathway], the variant may contribute to both the development of dyslipidemia and the progression of atherosclerotic processes [20]. Thus, although some individual studies have reported divergent results regarding the association of the rs670 polymorphism with metabolic disorders, the meta-analysis conducted by increasing statistical power confirms the clinical relevance of this genetic marker. In addition, analysis of the APOA1/C3/A4/A5 gene cluster revealed that variants in adjacent genes, such as APOA5 rs662799, also interact with environmental factors, such as smoking and alcohol consumption, to modulate the risk of MS. These findings reinforce the complexity of the genetic regulation of lipid metabolism [6].
In this context, the discovery of polymorphisms in the APOA1 promoter region represented an important milestone in understanding the regulation of its gene expression, given that in 1984, a CpG site in the promoter of this gene was identified as a hot spot for polymorphism using the restriction enzyme MspI, which was used to identify the rs670 polymorphism. As CpG sites are highly susceptible to methylation, they are particularly prone to spontaneous modifications. In this context, the deamination of 5-methylcytosine can promote the transition from cytosine (C) to thymine (T), which resulted in the characterization of the rs670 (C > T) polymorphism, located about 75 base pairs upstream of the transcription start region 9.
Over the years, however, the exact position of this polymorphism has been described in different ways. Initially, it was reported as a substitution of guanine (G) for adenine (A) in the DNA template strand at different positions (−78G > A, −76G > A, and −75G > A) [19, 20]. Subsequently, however, the definition was consolidated as a −75G > A substitution in the template strand and rs670 (C > T) in the coding strand [21, 22]. Thus, considering that it is located in the promoter, this polymorphism can alter the transcriptional activity of the gene, modulating the synthesis of Apo A-I. Therefore, its implications are relevant, since the variation in the expression of this Apo can directly impact the regulation of lipid metabolism.
Thus, the reduction in Apo A-I expression caused by genetic factors such as the rs670 polymorphism or by the regulatory action of the long non-coding RNA APOA1-AS may directly impact HDL particle formation [9, 23]. With less Apo A-I available, HDL synthesis decreases, resulting in particles with lower protein content and compromising their essential functions, such as reverse cholesterol transport, antioxidant and anti-inflammatory activities, and the ability to modulate glycemic control [24]. Consequently, this functional reduction contributes to greater cholesterol accumulation in macrophages, favoring the formation of foam cells and the progression of atherosclerosis [25].
In addition, Apo A-I plays an essential role in biological systems, for example, in conjunction with Apo C-I, being responsible for the activation of LCAT. It is also known that Apo A-I is essential in the reverse transport of cholesterol, promoted by HDL, being a molecule that protects against cardiovascular disease (CVD) and inflammatory cases, also being responsible for inhibiting the expression of adhesion molecules that are very important in pro-inflammatory stimulation, such as intercellular adhesion molecule (ICAM), vascular cell adhesion molecule (VCAM), and selectins, through the release of miRNA 223 [5, 24].
As previously mentioned, the pathophysiology of atherosclerosis is marked by the formation of foam cells, appearing in the capillary intima, through factors such as high uptake of oxidized low-density lipoprotein (LDL-ox) cholesterol and excessive cholesterol esterification, resulting in cholesterol esters stored in the form of intracellular fatty droplets, which form foam cells through the aggregation of activated macrophages [26, 27].
Recently, the influence of SNP on the prevalence of CVD has been investigated, and among the genetic variations, SNP rs670 was classified as a risk allele associated with the onset of CVD [18, 19], including atherosclerosis, which may be due to its impaired HDL functionality associated with the polymorphic allele, which may generate inefficient LCAT activation, favoring the formation of LDL-ox that causes foam cell differentiation [5].
It should also be noted that among the limiting factors of the study is the lack of heterogeneity among the studies, indicating that the effect of rs670 is relatively stable in different populations. However, it should be recognized that most of the studies included were conducted in Asian populations, which may limit the extrapolation of results to other ethnic groups. According to data provided by the ALFA (allele frequency aggregator) project, a database that provides allele frequencies for more than 1 million individuals from 12 different populations (including European, Asian, African, Latin American, and others), it was observed that the frequency of the polymorphic allele in the Asian population averages 19%. This justifies the higher prevalence of studies in the Asian population [19]. Also, herein, sex and dietary conditions were not considered for the analysis.
Thus, future investigations involving more diverse samples and in-depth functional analyses are necessary to clarify the mechanisms involved in a more comprehensive manner.
Furthermore, this meta-analysis demonstrated the influence of SNP rs670 on the occurrence of metabolic disorders. The presence of the rs670 polymorphism in the APOA1 gene also predisposes individuals to lipid disorders when compared to the control group, which may lead to a higher occurrence of cardiovascular events, such as the formation of atheromatous plaques, which may have a positive correlation with the presence of SNP rs670. Finally, future studies should seek to explore statistical analyses covering greater ethnic diversity, as well as possible gene-environment interactions that influence the phenotypic expression of this polymorphism.
Apo: apolipoproteins
CI: confidence interval
CpG: cytosine-phosphate-guanine
CVD: cardiovascular disease
GWAS: genome-wide association study
HDL: high-density lipoprotein
LCAT: lecithin-cholesterol acyltransferase
LDL-ox: oxidized low-density lipoprotein
MAF: minor allele frequency
MS: metabolic syndrome
RR: relative risk
SNP: single-nucleotide polymorphism
TTP: tristetraprolin
The authors would like to thank the Federal University of Ceará and Clinical Biochemistry Laboratory.
CFA: Conceptualization, Formal analysis, Methodology, Writing—original draft, Writing—review & editing. MOF: Conceptualization, Investigation, Writing—original draft. AGdSF: Data curation, Writing—review & editing. SMGO: Conceptualization, Writing—original draft. LMK: Formal analysis, Writing—original draft. EPM: Conceptualization, Formal analysis, Methodology, Writing—original draft, Writing—review & editing. GdAV: Data curation, Writing—review & editing. BRD: Formal analysis, Writing—original draft. IMMT: Conceptualization, Formal analysis. NMLP: Supervision, Validation. MDRdC: Conceptualization, Formal analysis, Methodology, Writing—original draft, Writing—review & editing. RRPPBdM: Supervision, Validation. TLS: Supervision, Validation. All authors read and approved the submitted version.
The author declares that there are no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
The datasets analyzed for this study can be found in PubMed (https://pubmed.ncbi.nlm.nih.gov/) and the LitVar2 database (https://www.ncbi.nlm.nih.gov/research/litvar2/). The datasets that support the findings of this study are available from the corresponding author upon reasonable request.
Not applicable.
© The Author(s) 2026.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2026. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 540
Download: 30
Times Cited: 0
