Affiliation:
Research and Development, Vedanadhi, Salem 636115, Tamil Nadu, India
Email: drvishnushivam@gmail.com
ORCID: https://orcid.org/0000-0002-1998-4199
Affiliation:
Research and Development, Vedanadhi, Salem 636115, Tamil Nadu, India
ORCID: https://orcid.org/0009-0001-9757-5841
Affiliation:
Research and Development, Vedanadhi, Salem 636115, Tamil Nadu, India
ORCID: https://orcid.org/0009-0003-6011-3445
Explor Endocr Metab Dis. 2025;2:101439 DOI: https://doi.org/10.37349/eemd.2025.101439
Received: May 26, 2025 Accepted: July 18, 2025 Published: August 07, 2025
Academic Editor: Aimin Xu, The University of Hong Kong, China
Background: Emerging evidence suggests that genetic variations in taste receptor genes may influence dietary behaviors, energy homeostasis, and metabolic risk, contributing to type 2 diabetes mellitus (T2DM) pathogenesis. The objective of this study is to evaluate the association between single nucleotide polymorphisms (SNPs) in taste receptor genes and T2DM.
Methods: This systematic review followed the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) 2020 guidelines and was registered with the International Prospective Register of Systematic Reviews (PROSPERO; CRD42022351880). A comprehensive literature search was conducted across PubMed, ScienceDirect, Cochrane Library, and Google Scholar through June 2025. Original studies examining SNPs in taste receptor genes among individuals with T2DM were included. Quality assessment was performed independently by using the Newcastle-Ottawa scale.
Results: Sixteen studies involving diverse populations met the inclusion criteria. Significant associations with T2DM were observed for SNPs in type 2 taste receptor gene family R member 3 (TAS2R3; rs11763979), TAS2R4 (rs2233998), TAS2R7, TAS2R9, TAS2R38, TAS2R50, cluster determinant 36 (CD36; rs1761667, rs3211956, rs7755), carbonic anhydrase VI gene (CA6; rs2274327), transient receptor potential vanilloid-1 (TRPV1; rs161364, rs8065080), transient receptor potential cation channel subfamily M gene member 5 (TRPM5; rs4929982), and TRPM8 (rs12472151). These polymorphisms may alter taste perception and gut hormone responses [e.g., glucagon-like peptide 1 (GLP-1)], affecting dietary intake, satiety, insulin secretion, and glucose regulation.
Discussion: The findings suggest that genetic variations in taste receptor genes may contribute to T2DM through behavioral and metabolic mechanisms. Incorporating gustatory phenotyping with genotypic profiling could enable personalized dietary strategies and inform novel therapeutic approaches targeting taste-mediated gut signaling. Further large-scale, multi-ethnic, and mechanistic studies are warranted to confirm these associations and elucidate their clinical implications.
Over the past few decades, the global prevalence of type 2 diabetes mellitus (T2DM) has increased markedly, with projections estimating up to 7,862 cases per 100,000 individuals [1]. This upward trend may be attributed to a complex interplay of genetic and environmental risk factors, including obesity, insulin resistance, metabolic dysfunction, dietary habits, and epigenetic modifications [1]. In addition to these well-established risk factors, emerging evidence suggests that behavioral psychology and impaired satiety signaling also play a significant role in the pathogenesis of T2DM [2–4].
Recent studies have identified that single nucleotide polymorphisms (SNPs) in taste genes are significantly associated with elevated risk of metabolic syndrome, diabetes mellitus, obesity, carcinogenesis, Alzheimer’s disease, Parkinson’s disease, thyroid dysfunction, and substance use disorders [3–5]. Taste perception and signal transduction across the six taste modalities—sweet, salt, sour, bitter, umami, and fat taste—are mediated by various taste receptor genes such as type 1 taste receptor gene family R (TAS1R), TAS2R, sodium channel epithelial 1 (SCNN1), cluster determinant 36 (CD36), transient receptor potential cation channel subfamily M gene (TRPM), guanine nucleotide binding protein subunit alpha transducing 3 (GNAT3), carbonic anhydrase VI gene (CA6), IZUMO sperm-egg fusion 1 gene (IZUMO1), metabotropic glutamate receptor 1 gene (GRM1), and polycystic kidney disease (PKD)-like genes (e.g., PKD1L3, PKD2L1, and PKD2L3) [4–7]. Each taste modality is regulated by specific taste receptor genes. For instance: sweet taste is primarily mediated by TAS1Rs, TRPMs, and GNAT3; bitter taste by TAS2Rs, TRPMs, and CA6; salt taste by transient receptor potential vanilloid-1 (TRPV1; nerve endings) and SCNN1s (subunits alpha, beta, gamma, and delta); sour taste by TAS1Rs and PKD-like genes; umami taste by TAS1Rs, TRPMs, GNAT3, and GRM1; and fat taste by CD36 and IZUMO1 [5–7]. Taste perception begins with the interaction of food containing particular taste stimuli with oral and extra-oral taste receptors, triggering intracellular calcium release into the cytoplasm, depolarization of afferent nerve fibers and signal transduction via cranial nerves (facial nerve, glossopharyngeal nerve, sensory vagal afferents, trigeminal nerve and the trigeminal ganglion) to the central processing centers (nucleus of solitary tract, ventral posteromedial thalamic nucleus, the operculum, insular and the somatosensory cortex) [8]. These brain regions also influence gut hormone secretion [e.g., ghrelin, glucagon-like peptide 1 (GLP-1), glucose-dependent insulin-releasing peptide], thereby regulating satiety and energy homeostasis [8]. Genetic variations in taste receptors may impair this signaling cascade, leading to altered taste perception, eating behavior, impaired energy homeostasis, and increased susceptibility to T2DM [5]. Notably, TAS1R expression in the gut is upregulated in response to hyperglycemia in individuals with T2DM [9]. This suggests that modulating taste receptor pathways may have therapeutic potential, particularly through agents that mimic GLP-1 receptor agonists [2, 8].
Several studies have reported significant taste impairments in individuals with diabetes mellitus [10, 11]. Chamoun et al. [12] demonstrated associations between psychophysical measures of taste and 94 SNPs across 11 taste receptor genes, particularly those related to sweet, salty, umami, and fat taste perception. A recent review further highlighted the critical role of taste receptor function in the pathophysiology of T2DM and energy homeostasis [13].
Despite the growing body of evidence, no qualitative meta-synthesis has systematically evaluated the association between SNPs in taste receptor genes and T2DM. This study aims to evaluate the evidence on the association of taste gene polymorphisms and T2DM.
This qualitative meta-synthesis was registered with the International Prospective Register of Systematic Reviews (PROSPERO; Registration No. CRD42022351880) [14] and conducted in accordance with the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) 2020 guidelines.
A comprehensive literature search was performed across PubMed, ScienceDirect, Cochrane Library databases, and Google Scholar using the keywords “Diabetes mellitus” AND (“taste receptor gene” OR “taste gene” OR “taste gene polymorphisms” OR “taste gene mutations”). Manual screening of reference lists and citation tracking of relevant articles was also conducted to ensure thorough coverage of the literature up to June 2025. Inclusion criteria: Original research articles investigating SNPs in taste receptor genes among patients with T2DM were included. Exclusion criteria: Articles not relevant to the study, including review articles, case reports, editorials, consensus statements, clinical guidelines, conference abstracts, and book chapters, were excluded. Titles and abstracts were screened to identify potentially relevant records. Full-text screening of the identified records was assessed for inclusion by one reviewer and independently reassessed by the reviewer. The PICO framework for this study are as follows: (1) population: individuals diagnosed with T2DM, (2) intervention/exposure: presence of SNPs or variant genotypes in taste receptor genes, (3) comparison: comparison between individuals with variant versus wild-type genotypes, and (4) outcome: association between taste receptor polymorphisms and T2DM.
Relevant study characteristics were systematically extracted, including first author, year of publication, study design, country of origin, sample size, age of participants, diagnostic criteria for T2DM, taste receptor genotyping, and other characteristic features, from studies that met the inclusion criteria. Methodological quality assessment of the included studies was carried out with the Newcastle-Ottawa scale (Table S1) [3, 4]. Data extraction was performed by one reviewer and cross-verified by another reviewer.
The PRISMA flowchart (Figure 1) illustrates the selection process. A total of 5,712 records were identified through database searches and manual screening. After title and abstract screening, followed by full-text review, sixteen studies met the eligibility criteria and were included in the qualitative meta-synthesis. Four studies were excluded due to the inclusion of populations other than T2DM (one study was conducted in individuals with prediabetes [15], and three studies were conducted in patients with gestational diabetes mellitus (GDM) [16–18]).
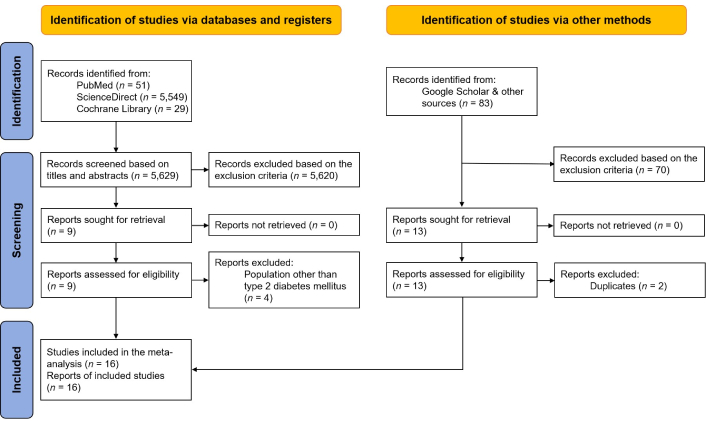
Flow chart summarizing the selection process. Adapted from [19], © 2021, The Author(s) (CC BY 4.0)
The characteristics of the included studies were summarized in Table 1 and Table S1. Diagnostic criteria for T2DM were based on the definitions used within each included study. Methodological quality assessment of the included studies by the Newcastle-Ottawa scale indicated that all sixteen studies were of good quality (Table S1).
General characteristics of the studies included in the qualitative meta-synthesis
| Study | Study design (country) | Population | Genotyping | Inference |
|---|---|---|---|---|
| Leprêtre et al. [20], 2004 | Cohort (France) | 454 | CD36 gene (entire sequence) | No significant association was found between CD36 gene and T2DM (P > 0.1) |
| Corpeleijn et al. [21], 2006 | Cohort (Netherlands) | 151 | CD36 gene SNP rs1527479 and 478 C/T substitution | CD36 gene SNP rs1527479 TT haplotype was significantly associated with T2DM (P = 0.035), fasting glucose concentration (P < 0.05) and insulin resistance (P < 0.05) |
| Dotson et al. [22], 2008 | Case-control (USA) | 503 | 70 SNPs in TAS1Rs and TAS2Rs gene subtypes | TAS2R3 gene SNP rs11763979 (P = 0.03), TAS2R7 gene SNPs rs2588350 (P = 0.0007) and rs619381 (P = 0.009), TAS2R9 gene SNP rs3741845 (P = 0.005), and TAS2R50 gene SNP rs6488334 (P = 0.04) were significantly associated with patients with T2DM |
| Banerjee et al. [23], 2010 | Case-control (India) | 400 | Two SNPs (rs1527483 and rs1761667) in the CD36 gene | CD36 SNP (G>A) rs1761667 GA was significantly associated with patients with T2DM (P < 0.001) |
| Wang et al. [24], 2012 | Case-control (China) | 113 | CD36 gene SNPs (rs1527483 and rs1049673) | CD36 SNP rs1049673 CG & GG haplotypes were significantly associated with impaired glucose tolerance (P = 0.023) and T2DM (P = 0.011) in patients with essential hypertension, respectively |
| Gautam et al. [25], 2015 | Case-control (India) | 100 | Six CD36 SNPs (rs1984112, rs1761667, rs1527479, rs3211938, rs1527483, and rs3212018) | CD36 SNPs rs1761667 (G>A) and rs3211938 (T>G) showed significant association with T2DM (GAATTC1, P < 0.001) |
| Tabur et al. [26], 2015 | Case-control (Turkey) | 308 | 25 TRPM1–8 gene SNPs rs28441327, rs11070811, rs2241493, rs111649153, rs1618355, rs1328142, rs3760663, rs34364959, rs4929982, rs886277, rs34551253, rs3986599, rs3750425, rs62569677, rs55924090, rs1016062, rs2362294, rs2362295, rs10490018, rs2052029, rs6431648, rs10803666, rs12472151, rs2215173, and rs6740118 | TRPM5 gene SNP rs4929982 A allele (P = 0.0019) and TRPM8 gene SNP rs12472151 C allele (P < 0.001) polymorphisms might be related to the individual susceptibility to metabolic syndrome (including T2DM) |
| Park et al. [27], 2016 | Cohort (South Korea) | 8,842 | 7 SNPs in TRPV1 gene such as SNPs rs161364, rs8065080, rs150908, rs222745, rs7217945, rs222741, and rs2737141 | TRPV1 gene SNPs rs161364 C allele (P = 0.0487) and rs8065080 C allele (P = 0.0378) were significantly associated with the prevalence of T2DM in dominant genetic models |
| Zhang et al. [28], 2018 | Case-control (China) | 546 | Four CD36 SNPs rs1194197, rs2151916, rs3211956, and rs7755 | Overweight/obesity individuals carrying SNP variant alleles of rs3211956 (GG+GT, P = 0.024) and rs7755 (AA+AG, P = 0.007) were associated with increased risk of T2DM compared to normal weight individuals carrying wild-type homozygous alleles |
| Fujii et al. [29], 2019 | Cross-sectional (Japan) | 495 | Two CD36 gene SNPs (rs1761667 and rs1527483) | CD36 gene SNP rs1761667 AA haplotype was associated with higher intake of total fat (P = 0.01) and monounsaturated fatty acids (P = 0.05) when compared to GG and GA haplotypes. In addition, the frequency of CD36 gene SNP rs1761667 GG haplotype was higher in T2DM |
| Mrag et al. [30], 2020 | Cohort (Tunisia) | 300 | CA6 gene SNP rs2274327 | The CA6 gene SNP rs2274327 T allele in its dominant model (TT+CT vs. CC, 67.7% vs. 32.3%) was increasingly associated with T2DM. Similarly, taste impairment in T2DM was significantly associated with CA6 gene SNP rs2274327 T allele in its dominant model (OR = 1.97 [95% CI = 1.21 to 3.23], P = 0.006) |
| Hatmal et al. [31], 2021 | Case-control (Jordan) | 350 | CD36 gene rs1761667 (G>A) and rs1527483 (C>T) were genotyped | No significant association was observed between CD36 polymorphisms and patients with T2DM or dyslipidemia (P > 0.1) |
| Touré et al. [32], 2022 | Cross-sectional (Senegal) | 100 | 2 tag SNPs in CD36 (rs3211867 and rs1761667) | No significant difference was observed between controls and T2DM subjects (P = 0.9) |
| Franzago et al. [33], 2023 | Cohort (Italy) | 23 | CD36 gene SNPs rs1984112 (A>G) and rs1761667 (G>A), BMAL1 gene SNP rs7950226 (G>A), and CLOCK gene SNPs rs1801260 (A>G), rs4864548 (A>G), and rs3736544 (G>A) | CD36 gene SNP rs1761667 (G>A) A allele in its dominant form (AA+GG genotype) was significantly associated with patients with T2DM (P = 0.001) |
| Lee and Shin [34], 2023 | Cohort (Korea) | 4,552 | TAS2R4 SNP rs2233998 | TAS2R4 SNP rs2233998 TT haplotype was significantly associated with the incidence of T2DM in women (HR [95% CI] = 1.48 [1.13–1.93], P = 0.0182) |
| Husami et al. [35], 2025 | Case-control (India) | 680 | 2,658 gene variants | TAS2R38 genetic variants were associated with an increased risk of T2DM (P < 0.05) |
case: patients with T2DM as defined in the respective studies; control: normal healthy individuals as defined in the respective studies; CD36: cluster determinant 36; T2DM: type 2 diabetes mellitus; SNP: single nucleotide polymorphism; TAS1R: type 1 taste receptor gene family R; TRPM1: transient receptor potential cation channel subfamily M gene member 1; TRPV1: transient receptor potential vanilloid-1; CA6: carbonic anhydrase VI gene; OR: odds ratio; CI: confidence intervals; BMAL1: brain and muscle aryl hydrocarbon receptor nuclear translocator-like protein-1; CLOCK: circadian locomotor output cycles kaput; HR: hazard ratio
The results revealed that SNPs in taste genes including TAS2R3 gene (SNP rs11763979), TAS2R4 gene (SNP rs2233998), TAS2R7 gene (SNPs rs2588350 and rs619381), TAS2R9 gene (SNP rs3741845), TAS2R38, TAS2R50 gene (SNP rs6488334), TRPV1 gene (SNPs rs161364 and rs8065080), CD36 gene (SNPs rs1761667, rs3211956, rs7755, rs1049673, and rs1527479), and CA6 gene (SNP rs2274327) were significantly associated with patients with T2DM. Additionally, TRPM5 gene (SNP rs4929982) and TRPM8 gene (SNP rs12472151) polymorphisms were significantly associated with metabolic syndrome, including T2DM.
The findings suggest that SNPs in several taste-related genes, including TAS2R3, TAS2R4, TAS2R7, TAS2R9, TAS2R38, TAS2R50, TRPV1, CD36, CA6, TRPM5, and TRPM8, may contribute to the etiology of T2DM and its associated metabolic complications. Although the precise mechanisms remain to be elucidated, current evidence supports the hypothesis that genetic variations in taste genes influence taste stimuli perception, individual food preferences, nutrient intake, and eating behavior, thereby increasing the risk of T2DM [22].
For instance, TAS2R9 is expressed in gut entero-endocrine L-cells and mediates GLP-1 secretion in response to sweet taste stimuli [22]. Canivenc-Lavier et al. [13] recently described local glucose-dependent GLP-1 secretion by taste bud cells and proposed taste receptors as potential targets for T2DM treatment [3]. Similarly, reduced CD36 expression due to genetic polymorphisms may influence fat taste sensitivity and intake behaviors, possibly exerting a protective effect due to reduced fat taste sensitivity [4, 33]. Variations in the TRPM5 gene are associated with reduced GLP-1 levels, impaired insulin release, and altered glucose tolerance, highlighting the metabolic relevance of taste receptor polymorphisms [3, 22].
This is in line with previous observations that the taste-dependent manner of GLP-1 secretion, glucose-stimulated insulin secretion, and insulin sensitivity in patients with T2DM might be associated with the polymorphisms in taste receptor genes [3, 22, 26, 33]. These genetic variations may affect hunger signaling, gut motility, and taste-driven satiety hormone release [3, 4, 13]. For example, individuals carrying the TAS2R38 proline-alanine-valine (PAV) allele [a 6-n-propylthiouracil (PROP) taster genotype] exhibit heightened sensitivity to bitter compounds, which may impact food choices and contribute to dietary avoidance of bitter but nutritionally beneficial foods [4, 36–38]. Such phenotypic taste differences can now be identified using validated clinical gustatory tests (e.g., taste strips, solutions), which could help screen individuals at risk of metabolic diseases, including obesity and diabetes [3, 4, 36]. Moreover, pharmacologic agents that modulate the taste receptor expression based on an individual’s genotype may offer a novel therapeutic avenue [37]. Nevertheless, further large-scale molecular and clinical studies are warranted to validate these associations and uncover the underlying mechanisms driving gene-diet interactions in T2DM.
Overall, this study provides the first systematic synthesis of the association between taste receptor gene polymorphisms and T2DM. However, several limitations exist: (1) high methodological heterogeneity among included studies; (2) potential selection and publication biases; (3) lack of randomized controlled trials; and (4) insufficient data for meta-analysis. Allele frequency variations across ethnicities further limit generalizability. Due to insufficient data to conduct a statistical analysis for all the genes included in the present study, the meta-analysis was not feasible. To address the potential heterogeneity in genotyping methods, variability in study designs, population characteristics, and definitions of T2DM in the included studies, Table 1 and Table S1 summarize the comparison of the characteristics of the included studies, including study design, sample size, country or location of study, and main findings. The risk of bias was assessed using the Newcastle-Ottawa scale, and narrative synthesis and reporting followed the PRISMA 2020 guidelines. Further, citation searches and gray literature searches were performed and a predetermined inclusion and exclusion criteria based on the PROSPERO protocol was used to select the studies for inclusion.
Further molecular and clinical studies, particularly randomized controlled trials and large, multi-ethnic cohort studies, are warranted to validate the role of taste receptor gene polymorphisms in the pathophysiology of T2DM. Gustatory testing to assess taste phenotypes, when paired with genotypic profiling, may offer a cost-effective approach to identify at-risk individuals and guide personalized dietary interventions. Exploration of taste modulators and GLP-1 analogs targeting specific taste receptors may open new therapeutic avenues in obesity and T2DM management. Moreover, SNPs in taste genes could serve as genetic markers for early detection and risk stratification.
In summary, this review provides evidence that SNPs in taste receptor genes are associated with T2DM. Altered taste perception and signal transduction may influence eating behavior, energy homeostasis, and glucose metabolism. Identifying taste phenotypes and targeting taste receptor gene expression may represent promising strategies for the prevention and treatment of T2DM. However, larger, well-designed studies are needed to confirm these associations and facilitate their clinical translation.
CA6: carbonic anhydrase VI gene
CD36: cluster determinant 36
GLP-1: glucagon-like peptide 1
GNAT3: guanine nucleotide binding protein subunit alpha transducing 3
GRM1: metabotropic glutamate receptor 1 gene
IZUMO1: IZUMO sperm-egg fusion 1 gene
PKD: polycystic kidney disease
PRISMA: Preferred Reporting Items for Systematic reviews and Meta-Analyses
PROSPERO: International Prospective Register of Systematic Reviews
SCNN1: sodium channel epithelial 1
SNPs: single nucleotide polymorphisms
T2DM: type 2 diabetes mellitus
TAS1R: type 1 taste receptor gene family R
TRPM: transient receptor potential cation channel subfamily M gene
TRPV1: transient receptor potential vanilloid-1
The supplementary materials for this article are available at: https://www.explorationpub.com/uploads/Article/file/101439_sup_1.pdf.
VS: Conceptualization, Visualization, Methodology, Investigation, Data curation, Writing—original draft, Writing—review & editing. SK: Writing—review & editing. VH: Writing—review & editing.
Part of the manuscript has been presented as a poster presentation in the International Diabetes Federation Congress 2025 and the preparation of this manuscript has not been influenced by the presentation or any other factors.
Not applicable.
Not applicable.
Not applicable.
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
