Affiliation:
1Chemistry Department, Universidad Nacional de Colombia, Bogotá 111321, Colombia
Email: murquizam@unal.edu.co
ORCID: https://orcid.org/0000-0001-5478-606X
Affiliation:
1Chemistry Department, Universidad Nacional de Colombia, Bogotá 111321, Colombia
ORCID: https://orcid.org/0009-0007-2398-0813
Affiliation:
2Curauma Biotechnology Center, Pontificia Universidad Católica de Valparaíso, Valparaíso 2362807, Chile
ORCID: https://orcid.org/0000-0002-7829-0568
Explor Drug Sci. 2025;3:1008114 DOI: https://doi.org/10.37349/eds.2025.1008114
Received: March 09, 2025 Accepted: May 13, 2025 Published: June 25, 2025
Academic Editor: Cheorl Ho Kim, Sungkyunkwan University, Samsung Advances Institute of Health Science and Technology (SAIHST), Republic of Korea
Merozoite invasion of erythrocytes relies on molecular interactions between parasite and host proteins, making these proteins potential therapeutic targets. This review summarizes research on Plasmodium falciparum merozoite invasion conducted at the Instituto de Inmunología, San Juan de Dios Hospital, between 1990 and 2000. Erythrocyte-binding analyses of P. falciparum proteins merozoite surface protein 1 (MSP-1), MSP-2, acid basic repeat antigen (ABRA) (MSP-9), apical membrane antigen-1 (AMA-1), rhoptry-associated protein 1 (RAP-1), glycophorin-binding protein 130 (GBP-130), serine repeat antigen 5 (SERA-5), erythrocyte-binding antigen 140 (EBA-140), and EBA-175 identified 50 high-activity binding peptides (HABPs) with nanomolar-range dissociation constants. Most of these peptides inhibit merozoite invasion and belong to erythrocyte-binding regions of their respective proteins. Several HABPs overlap with epitopes recognized by inhibitory monoclonal antibodies (mAbs). MSP-1 HABP-5501 contains epitopes for mAbs 12.8, 12.10, MaliM03 fragment antigen binding (Fab), and 42D6 Fab, all of which block invasion. MSP-2 HABPs include epitopes targeted by opsonizing antibodies, while MSP-9 HABPs (2148–2153) interact with Band 3 during invasion. AMA-1 HABPs (4315, 4316, and 4325) contribute to the 1F9 epitope, a key target of immune recognition. RAP-1 HABP-26188 elicits inhibitory antibodies, including cross-reactive mAbs SP5.2 and SP8.18, the latter displaying parasite growth inhibition. GBP-130 HABP-2220 binds glycophorin and induces invasion-blocking antibodies. SERA-5 HABP-6725 contains sequences targeted by native SERA-5 antibodies, while HABPs 6727 and 6733 are recognized by antibodies from natural infection and vaccination. EBA-140 HABPs (26135, 26144, and 26147) are located in Region II (RII) and are recognized by mAbs with moderate to strong neutralizing activity. EBA-175 HABPs (1779, 1783, 1814, 1815, and 1818) are in recombinant fragments recognized by antibodies eluted from immune complexes. HABPs 1779 and 1783, located in RII, bind erythrocytes independently of sialic acid, inhibit rRII-EBA binding, and interact with α(2,3)-sialyllactose. HABP-1783 also contains the target site of mAb R217, which potently blocks EBA-175 binding to glycophorin A and inhibits invasion.
Malaria, a parasitic disease, is a significant public health concern, particularly in tropical regions, according to the World Malaria Report 2023 issued by the World Health Organization (WHO) [1]. Malaria incidence rates have globally decreased by an average of 0.80% per year from 1990 to 2019. However, since 2015, incidence has unexpectedly increased in certain regions, particularly in Sub-Saharan Africa, where children under five years old are disproportionately affected [2]. Additionally, anti-malarial drug resistance has been reported in the Greater Mekong Subregion [3], and there is strong evidence that climate change—especially variations in temperature and precipitation—is influencing the epidemiology of malaria [4]. The development of effective strategies to control malaria is crucial, particularly in vulnerable regions where the disease burden remains high. The life cycle of Plasmodium spp., the parasite responsible for malaria, alternates between Anopheles mosquitoes and vertebrate hosts, requiring specialized forms to invade specific cell types at different stages [5].
In mammals, particularly humans, sporozoites are injected during an Anopheles mosquito bite, migrate to the liver, and infect hepatocytes. Following hepatic development, merozoites are released into the bloodstream, where they invade erythrocytes and continue the asexual replication cycle (Figure 1) [6]. This invasion process is highly specialized, as Plasmodium spp. has evolved unique mechanisms to penetrate and survive within host cells, particularly erythrocytes, making it a key target for treatment strategies [7]. Merozoite invasion of erythrocytes involves numerous parasite and host proteins and follows distinct pathways that can be broadly classified into sialic acid (SA) dependent and SA independent (glycophorin-independent) pathways [8].
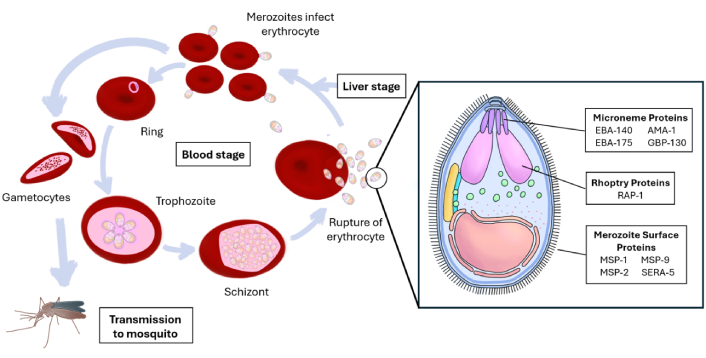
The Plasmodium falciparum blood stage (asexual replication cycle), emphasizing the merozoite structure with important proteins involved in the invasion of RBCs. RBCs: red blood cells; MSP-1: merozoite surface protein 1; EBA-140: erythrocyte-binding antigen 140; AMA-1: apical membrane antigen-1; GBP-130: glycophorin-binding protein 130; RAP-1: rhoptry-associated protein 1
This review focuses on the molecular interactions between Plasmodium falciparum merozoite proteins and erythrocyte proteins that facilitate red blood cell (RBC) invasion. Due to the complexity of the malaria parasite, covering all molecular interactions in a single review is challenging. Therefore, we summarize research on P. falciparum merozoite invasion conducted at the Instituto de Inmunología, San Juan de Dios Hospital, Bogotá, Colombia, from 1990 to 2000, before its transition into La Fundación Instituto de Inmunología de Colombia (FIDIC). This review represents only the first part of the peptide-based research carried out at the institute under the direction of Professor Manuel E. Patarroyo.
This research was conducted at a time when no structural information on these proteins was available. The findings presented in this review were obtained using over a thousand synthetic peptides synthesized via the t-Boc methodology, utilizing devices designed and developed in Bogotá, Colombia. All peptides were radiolabeled with iodine-125, a gamma emitter, to study their interactions with erythrocytes.
The P. falciparum genome contains 14 chromosomes with approximately 5,300 protein-encoding genes [9]. Several of these proteins are involved in merozoite invasion of erythrocytes and are located mainly on the merozoite surface and in the apical organelles. Most of them interact with erythrocyte proteins during the invasion process. At least 15–20 different merozoite proteins are involved in the erythrocyte invasion [9].
The merozoite surface is covered in a “fuzzy” fibrillar coat of proteins, including merozoite surface protein 1 (MSP-1), the most abundant surface protein, forming a complex with MSP-6 and MSP-7 [10] and MSP-2, MSP-3, MSP-4, MSP-5, MSP-8, MSP-9, MSP-10 [11] and PfMSA180 a recently identified protein involved in the invasion process [12]. Moreover, MSPDBL1 and MSPDBL2 bind to MSP-1 to form complexes [9].
The merozoite contains specialized organelles called rhoptries and micronemes at its apex, which store and release invasion-related proteins [13]. Amongst these erythrocyte-binding antigen (EBA) family involved in specific receptor-ligand interactions during invasion [7]; and reticulocyte binding-like homologous [P. falciparum reticulocyte binding-like homologous (PfRH)] proteins, working alongside EBA proteins in alternate invasion pathways [7] and rhoptry-associated proteins (RAPs) as part of the invasion machinery [13].
The merozoite invasion process of erythrocytes begins with initial attachment, followed by reorientation, tight junction formation, and internalization, each step requiring specific protein interactions [7]. P. falciparum merozoites can initially contact erythrocytes at any point on their surface [12], rolling over the erythrocyte membrane to establish receptor-ligand interactions between merozoite and erythrocyte surface proteins. They then rapidly reorient, positioning their apical end toward the erythrocyte membrane [13]. At this stage, specialized apical organelles rhoptries, micronemes, and dense granules release their contents in a regulated sequence [13]. A tight junction then forms between the merozoite and erythrocyte, creating an electron-dense thickening on the erythrocyte membrane [12]. The merozoite actin-myosin motor is activated at this contact point [13], propelling the parasite into the host cell. The tight junction moves along the merozoite surface as the parasite enters the erythrocyte [12]. Once inside, the parasite forms a parasitophorous vacuole (PV), where it will reside and develop [9]. The entire invasion process is extremely rapid, typically completed in less than two minutes [12].
Some of the MSPs involved in erythrocyte invasion are:
MSP-1: Major surface protein; processed into fragments essential for invasion.
MSP-2 and MSP-3: Involved in immune evasion and binding to erythrocytes.
MSP-6 and MSP-7: Function in complex with MSP-1 to enhance invasion.
MSP-10: Another key surface protein with a role in invasion.
MSP-1, MSP-3, MSP-7, serine repeat antigen 4 (SERA-4), and SERA-5 are cleaved and shed at the tight junction. Glycosylphosphatidylinositol (GPI)-anchored proteins MSP-2 and MSP-4 are carried into the erythrocyte without detectable processing. MSP-2 rapidly degrades within 10 min post-invasion, while MSP-4 is maintained for hours.
Carbohydrates are important for the invasion of merozoite, as previously mentioned, the invasion of RBCs can occur through SA-dependent and SA-independent pathways, depending on the ligand-receptor interactions involved [8]. SAs are terminal carbohydrate moieties on cell glycocalyx, most of them are linked to glycoproteins [14], it was shown that protein EBA-175 ligand recognizes glycophorin A (GPA), the EBA-140 ligand binds to glycophorin C (GPC) [15]. Additionally, glycophorin-binding protein 130 (GBP-130) may also interact with sialylated receptors, though its binding mechanism is less well-defined [16].
The SA-dependent pathway plays a vital role in merozoite invasion by targeting abundant GPA/GPC receptors on RBCs for efficient adhesion. This strategy also aids immune evasion by mimicking host-cell interactions. Therapeutically, disrupting SA-dependent interactions offers promising avenues for vaccine and drug development [8]. Additionally, P. falciparum exhibits remarkable adaptability by maintaining redundant invasion strategies; if SA-dependent pathways are compromised, the parasite can switch to SA-independent mechanisms [e.g., MSP-1, MSP-2, and apical membrane antigen-1 (AMA-1)] to ensure successful RBC entry [17, 18].
Carbohydrates are also associated with GPI-anchored proteins (e.g., MSP-1 and MSP-2), which mediate initial attachment of the merozoite to the RBCs surface [19]. For instance, MSP-1, one of the most abundant GPI-anchored proteins, is involved in binding to host cell receptors, facilitating the subsequent invasion process [20]. These proteins undergo extensive proteolytic processing which modifies their structure, allowing them to interact with host cell receptors. GPI-anchored proteins exhibit substantial polymorphism, which can affect their immunogenicity and the ability of vaccines to provide broad protection against malaria. This variability necessitates careful consideration in vaccine design [19–21].
As of 2025, malaria vaccine development is based on candidates targeting all parasite life stages. The WHO-approved RTS,S/AS01 and R21/Matrix-M (showing 75% efficacy in Phase 3) both target circumsporozoite protein, while blood-stage candidates like RH5.1/Matrix-M demonstrate 55% six-month efficacy in African children [22–24]. However, challenges persist with blood-stage vaccines due to antigenic polymorphism (e.g., AMA-1 variants showing strain-specific efficacy) and redundant invasion pathways. Other promising approaches include whole-sporozoite vaccines (PfSPZ) achieving 90% sterile protection in challenge trials, and transmission-blocking candidates like ProC6C targeting mosquito-stage antigens. In the case of the RTS,S/AS01 malaria vaccine represents a significant advancement but faces several challenges, including moderate efficacy (36–50% in clinical trials) and waning protection over time. Vaccine efficacy declines from approximately 55% in the first year to 28% by four years post-vaccination, necessitating booster doses to maintain protection [22–25]. Furthermore, co-administration with other candidates like FMP1/AS02 revealed immunological interference, reducing anti-RTS,S antibody titers by 30–40% without compromising efficacy in controlled human malaria infection models [26]. The FMP1/AS02 vaccine itself showed no significant protection in Kenyan children despite promising sporozoite challenge data, underscoring the difficulty of translating immunogenicity to real-world efficacy in endemic settings.
MSP-1 is the most abundant P. falciparum MSP, comprising 40% of the GPI-anchored coat [5]. Its essential role in merozoite survival is supported by the fact that attempts to disrupt the MSP-1 gene have failed [12]. MSP-1 mediates merozoite attachment to RBCs, initiating invasion through weak receptor-ligand interactions and heparin-like receptors. The carboxyl-terminus of MSP-1 interacts with RBC Band 3, while the amino-terminus binds to GPA. Mapping of the binding domains has confirmed a direct interaction between MSP-1 and GPA within a region known to strongly inhibit P. falciparum invasion of human RBCs. Additionally, a genetically modified mouse model lacking the GPA-Band 3 complex in RBCs exhibits complete resistance to malaria infection in vivo. These findings suggest that the MSP-1-GPA-Band 3 complex plays a crucial role in the malaria parasite invasion [27]. On the other hand, MSP-1 forms invasion complexes with MSP-3, MSP-6, MSP-7, MSPDBL1, and MSPDBL2, which bind independently but contribute to erythrocyte recognition and invasion, and directly bind to human erythrocytes [28].
MSP-1 undergoes proteolytic processing, generating fragments of 83, 38, 33, and 19 kDa, with MSP-119 remaining membrane-anchored and entering RBCs during invasion [12]. These fragments remain assembled within the MSP-1 structure, which features coiled-coil helical domains with a central cavity, structurally resembling proteins involved in membrane and cytoskeletal interactions [29]. MSP1-42, a key protein fragment in merozoite invasion, binds specifically to heparin, with similar interactions observed for its MSP1-33 fragment but not in MSP1-19. Notably, heparin-like molecules optimally inhibit invasion when highly sulfated (≥ 2 sulfate groups) [30]. Additionally, a 115-amino acid region (p115MSP-1) within the p38 domain of MSP-1 binds specifically to human erythrocytes, independent of GPA, and significantly inhibits P. falciparum invasion in vitro. This region is recognized by sera from individuals in malaria-endemic regions of Africa, and polyclonal antibodies against p115MSP-1 prevent parasite invasion by nearly 90% in vitro [31].
MSP-1 is a major target for opsonizing antibodies [32]. Moreover, MSP-1 contains epitopes that elicit antibodies capable of inhibiting merozoite invasion, which are distributed across its processing products: p83, p30, p38, and p42. These antibodies can act synergistically to enhance their inhibitory effects. Additionally, rabbit antibodies against MSP-1 processing products can inhibit both erythrocyte invasion and intraerythrocytic parasite growth by targeting multiple MSP-1 epitopes [33]. Antibodies targeting MSP-1 and its complexes inhibit P. falciparum invasion by preventing the essential protein shedding required for erythrocyte entry [33]. In particular, antibodies targeting the MSP-1 p83 fragment disrupt all MSP-1 complexes, blocking merozoite attachment and significantly inhibiting parasite growth in vitro [29]. On the other hand, antibodies specific to MSP-119 inhibit parasite growth through multiple mechanisms, including blocking MSP-1 secondary processing and directly preventing invasion. Invasion-inhibitory antibodies targeting MSP-119 significantly reduce infection risk [34]. Interestingly, antibodies against 3D7 MSP-1 cross-inhibit the replication of the FCB-1 strain [33].
Nine high-activity binding peptides (HABPs) within MSP-1 that bind to erythrocytes were identified at the Instituto de Inmunología, San Juan de Dios Hospital (Table 1). These include peptides 1513 and 1522 in the 83 kDa fragment; 1577 and 1582 in the 38 kDa fragment; 1585, 1589, 1590, and 1591 in the 33 kDa fragment; and 5501 in the 19 kDa fragment. These peptides were discovered from a library of 78 synthetic peptides covering the entire MSP-1 molecule. They exhibit binding affinities to RBCs ranging from 140 nM to 250 nM, and some have been shown to inhibit merozoite invasion of erythrocytes [35]. Additionally, certain peptides may interact with erythrocyte receptors such as Band 3 and GPA [36].
Peptides of MSP-1, their sequence and in vitro invasion inhibition with 200 μM
| Peptide | AA | Sequence* | Invasion inhibition (% ± SD) |
|---|---|---|---|
| 1513 | 20 | 42GYSLFQKEKM | 60 ± 10 |
| 1522 | 20 | 202QIP | 60 ± 14 |
| 1577 | 20 | 1122F | 43 ± 3 |
| 1582 | 20 | 1222 | 50 ± 2 |
| 1585 | 20 | 1282 | 3 ± 8 |
| 1589 | 20 | 1362L | 29 ± 1 |
| 1590 | 20 | 1382 | 10 ± 5 |
| 1591 | 20 | 1402 | 0 ± 7 |
| 5501 | 20 | QGMLNISQHQCVKKQCPQNS | 30 ± 4 |
* The bolded residues are variable residues across different strains; green is for K1, purple is for two or more strains (Dd2, 3D7, K1, HB3). AA: aminoacids; MSP-1: merozoite surface protein 1
HABPs 1513, 1522, 1577, 1585, 1589, 1590, and 1591 are predominantly located in α-helices that are largely surface-exposed in the three-dimensional structure of MSP-1 [35] (PDB 6ZBH). In contrast, HABP-5501 is situated within an exposed loop in the EGF-1 domain structure [37, 38] (PDB 6XQW and 1OB1). Notably, HABPs 1585, 1589, 1590, and 1591 form a contiguous surface-exposed region that could serve as a target for antibodies (Figure 2). Certain HABPs exhibit immunogenic properties, inducing antibodies that recognize both MSP-1 and the merozoite surface. For instance, peptide 1513 elicits antibodies capable of recognizing native MSP-1 and inhibiting merozoite invasion [39–42]. Additionally, a segment of peptide 1513’s sequence was incorporated into the SPf66 vaccine, which has demonstrated protective efficacy ranging from 31% to 60.2%, depending on the population and geographic factors [43].
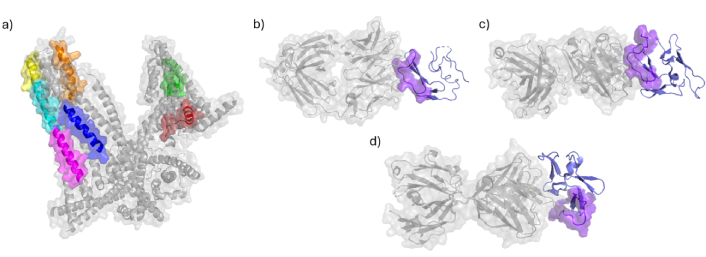
Peptides located on MSP-1 protein and its complexes. a) Structure of MSP-1 protein determined by electron microscopy (PDB ID: 6ZBH [44]) displays peptides 1513 in green and 1522 in red, 1577 in blue, 1582 was not crystallise, 1585 in orange, 1589 in magenta, 1590 in cyan and 1591 in yellow; b) crystal structure MaliM03 Fab in complex with PfMSP1-19 (PDB ID: 6XQW [45]) determined by X-ray diffraction displays peptide 5501 in purple; c) crystal structure of a Fab complex with PfMSP1-19 (PDB ID: 1OB1 [46]) determined by X-ray diffraction displays peptide 5501 in purple; d) crystal structure of potently neutralizing human monoclonal antibody 42D6 Fab in complex with MSP1-19 (PDB ID: 8DFG [47]) determined by X-ray diffraction displays peptide 5501 in purple. MSP-1: merozoite surface protein 1; Fab: fragment antigen binding
The first EGF-like domain of the 19 kDa fragment, where peptide 5501 is located, is a critical target of MSP-1-specific monoclonal antibodies (mAbs). These mAbs are classified as inhibitory (preventing MSP-142 processing and erythrocyte invasion), blocking (interfering with the binding and function of inhibitory mAbs), or neutral (neither inhibitory nor blocking). Inhibitory mAbs 12.8 and 12.10, as well as blocking mAbs 1E1 and 7.5, recognize key residues within peptide 5501 (QGMLNISQHQCVKKQCPQNS; inhibitory residues underlined, blocking antibody targets in bold). Specifically, peptide CPQNSGCFR is recognized by mAb 12.8, while FQDMLNIS is targeted by mAb 1E1 (underlined and bold residues corresponding to peptide 5501) [48]. The mAbs 12.8 and 12.10 specifically target the 19 kDa subunit of MSP-1 (MSP-119) and have been shown to prevent both proteolytic cleavage of MSP-1 and erythrocyte invasion [49, 50]. Interestingly, human chimeric antibodies derived from mouse mAbs 12.8 and 12.10 retain their ability to effectively inhibit merozoite invasion and can elicit an immune response in humans [50].
Moreover, the structure of the fragment antigen binding (Fab) G17.12, which does not inhibit merozoite invasion, in complex with the 19 kDa fragment, reveals VKKQ as part of the contact region [46] (PDB 1OB1). In contrast, the MaliM03 Fab, which effectively inhibits P. falciparum merozoite invasion, interacts with the 19 kDa fragment through the QHQCVKKQCP region [37] (PDB 6XQW). The potency of inhibition depends on the nature of these interactions. For instance, the potently neutralizing human monoclonal antibody 42D6 Fab, in complex with MSP1-19, recognizes the QHQC motif within this peptide sequence, leading to strong inhibition of merozoite invasion [51] (PDB 8DFG) (Figure 2).
MSP-2 is a 30-kDa GPI-anchored protein on P. falciparum merozoites and a potential blood-stage vaccine candidate [51, 52]. PfMSP2 is essential for parasite growth and directly involved in merozoite invasion of erythrocytes, as it binds to human RBCs with high affinity facilitating merozoite entry and it is crucial for successful invasion during malaria infection [53, 54]. Synthetic peptides targeting MSP-2 can inhibit invasion, further supporting its role in this process [9].
MSP-2 is an intrinsically unstructured protein (IUP) that forms amyloid fibrils in its recombinant form, probably being part of a possible fibrillar merozoite surface coat. However, its native conformation may differ due to membrane anchoring and protein interactions [55]. Once inside the erythrocyte, PfMSP2 degrades within 10 min, suggesting a role during and immediately after invasion [56]. Monomeric and fibrillar forms of MSP-2 may elicit distinct immune responses [57]. Indeed, antibodies targeting PfMSP2 inhibit parasite growth in vitro and significantly block erythrocyte invasion in both Babesia bovis and P. falciparum [58, 59].
Due to its key role in invasion and its ability to generate immune responses against the parasite, MSP-2 is an important target for vaccine development [60]. However, the antibody response to MSP-2 is short-lived without re-exposure [53], suggesting that MSP-2-specific antibodies can serve as sensitive markers of recent parasite exposure [57]. MSP-2 is highly polymorphic, with conserved N- and C-terminal domains and two major allelic families 3D7 and FC27 [61]. Additionally, genetic variations in MSP-2 have been associated with disease severity [61].
Neutralizing anti-MSP-2 antibodies are associated with malaria resistance [62], and higher antibody affinity for the 3D7 allele correlates with a reduced risk of malaria [63]. Additionally, chimeric MSP-2 constructs have been shown to induce broad immune protection [64]. Evidence also supports strain-transcending opsonizing immunity mediated by anti-MSP-2 antibodies [65]. However, some anti-MSP-2 antibodies are internalized rather than inhibiting invasion, suggesting a complex interplay between immune recognition and parasite evasion [56].
A screening assay using peptides spanning the entire PfMSP2 sequence identified three HABPs (4044, 4045, and 4053) (Table 2), with significant specific binding to human erythrocytes [57]. HABPs 4044 and 4045 are located in the N-terminal region, while HABP 4053 is in the central region. Peptides 4044 and 4045 share the sequence INNAYNMSIR, with binding affinities of 72 nM and 49 nM, respectively. Their binding is affected by neuraminidase and trypsin but remains resistant to chymotrypsin degradation. Both peptides inhibit in vitro erythrocyte invasion by up to 95%, and immunization with a related octapeptide has demonstrated protective effects.
Peptides of MSP-2, their sequence and in vitro invasion inhibition with 200 μM
| Peptide | AA | Sequence* | Invasion inhibition (% ± SD) |
|---|---|---|---|
| 4044 | 20 | 21KN | 96 ± 12 |
| 4045 | 20 | 31INNAYNMSIRRMA | 31 ± 1 |
| 4053 | 20 | 111 | 95 ± 5 |
* The bolded residues are variable residues across different strains; green is for K1, purple is for two or more strains (3D7, 7G8, K1). MSP-2: merozoite surface protein 2; AA: aminoacids
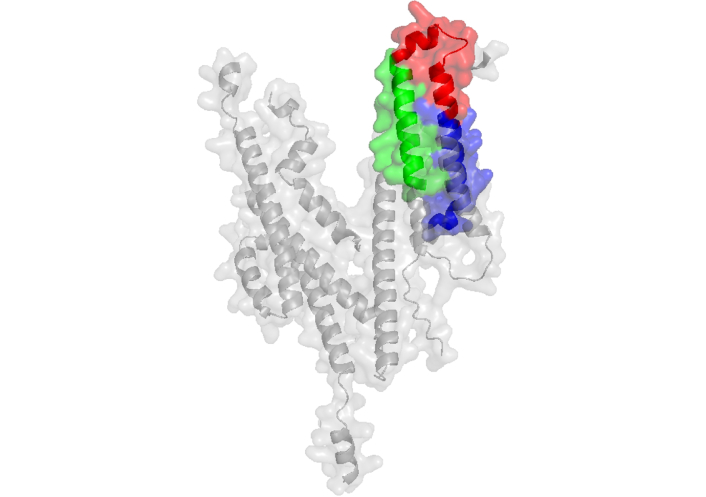
Immunization of rabbits with recombinant MSP-2 elicits high-titer anti-PfMSP2 antibodies capable of opsonizing P. falciparum merozoites for phagocytosis. These antibodies recognize rPfMSP2 from both 3D7 and FC27 alleles, as well as native PfMSP2 and previously identified linear epitopes within these three HABPs [63]. Additionally, mAbs 6D8 and 11E1 recognize the epitopes FINNAYNMSIRRS (present in HABPs 4044 and 4045) and NPKGKGEVQEPNQ (present in HABP 4053), respectively, of MSP-2 [66]. The three-dimensional structure of monoclonal antibody 6D8 in complex with MSP-2 specifically binds to the sequence contained in HABP 4044 [67] (PDB 4R3S0). Moreover, the N-terminal region of MSP-2 (MSP-21–25), where HABP 4044 is located, binds lipids and adopts an extended helical conformation spanning at least residues 10–22 [68] (Figure 3). These findings strongly suggest that most of the sequences of these HABPs are exposed not only on the native protein but also on the merozoite surface, where they can be targeted by neutralizing antibodies, as previously reported [66–68].
Peptides located on MSP-2 protein and its complex. a) Structure of MSP-2 protein determined by AlphaFold Server displays peptides 4044 in green and 4045 in red, 4053 peptide is in the region without a defined structure; b) crystal structure of anti-MSP-2 Fv fragment in complex with MSP-211–23 (PDB ID: 4R3S [69]) determined by X-ray diffraction displays peptide 4045 in red. MSP-2: merozoite surface protein 2
The P. falciparum ABRA, also known as MSP-9, is located on the merozoite surface and within the PV of infected erythrocytes. MSP-9 plays a crucial role in erythrocyte invasion and is physically associated with other essential invasion proteins [70, 71]. It directly interacts with the erythrocyte Band 3 receptor, binding specifically through two regions (MSP-9/Δ1 and MSP-9/Δ2) to the 5ABC region of Band 3, thereby facilitating P. falciparum merozoite invasion of erythrocytes [72]. Additionally, native MSP-9 forms a stable complex with MSP1-42, which targets erythrocyte receptors during invasion [28, 72]. Blocking MSP-9 binding using recombinant polypeptides significantly reduces P. falciparum reinvasion [72].
The immune response against MSP-9 not only inhibits parasite replication but also modulates inflammatory responses, with significant implications for vaccine development [73]. Vaccination with virus-like particles presenting MSP-9 elicits a strong antibody response and enhances T cell activation, generating anti-MSP-9 antibodies that effectively neutralize the parasite during invasion [73].
A series of 20-mer peptides derived from the P. falciparum ABRA/MSP-9, spanning the entire protein, have been evaluated in erythrocyte binding assays, leading to the identification of peptides 2148, 2149, 2150, 2152, and 2153 (Table 3) [74]. These peptides, originating from the N-terminal region of ABRA (residues 121–240), exhibit high-affinity binding to erythrocytes (Kd = 70–180 nM). Peptides 2148 and 2149 significantly inhibited merozoite invasion, while peptide 2149 and its analogs also displayed hemolytic and antimicrobial activity. Synthetic ABRA peptides elicit antibodies capable of blocking in vitro erythrocyte invasion by merozoites. Notably, peptides 2148 and 2149 inhibited invasion by over 90% without affecting intraerythrocytic parasite development. The binding of these peptides is reduced by up to 60% following chymotrypsin treatment of erythrocytes, suggesting that their target receptor is protease-sensitive. Interestingly, peptides 2148–2153 are located within the first of two MSP-9 regions (amino acids 77–183 and 364–528) that directly interact with the 5ABC segment of Band 3 during P. falciparum erythrocyte invasion. These regions form a co-ligand complex with MSP1-42, which binds to the invasion receptor 5ABC [72]. Antibodies against these MSP-9 regions may inhibit merozoite reinvasion, while the hemolytic activity of peptide 2149 suggests a potential interaction with the erythrocyte membrane [72]. Additionally, synthetic ABRA peptides inoculated into rabbits elicited IgG antibodies that blocked in vitro merozoite invasion of human erythrocytes by up to 90% [75]. According to the theoretical structure of MSP-9 predicted by AlphaFold Server, HABPs 2148, 2149, and 2150 form α-helices that are potentially surface-exposed. This structural prediction aligns with experimental evidence suggesting that they are targets of neutralizing antibodies (Figure 4).
Peptides of MSP-9, their sequence and in vitro invasion inhibition with 200 μM
| Peptide | AA | Sequence* | Invasion inhibition (% ± SD) |
|---|---|---|---|
| 2148 | 20 | 121LQSHKKLIKALKKNIES | 95 ± 14 |
| 2149 | 20 | 141KKHLIYKNKSYNPLLLSCVK160 | 97 ± 12 |
| 2150 | 20 | 161KMNMLKENVDYIQKNQNLFK180 | 25 ± 17 |
| 2152 | 20 | 201YKSQGHKKETSQNQNENNDN220 | 16 ± 3 |
| 2153 | 20 | 221QKYQEVNDEDDVNDEEDTND240 | 0 ± 0 |
* The bolded residues are variable residues across different strains; purple is for changes in 3D7 and 7G8 strains. MSP-9: merozoite surface protein 9; AA: aminoacids
Structure of MSP-9 protein determined by AlphaFold Server displays peptides 2148 in green, 2149 in red, 2150 in blue, 2152 and 2153 peptides are in regions without a defined structure. MSP-9: merozoite surface protein 9
AMA-1 is a conserved transmembrane protein located in the apical complex of P. falciparum merozoites. Its role in erythrocyte invasion is supported by its interactions with host cells and its ability to induce neutralizing antibodies. AMA-1 undergoes proteolytic processing into several fragments, with its ectodomain containing critical binding sites [76]. This protein facilitates strong attachment and mediates the formation of a tight junction with the host cell membrane [77].
AMA-1 forms a crucial complex with RON2, RON4, and RON5 (rhoptry neck proteins), collectively known as the AMA-1/RON complex, which is essential for tight junction formation between the merozoite and the erythrocyte [12, 78, 79]. Additionally, AMA-1 associates with MSP-1/6/7, a complex involved in merozoite invasion of erythrocytes; however, these two systems function independently, as antibodies targeting MSP-1 do not affect AMA-1 processing and vice versa [80].
Antibodies against AMA-1 effectively inhibit merozoite invasion by blocking its secondary processing and surface redistribution [81]. Studies have also identified critical residues in AMA-1 that are necessary for antibody binding, which are important for vaccine development [82]. The avidity of antibodies against AMA-1 is influenced by infection frequency, with higher malaria transmission linked to lower avidity levels, which may impact immune protection [83].
From a set of thirty-one non-overlapping synthetic peptides covering the full length of the AMA-1 protein, eight HABPs to erythrocytes have been identified, including 4313, 4321, and 4325 (Table 4) [84]. These peptides exhibit binding constants around 103 nM, and all conserved peptides blocked merozoite invasion and development, suggesting their involvement in P. falciparum invasion. Some of these peptides also share homology with the erythrocyte binding domains of MAEBL.
Peptides of AMA-1, their sequence and in vitro invasion inhibition with 200 μM
| Peptide | AA | Sequence* | Invasion inhibition (% ± SD) |
|---|---|---|---|
| 4313 | 20 | 134DAEVAGTQYRLPSGKCPVFG153 | 72 ± 5 |
| 4315 | 20 | 174Q | ND |
| 4316 | 20 | 194TLD | ND |
| 4321 | 20 | 294VVDNWE | ND |
| 4322 | 20 | 314LWVDGNCEDIPHVNEF | ND |
| 4325 | 20 | 374MIKSAFLPTGAFKADRYKS | 111 ± 13 |
| 4328 | 20 | 434P | ND |
| 4337 | 20 | 603WGEEKRASHTTPVLMEKPYY622 | 104 ± 7 |
* The bolded residues are variable residues across different strains; red is for Dd2, orange is for 3D7, yellow is for 7G8, purple is for two or more strains (Dd2, 3D7, 7G8). AMA-1: apical membrane antigen-1; ND: not determined; AA: aminoacids
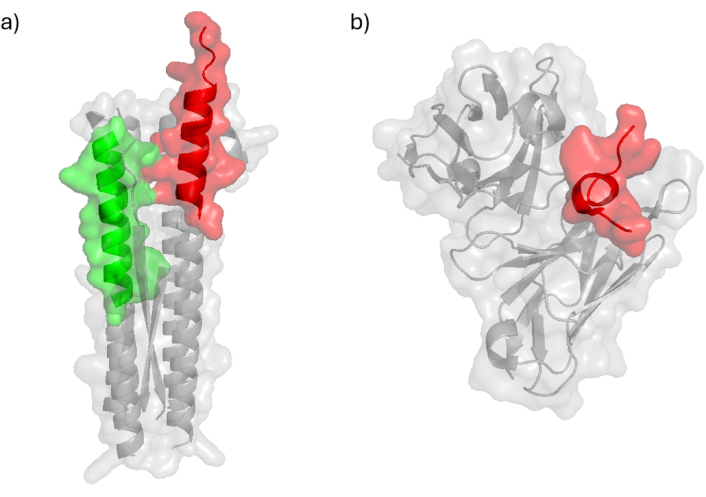
High-binding activity peptides 4313, 4315, 4316, and 4321 are located in domain I, a major target for antibodies capable of inhibiting erythrocyte invasion (Figure 5). Additionally, peptide 4322 overlaps with a known B-cell epitope [85]. Peptide 4322 is one of the most exposed regions, forming a hairpin structure, which aligns with findings supporting its immunogenic role. Notably, peptide 4322 (LWVDGNCEDIPHVNEFSAID) contains an underlined amino acid sequence previously identified as a B-cell epitope capable of inducing antibodies that block parasite growth and incorporated into a multi-component malaria vaccine candidate [85].
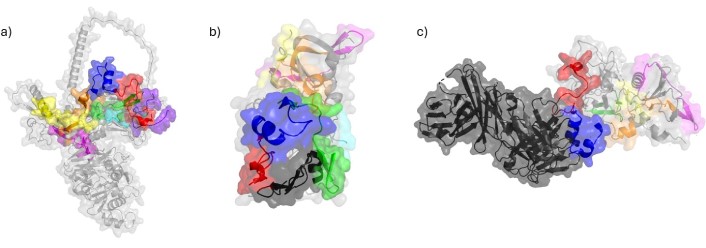
Peptides located on AMA-1 protein and its complexes. a) Structure of AMA-1 protein (PDB ID: 4R19 [86] & AlphaFold ID: AF-P50492-F1 [87]) displays peptides 4313 in green, 4315 in red, 4316 in blue, 4321 in orange, 4322 in magenta, 4325 in cyan, 4328 in yellow and 4337 in purple; b) crystal structure of AMA-1 in complex with a PfRON2 peptide (PDB ID: 3ZWZ [88]) determined by X-ray diffraction displays RON2 peptide in black; c) structure of AMA-1 in complex with a growth-inhibitory antibody (PDB ID:2Q8A [89, 90]) determined by X-ray diffraction with antibody in black. AMA-1: apical membrane antigen-1
According to the crystal structure of AMA-1, HABPs 4313, 4325, and 4337 form a binding region that is surface-exposed and located opposite the region that interacts with the RON complex proteins [91] (PDB 3ZWZ). Peptides 4325, 4315 and 4316 contribute to the binding site of the inhibitory mAb 1F9 on the AMA-1 surface, as observed in the crystal structure of AMA-1 in complex with the Fab fragment 1F9. Indeed, 1F9 binds to the solvent-exposed hydrophobic patch of AMA-1. Mutations in polymorphic AMA-1 residues within the 1F9 epitope disrupt 1F9 binding and significantly reduce the affinity of human antibodies. Remarkable, 1F9 binding to AMA-1 competes with naturally acquired human antibodies, confirming that the 1F9 epitope is a frequent target of immune recognition [89] (PDBs 2Q8A and 2Q8B) (Figure 5).
Rhoptry organelle antigens have been implicated in the invasion process and proposed as vaccine candidates for P. falciparum-induced malaria. Rhoptry-associated protein 1 (RAP-1) is a merozoite antigen located within P. falciparum rhoptries, with highly conserved nucleotide and amino acid sequences. RAP-1 has been detected on the surface of both normal erythrocytes and ring-stage-infected erythrocytes. Together with RSP-2/RAP-2, RAP-1 forms an integral part of the rhoptry complex involved in tagging normal erythrocytes and erythroid precursor cells, a process linked to erythrocyte destruction. Both RAP-1 and RAP-2 bind to the surface of normal erythrocytes [92].
Antibodies against RAP-1 contribute to natural immunity against malaria, alongside responses to MSPs [93]. A significant proportion (86%) of individuals in Papua New Guinea over the age of 30 possess antibodies recognizing both P. falciparum RAP-1 and RAP-2 [94]. The detection of RAP-1 in sera from protected animals, along with the ability of RAP-1-specific antibodies to inhibit parasite invasion, supports RAP-1 as a promising vaccine candidate [95, 96]. Notably, mAbs targeting RAP-1 have successfully inhibited P. falciparum growth in vitro and conferred protective immunity in monkey models [95]. Furthermore, mAbs developed against RSP-2/RAP-2 and RAP-1 have demonstrated the ability to inhibit parasite invasion in vitro [97].
A set of thirty-seven 20-mer and two 21-mer synthetic peptides corresponding to the complete P. falciparum RAP-1 protein sequence led to the identification of HABPs 26188, 26201, 26202, 26203, and 26204 (Table 5). These HABPs exhibit affinity constants (Kd) ranging between 700 nM and 900 nM. Some of these peptides have been shown to inhibit in vitro merozoite invasion and specifically bind to a 72 kDa protein on the RBC membrane. Two erythrocyte-specific RAP-1 binding regions have been identified: one within the cysteine-rich region (residues C461–K540) and another within the N-terminal region of the p67 product (residues T201–Y220). HABPs 26188 and 26203 significantly inhibit merozoite invasion (Figure 6) [98].
Peptides of RAP-1, their sequence and in vitro invasion inhibition with 200 μM
| Peptide | AA | Sequence* | Invasion inhibition (% ± SD) |
|---|---|---|---|
| 26188 | 20 | 201TLTPLEELYPTNVNLFNYKY220 | 97 ± 1 |
| 26201 | 20 | 461CLLNPKTLEEFLKKKEIKDL480 | 64 ± 3 |
| 26202 | 20 | 481MGGDDLIKYKENFDNFMSIS500 | 21 ± 1 |
| 26203 | 20 | 501ITCHIESLIYDDIEASQDIA520 | 93 ± 3 |
| 26204 | 20 | 521AVLKIAKSKLHVITSGLSYK540 | 25 ± 2 |
* There was no information about sequences for different strains. RAP-1: rhoptry-associated protein 1; AA: aminoacids
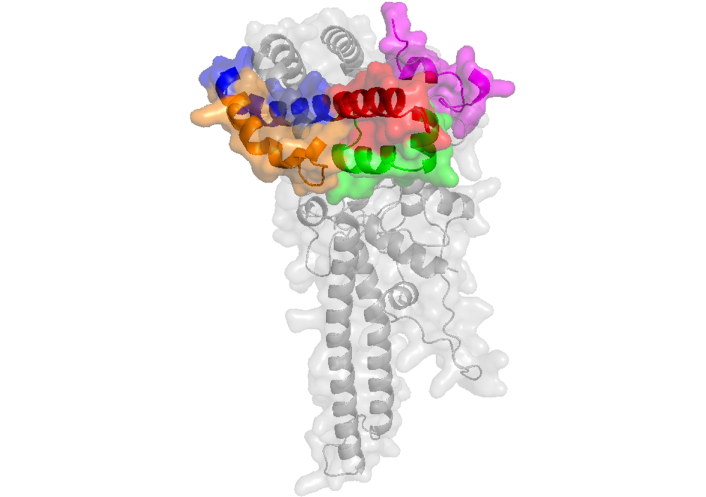
Structure of RAP-1 protein (AlphaFold ID: AF-Q9U414-F1 [99]) displays peptides 26201 in green, 26202 in red, 26203 in blue, 26204 in orange and 26188 in magenta. RAP-1: rhoptry-associated protein 1
Notably, the underlined sequence within peptide 26188 (TLTPLEELYPT) corresponds to an epitope that elicits inhibitory antibodies against parasite infection [98, 100]. Additionally, two mAbs, SP5.2 and SP8.18, generated through immunization with peptides from the synthetic malaria vaccine SPf66, specifically targeting its building block 35.1, have been found to cross-react with a RAP-1-derived sequence. These mAbs specifically recognize the sequence within peptide 26188 (LEELYPTNVNLFNYK). Cross-reactivity with RAP-1 is blocked by peptide 35.1, and the anti-35.1 mAb SP8.18 exhibits parasite growth-inhibitory activity, which is further enhanced in the presence of non-inhibitory anti-RAP-1 mAbs [101]. According to the AlphaFold ID: AF-Q9U414-F1 theoretical structure, HABPs 26201, 26202, 26203, and 26204 adopt helical conformations and form a surface-exposed patch on the protein. In contrast, HABP 26188 forms a prominent loop on the opposite side of this region, which could explain why this sequence is part of the epitope recognized by antibodies generated through immunization with the intact protein. In conclusion, most of the HABP sequences are exposed in this theoretical protein structure (Figure 6).
P. falciparum binds to host cells, potentially involving GBP-130 [102]. GBP-130 has been identified as a GBP that interacts with human erythrocytes [103]. GBP-130 binds weakly to human erythrocytes, with its binding sites located in the repeat region. Antibodies targeting this region have been shown to block merozoite invasion.
GBP-130 is also targeted by antibodies that prevent merozoite dispersal, leading to the formation of immune clusters of merozoites (ICM) and subsequent inhibition of parasite growth [104]. Additionally, GBP-130 generates neutralizing antibodies that protect against merozoite entry [102]. PfGBP-130 is recognized by antibodies from children who have developed resistance to P. falciparum infection but not by those who remain susceptible. Furthermore, mRNA encoding the full ectodomain of PfGBP-130 (amino acids 89–824) has been shown to generate parasite growth-inhibitory antibodies [105]. Immunization of mice with PfGBP-130-A (amino acids 111–374), formulated as a DNA plasmid or lipid-encapsulated mRNA, or mRNA encoding the full ectodomain of PfGBP-130 (amino acids 89–824), induced antibodies that inhibited RBC invasion [16].
A screening using synthetic peptides spanning the full length of this protein identified a HABP at residues 701–720, named 2220, which binds to human erythrocytes with a dissociation constant of 150 nM (Table 6). Notably, HABP 2220 does not bind to SA, as neuraminidase treatment increases its binding to erythrocytes, while trypsin treatment reduces it. HABP 2220 has been shown to inhibit P. falciparum merozoite invasion by 91% and exhibits higher specific binding to human erythrocytes compared to erythrocytes from other species [105]. Interestingly, peptide 2220 belongs to a repeat region that binds glycophorin and elicits antibodies capable of blocking merozoite invasion of erythrocytes [106]. According to the theoretical structure reported by AlphaFold (ID: AF-P02895-F1), this peptide is located within a beta-sheet region exposed on the protein surface (Figure 7).
Peptides of GBP-130, their sequence and in vitro invasion inhibition with 200 μM
| Peptide | AA | Sequence* | Invasion inhibition (% ± SD) |
|---|---|---|---|
| 2220 | 20 | 701FYKILTNTDPNDEVER | 91 ± 7 |
* The bolded residues are variable residues across different strains; purple is for changes in Dd2 and 3D7 strains. GBP-130: glycophorin-binding protein 130; AA: aminoacids
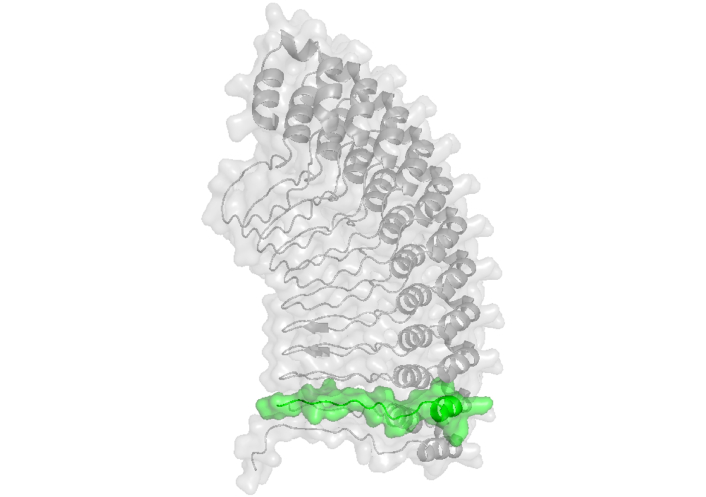
Structure of GBP-130 protein (AlphaFold ID: AF-P02895-F1 [107]) displays peptide 2220 in green. GBP-130: glycophorin-binding protein 130
Pf-SERA5 is a blood-stage antigen expressed during the late trophozoite and schizont stages as a 120 kDa precursor, which is secreted into the PV following the removal of its signal peptide [108]. Pf-SERA5 is cleaved by an essential P. falciparum protease into 47 kDa, 56 kDa, and 18 kDa fragments [109]. The 47 kDa fragment is linked to the 18 kDa fragment via a disulfide bond and localizes in the merozoite surface [56, 110]. The 56 kDa fragment undergoes further cleavage, generating 50 kDa and 6 kDa fragments just before parasite egress [109–111]. Pf-SERA5 plays a pivotal role in merozoite egress from erythrocytes, where its phosphorylation activates its proteolytic activity [112].
Despite its high transcription levels during blood-stage infection, low seropositivity for Pf-SERA5 in endemic areas suggests that it may bind to host proteins to evade immune recognition rather than directly binding to erythrocytes [113]. However, Pf-SERA5 has been shown to bind directly to human erythrocyte membranes and is synthesized as a 111-kDa antigen between 32 h and 36 h into the erythrocytic cycle.
Antibodies recognizing SERA form ICM, preventing merozoite dispersion and inhibiting parasite multiplication [104]. Moreover, antibodies against the N-terminal region of the SERA protein have been shown to inhibit parasite growth and block merozoite release through agglutination [114, 115]. mAbs targeting the SERA protein have also been demonstrated to inhibit in vitro parasite invasion [116]. According to Inselburg et al. [117], antibodies targeting SERA amino acids 24–285 or the N-terminal region (amino acids 24–506) significantly reduce parasitemia in Aotus monkeys.
From 49 non-overlapping peptides spanning the complete sequence of the SERA-5 protein, seven HABPs with strong RBC-binding activity have been identified (Table 7). Six of these peptides are located in conserved regions and have been demonstrated to inhibit in vitro merozoite invasion of erythrocytes. The affinity constants for these high-binding peptides range from 150 nM to 1,100 nM. Notably, enzyme treatment of erythrocytes does not alter the binding specificity of these peptides. Furthermore, all SERA-derived HABPs inhibit both parasite invasion and intraerythrocytic development in vitro [118]. Serum from mice immunized with the FESNSGSLEKKKYVKLPSNG peptide recognized SERA [12]. This peptide shares a sequence (bolded) with high-binding peptide 6725. Additionally, peptides 6727 and 6733 are recognized by antibodies elicited during natural infection in African patients, as well as by antibodies induced through vaccination with the N-terminal recombinant protein [119]. According to the crystal structure of the central protease-like domain of P. falciparum SERA-5 (which contains only HABPs 6746 and 6754), HABP 6746 is a surface-exposed random coil within this domain, while HABP 6754 adopts a loop-helix structure, with the loop region being surface-exposed (Figure 8).
Peptides of SERA-5, their sequence and in vitro invasion inhibition with 200 μM
| Peptide | AA | Sequence* | Invasion inhibition (% ± SD) |
|---|---|---|---|
| 6727 | 20 | 181YVRGDTEPISDSSSSSSSSSS200 | ND |
| 6733 | 20 | 321YA | 98 ± 5 |
| 6737 | 20 | 401YDNILVKMFKTNENNDKSELI420 | 85 ± 2 |
| 6746 | 20 | 581DQGNCDTSWIFASKYHLETI600 | 100 ± 2 |
| 6754 | 20 | 741KKV | 72 ± 5 |
| 6762 | 20 | 901NEVSERVHVYHILKHIKDGK920 | 100 ± 4 |
* The bolded residues are variable residues across different strains; orange is for changes in 3D7 strain. SERA-5: serine repeat antigen 5; ND: not determined; AA: aminoacids
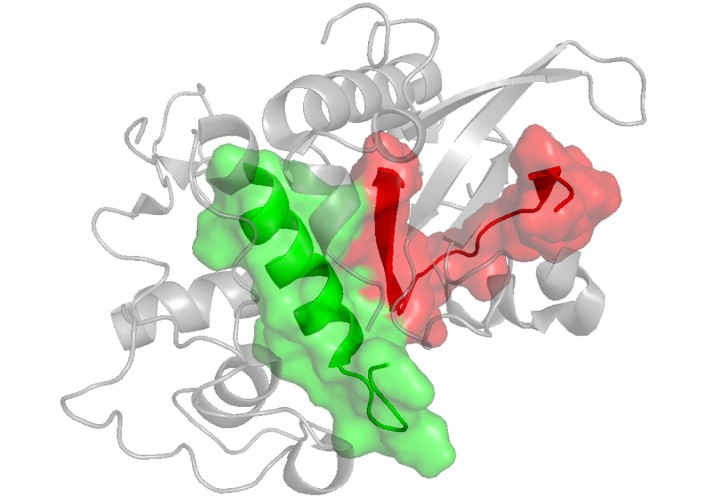
Crystal structure of SERA-5 (PDB ID: 2WBF [120]) displays peptides 6746 in green and 6754 in red. SERA-5: serine repeat antigen 5
EBA-140, also known as BAEBL or PfEBP-2, is a P. falciparum ligand that specifically binds to GPC on human erythrocytes, distinguishing it from EBA-175, which binds to GPA [121, 122]. The specificity of EBA-140 for GPC represents an adaptive response to host evolution [123]. EBA-140 interacts cooperatively with reticulocyte-binding-like (RBL) proteins, enhancing merozoite invasion efficiency [124]. This ligand plays a critical role in tight junction formation during invasion, serving as a key binding molecule at the erythrocyte interface [125].
Interestingly, both EBA-140 and EBA-175 can also bind to CD44, indicating that while their primary receptors are GPC and GPA, respectively, they may also interact with additional receptors [126]. The Region II (RII) domain of these EBA proteins is essential for receptor binding, with certain polymorphisms affecting affinity but not specificity [122, 127]. Additionally, RII of EBA-140 has been shown to interact with heparin and possibly with cell surface glycosaminoglycans (GAGs). Heparitinase treatment of erythrocytes inhibits RII-EBA-140 binding [128].
Antibodies targeting conserved regions of EBA-140 have demonstrated strong growth-inhibitory activity against diverse P. falciparum strains [129]. High levels of EBA-140-specific antibodies correlate with lower reinfection rates, suggesting their effectiveness in inhibiting merozoite invasion [125]. However, the ability of EBA-140 and other EBA family members to elicit invasion-inhibitory antibodies varies over time [130].
A total of sixty-one synthetic peptides covering the complete EBA-140 sequence have been analyzed, identifying HABPs 26135, 26144, 26147, 26160, 26170, and 26177, which exhibit high erythrocyte-binding activity, with affinity constants ranging from 350 nM to 750 nM (Table 8). These HABPs have conserved amino acid sequences and are capable of inhibiting invasion by 11–69% at 200 μM, with HABPs 26160 and 26170 showing the highest inhibition rates (69 ± 2% and 39 ± 2%, respectively). These HABPs specifically bind to erythrocyte proteins with apparent molecular weights of approximately 25, 52, and 75 kDa [131].
Peptides of EBA-140, their sequence and in vitro invasion inhibition with 200 μM
| Peptide | AA | Sequence* | Invasion inhibition (% ± SD) |
|---|---|---|---|
| 26135 | 20 | 361SYTSFMKKSKTQMEVLTNLY380 | 30 ± 2 |
| 26144 | 20 | 541DLADIIKGSDIIKDYYGKKM560 | 11 ± 9 |
| 26147 | 20 | 601LKNKETCKDYDKFQKIPQFL620 | 23 ± 7 |
| 26160 | 20 | 861GHSESSLNRTTNAQDIKIGRY880 | 69 ± 2 |
| 26170 | 20 | 1061CNNEYSMEYCTYSDERNSSP1080 | 39 ± 2 |
| 26177 | 20 | 1191VQETNISDYSEYNYNEKNMY1210 | 26 ± 0 |
* None of the residues changed in 3D7 and HB3 strains. EBA-140: erythrocyte-binding antigen 140; AA: aminoacids
Notably, HABPs 26135, 26144, and 26147 are located in RII of EBA-140: HABP 26135 is within the F1 domain, while HABPs 26144 and 26147 reside within the F2 domain. Immunization with EBA-140 induced antibodies that recognize the amino acid sequences of these peptides. Specifically, mAbs 4E7, 4B7, and 1G6 recognize peptide 26144 and exhibit moderate neutralization activity, while mAbs 9G3 and 1E2, which recognize HABP-26135, display stronger neutralization activity [125].
According to the crystal structure of domain II from EBA-140, HABPs 26135, 26144, and 26147 are predominantly helical. HABP 26135 forms a fully helical structure that is surface-exposed on this domain, while HABPs 26144 and 26147 contain surface-exposed random coil fragments (Figure 9).
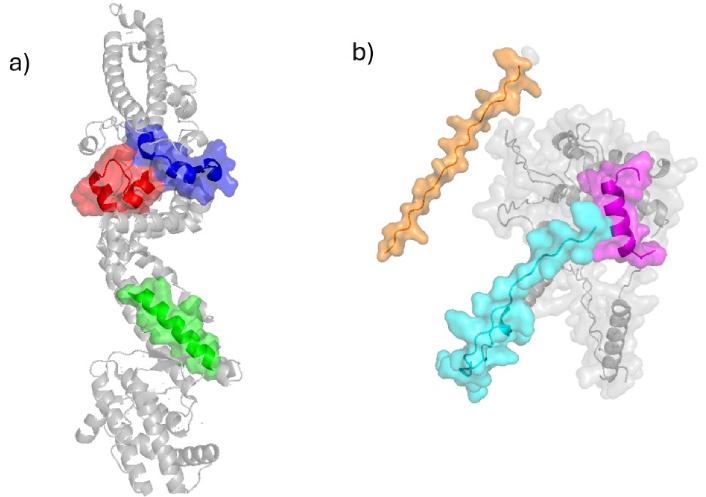
Peptides located on EBA-140 protein. a) Crystal structure of EBA-140 Region II (PDB ID: 4JNO [132]) displays peptides 26135 in green, 26144 in red, and 26147 in blue; b) structure of EBA-140 determined by AlphaFold Server displays peptides 26160 in orange, 26170 in magenta, and 26177 in cyan. EBA-140: erythrocyte-binding antigen 140
EBA-175 is a microneme protein that functions as a ligand for merozoite binding to erythrocytes. It is highly conserved and binds to GPA on the erythrocyte surface in a SA-dependent manner. Antibodies targeting RII of EBA-175, the domain responsible for binding GPA, inhibit merozoite invasion of erythrocytes [133]. Interestingly, antibodies generated against EBA-175 RII potently block the binding of native EBA-175 to erythrocytes [134–136] and even inhibit the invasion of P. falciparum strains that do not require SAs for erythrocyte invasion in vitro [137].
Moreover, conserved regions of EBA-175 stimulate a strong production of neutralizing antibodies, which can inhibit merozoite invasion across various P. falciparum strains [129]. The presence of invasion-inhibitory antibodies against EBA-175 correlates with a reduced risk of malaria symptoms, demonstrating their protective function during infection [137]. However, variability in antibody responses to EBA-175 over time underscores the challenge of achieving long-lasting immunity [130].
Six HABPs from the EBA-175 CAMP strain, 1758, 1779, 1783, 1814, 1815, and 1818, have been identified using synthetic peptides spanning the entire protein (Table 9). These peptides exhibit affinity constants ranging from 60 nM to 180 nM. Some of these high-binding peptides can inhibit in vitro merozoite invasion and block the binding of recombinant RII-EBA to RBCs [138].
Peptides of EBA-175, their sequence and in vitro invasion inhibition with 200 μM
| Peptide | AA | Sequence* | Invasion inhibition (% ± SD) |
|---|---|---|---|
| 1758 | 20 | 80K | 85 ± 10 |
| 1779 | 20 | 500NI | 24 ± 9 |
| 1783 | 20 | 580HRNK | 34 ± 10 |
| 1814 | 20 | 1200DRNSNTLHLKD | 3 ± 5 |
| 1815 | 20 | 1220 | 0 ± 10 |
| 1818 | 20 | 1280NNFNNIPSRYNLYDKKLDL1299 | 0 ± 18 |
* The bolded residues are variable residues across different strains; orange is for 3D7, yellow is for 7G8, purple is for two or more strains (3D7, 7G8, HB3). EBA-175: erythrocyte-binding antigen 175; AA: aminoacids
In Region I, the conserved high-binding peptide 1758 (KSYGTPDNIDKNMSLIHKHN) has been identified, which binds independently of SA, suggesting an alternative mechanism of interaction. Interestingly, an EBA-175 recombinant fragment (residues 1054–1319) is recognized by antibodies eluted from ICMs, which preferentially recognize functionally important epitopes exposed on live merozoites. This fragment contains HABPs 1814, 1815, and 1818 (Figure 10) [139].
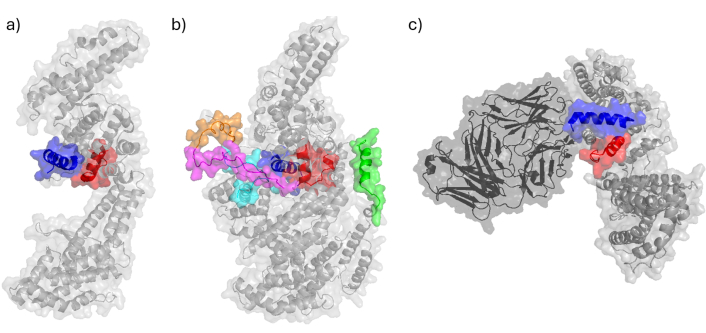
Peptides located on EBA-175 protein and its complexes. a) Crystal structure of EBA-175 Region II (RII) (PDB ID: 1ZRL [140]) has been determined by X-ray diffraction and displays peptides 1779 in red and 1783 in blue; b) the structure of EBA-175 protein (AlphaFold ID: AF-P19214-F1 [141]) displays peptides 1758 in green, 1779 in red, 1783 in blue, 1814 in orange, 1815 in magenta and 1818 in cyan; c) crystal structure of EBA-175 RII in complex with a Fab fragment from inhibitory antibody R217 (PDB ID: 4QEX [142]) determined by X-ray diffraction displays peptides 1779 in red and 1783 in blue. EBA-175: erythrocyte-binding antigen 175; Fab: fragment antigen binding
In RII, peptides 1779 and 1783, which are critical for invasion, have been found to effectively inhibit in vitro merozoite invasion. More precisely, they are located within the F2 region (residues 430–747), which belongs to the N-terminal portion of the 65 kDa fragment and binds independently of SA to RBCs [139, 143]. Notably, HABPs 1779 and 1783 effectively inhibited the binding of recombinant RII-EBA (rRII-EBA) to RBCs [139, 143]. Additionally, antibodies eluted from ICMs recognize a recombinant fragment (residues 501–753), which includes peptides 1779 and 1783, but do not bind to the fragment containing peptide CP-1 [139, 143].
Structural analyses indicate that these two peptides form helical structures that are spatially close in the three-dimensional structure of the RII-EBA fragment. They are also part of the binding site for α(2,3)-sialyllactose, with residues 5 and 6 of the sugar moiety interacting with the KQNDK motif, which corresponds to the KKNDK sequence in HABP 1783 [144] (PDB IDs: 1ZRO, 1ZRL).
A study using five mAbs specific for the F1 and F2 regions of EBA-175 find that three mAbs targeting F2 potently block EBA-175 binding to erythrocytes and inhibit merozoite invasion (IC50: 10 μg/mL to 100 μg/mL IgG in growth inhibition assays) [145]. Four of these mAbs recognize conformation-dependent epitopes within F1 or F2. When used in combination, F1- and F2-specific mAbs synergistically block the binding of native EBA-175 to erythrocytes and inhibit parasite invasion in vitro. Among them, mAb R217 is the most potent [145], and it directly blocks GPA engagement (Figure 10) [146].
Structural analyses reveal that R217 binds to a conformational epitope, which includes the F2 β-finger (residues 477–485, revised) and a loop and helix (residues 561–567, revised). These regions contain residues that are proposed to directly interact with glycans from GpA, playing a critical role in erythrocyte binding [144]. Residues K485, N561, and R566 are proposed glycan-binding sites, and mutations in these residues reduce erythrocyte binding [144]. Furthermore, R217 prevents the N-terminal region of peptide 1783 from interacting with the cell receptor, as observed in the native EBA-175 structure (PDB ID: 4QEX) (Figure 10).
Most HABPs discussed in this review are derived from erythrocyte-binding regions that serve as targets for neutralizing antibodies. These peptides correspond to critical contact regions in reported protein complexes and have been identified through multiple techniques, including: phage display libraries, surface plasmon resonance, flow cytometry, fluorescence microscopy, ELISA, and pull-down assays [147–149]. For instance, optical tweezers studies have demonstrated that while PfEBA and PfRH protein families are essential for erythrocyte attachment, MSP-1 plays a relatively minor role, with deletion experiments showing no significant impact on binding strength [147].
The development of P. falciparum merozoite protein-based vaccines faces significant challenges due to: (1) extensive antigenic polymorphism in key targets such as MSP-1, particularly within immunogenic block 2 regions (K1, MAD20, and RO33 alleles); (2) the parasite’s capacity to utilize alternative invasion pathways when primary routes are blocked; and (3) the transient exposure of critical invasion ligands during erythrocyte entry [125]. These biological features, combined with redundant invasion mechanisms, create substantial obstacles for antibody-mediated neutralization. Consequently, effective vaccine strategies must simultaneously target multiple conserved epitopes to overcome these evasion tactics and achieve broad protection or regionally tailored multivalent formulations to overcome parasite diversity and achieve consistent protection across malaria-endemic regions [150].
The rapid evolution of antigens like AMA-1 leads to strain-specific immunity, while the parasite’s ability to switch invasion mechanisms complicates target selection. To address these challenges, multi-antigen vaccines are being developed that combine targets such as AMA-1-RON2 or MSP-1-MSP-3, demonstrating synergistic protective effects in preclinical trials [151]. These approaches aim to broaden protection by targeting multiple invasion pathways while enhancing both humoral and cellular immune responses [152].
For example, the extensive genetic diversity of merozoite antigens, evidenced by high Shannon entropy scores (> 0.2) in immunogenic regions and binding interfaces, poses major challenges for vaccine design. Current vaccines based on reference strains like 3D7 often target rare haplotypes, potentially explaining their limited efficacy against diverse circulating strains. Geographical variation in antigen diversity suggests local adaptation to human and vector populations, as demonstrated by differential antibody recognition patterns between travelers and endemic populations [153]. In the case of MSP-1 vaccine development has been found strain-specific immunity due to antigenic diversity, limited cross-protection, and variable immunogenicity of different vaccine formulations (subunit vs. full-length). While anti-MSP-1 antibodies correlate with protection, their functional quality involving Fc-mediated effector mechanisms remains poorly characterized. Recent advances are addressing these limitations through structural-guided design of full-length MSP-1 immunogens that preserve native conformation, novel adjuvants like TLR4 agonists to enhance antibody effector functions, and multivalent approaches combining MSP-1 with other blood-stage antigens such as RH5. Clinical evaluation using controlled human malaria infection models and systems serology approaches are helping bridge the gap between preclinical promise and clinical efficacy [154].
An effective blood-stage malaria vaccine should primarily elicit high-affinity neutralizing antibodies targeting conserved erythrocyte-binding regions, with sufficient durability to maintain long-term circulating protection. Current research reveals complex antibody persistence patterns, with some responses being short-lived while others demonstrate extended durability. A significant challenge arises from immunodominant epitopes that are often non-protective and localized to polymorphic antigen regions. For instance, endemic populations develop higher-affinity antibodies against AMA-1 compared to MSP-2, particularly favoring the MSP-2-3D7 allele [63]. Surface plasmon resonance studies demonstrate substantial variation in antibody affinities across antigen types, allelic variants, and individual exposure histories. Longitudinal data indicate 67–79% of individuals maintain stable antibody affinities over time, while others exhibit fluctuation patterns that correlate with parasite exposure intensity [18]. Crucially, anti-PfRh2 antibody affinity serves as a strong predictor of invasion inhibition capacity, highlighting the functional significance of antibody quality beyond mere binding metrics.
HABPs represent a promising approach for malaria vaccine development due to their interspecies conservation and demonstrated invasion-inhibitory properties. These synthetic peptides contain predicted B-cell epitopes that are recognized by SERA from Plasmodium-infected mice [155], with similar immunogenicity observed in other animal models and humans (as discussed below). This cross-species reactivity suggests their potential to elicit broad protective immunity. To optimize vaccine efficacy, development efforts should prioritize: (1) multivalent formulations combining liver- and blood-stage antigens; (2) novel adjuvant systems to enhance balanced Th1/Th2 responses; and (3) advanced delivery platforms that address both antigenic diversity and the need for durable immunity in endemic populations.
Effective peptide vaccines must induce coordinated humoral and cellular immunity, as demonstrated by early viral peptide studies showing MHC-I-restricted CD8+ T cell protection. The canine parvovirus peptide vaccine achieved T-cell-dependent efficacy comparable to whole-virus vaccines, while cancer immunotherapy research has shown that longer peptides (> 20 amino acids) enhance antigen-presenting cell (APC) processing and CD4+ T cell help compared to minimal epitopes [156]. Current strategies focus on overcoming antigenic variation through conserved multi-epitope designs (e.g., P. falciparum MSP-RH5-AMA-1 combinations) and optimizing delivery systems such as nanoparticle formulations and mRNA conjugates to improve immunogenicity.
Phage display technology has transformed malaria antigen discovery, enabling identification of novel erythrocyte-binding proteins [157] and inhibitory peptides targeting AMA-1 [158]. Proteomic approaches have characterized vital merozoite protein complexes, revealing stage-specific interactions between MSP-1 and rhoptry proteins like PfRhopH3 that are shed during invasion [159]. These methods, combined with structural analyses and functional assays, provide a robust toolkit for vaccine antigen selection and validation. The integration of computational predictions with experimental data is significantly advancing the development of next-generation multi-antigen vaccines against P. falciparum.
Recombinant EBA-140 RII and its F2 domain fragment were successfully characterized using anti-myc monoclonal antibodies in immunoblotting assays. Polyclonal rabbit antibodies generated against full-length EBA RII with MPL adjuvant showed cross-reactivity with both baculovirus-expressed RII and bacterially produced F2 fragments. Crucially, these antibodies effectively blocked EBA-140 binding to erythrocytes, confirming this region as a promising vaccine target [160]. Key blood-stage antigens such as AMA-1, MSPs, and RON2 have been prioritized based on their essential roles in invasion and capacity to induce neutralizing antibodies in preclinical studies. Structural analyses have identified conserved epitopes in these antigens that may help overcome strain-specific immunity limitations.
The development of minimal, subunit-based, multi-epitope vaccines against P. falciparum involves a systematic approach focusing on modified HABPs (mHABPs) derived from conserved regions of key invasion proteins (MSP, RH5, and AMA-1). These engineered mHABPs demonstrate: (1) high immunogenicity in immunoassays; (2) protective efficacy in Aotus monkey challenge models; and (3) confirmed MHC-II binding affinity through computational modeling (NetMHCIIpan 4.0), leveraging the 50% allelic similarity between Aotus and human MHC systems [103, 161]. Structural characterization via 1H-NMR has revealed that targeted residue modifications significantly alter peptide secondary structure, directly modulating immunological presentation [162]. These findings position mHABPs as promising candidates for multi-stage vaccine development, though clinical validation remains essential.
MHC-II-restricted CD4+ T cell activation represents a crucial determinant of vaccine efficacy, driving robust adaptive immunity. Compared to traditional whole-pathogen vaccines, peptide-based approaches offer distinct advantages through:
Precise 20–30 residue epitopes minimizing allergenic potential.
Customizable structural optimization maintaining native antigen conformation.
Targeted engagement of both MHC-I (CD8+ T cells) and MHC-II (CD4+ T cells).
Key design parameters must be incorporated.
Immunodominant epitope selection using predictive tools (i.e., NetMHCIIpan).
Population coverage analysis accounting for MHC polymorphism.
Integrated computational-experimental validation (e.g., ABCpred for B-cell epitopes combined with phage display/MS verification).
The rational design process employs a multidisciplinary framework.
Computational prediction of conserved, immunogenic epitopes.
Structural optimization through NMR-guided modifications.
Immunological validation in relevant animal models.
Clinical translation of lead candidates.
This approach combines the precision of structural biology with advanced immunoinformatics to overcome historical challenges in malaria vaccine development, particularly regarding antigenic diversity and population-level coverage. The integration of conserved mHABPs with optimized delivery systems represents a promising path toward developing effective multi-epitope vaccines against P. falciparum.
These innovative approaches, when combined with next-generation adjuvant systems, aim to achieve durable protection against malaria while maintaining the inherent safety and cost-effectiveness of peptide-based platforms. The integration of conserved HABPs with multivalent designs and advanced delivery technologies represents a promising path toward developing effective malaria vaccines capable of addressing the challenges posed by parasite diversity and immune evasion mechanisms.
Fifty erythrocyte HABPs were identified in a study using synthetic peptides spanning the full sequences of P. falciparum merozoite proteins, MSP-1, MSP-2, ABRA (MSP-9), AMA-1, RAP-1, GBP-130, SERA-5, EBA-140, and EBA-175, conducted at the Instituto de Inmunología, San Juan de Dios Hospital, Bogotá, Colombia, between 1990 and 2000, prior to the hospital’s closure. These HABPs exhibit nanomolar-range dissociation constants (Kd) for erythrocytes, with many localized within key erythrocyte-binding regions crucial for merozoite invasion. Most of these peptides effectively inhibit in vitro merozoite invasion.
Several HABPs share conserved amino acid sequences, suggesting their essential role in parasite survival. Notably, all HABPs were identified before the three-dimensional structures of these proteins were resolved. Subsequent structural analyses revealed that most HABPs are surface-exposed, accessible to host receptors or antibodies. Indeed, multiple HABPs across different proteins serve as targets for invasion-inhibitory antibodies. Structural studies of antibody-protein complexes further underscore the critical role of these HABP sequences in merozoite invasion.
This review presents the first part of the research on synthetic peptides from malaria proteins conducted at the Instituto de Inmunología, now the Fundación Instituto de Inmunología de Colombia (FIDIC), with a focus on the identification of merozoite protein regions involved in merozoite invasion using synthetic peptides. Some of these HABPs represent promising tools for elucidating the molecular mechanisms of merozoite invasion and hold potential for malaria control strategies, including vaccine development.
ABRA: acid basic repeat antigen
AMA-1: apical membrane antigen-1
EBA-140: erythrocyte-binding antigen 140
Fab: fragment antigen binding
GBP-130: glycophorin-binding protein 130
GPA: glycophorin A
GPC: glycophorin C
GPI: glycosylphosphatidylinositol
HABPs: high-activity binding peptides
ICM: immune clusters of merozoites
mAbs: monoclonal antibodies
mHABPs: modified high-activity binding peptides
MSP-1: merozoite surface protein 1
PfRH: Plasmodium falciparum reticulocyte binding-like homologous
PV: parasitophorous vacuole
RAP-1: rhoptry-associated protein 1
RBCs: red blood cells
RII: Region II
SA: sialic acid
SERA-5: serine repeat antigen 5
This review is dedicated to the memory of our esteemed mentor, Prof. Manuel E. Patarroyo, whose visionary leadership, unwavering dedication to scientific inquiry, and invaluable mentorship were instrumental in shaping the research that we have summarized and reviewed here. His profound contributions to immunology and malaria research have left an enduring legacy that continues to inspire future generations of scientists. Sadly, Prof. Patarroyo passed away during the preparation of this manuscript, but his impact on the field and his relentless pursuit of knowledge will always be remembered. This dedication also extends to the many scientists from FIDIC and the former Instituto de Inmunología, who, despite numerous challenges, persevered with unwavering commitment to advancing this research. Their dedication, resilience, and collective efforts have significantly contributed to our understanding of Plasmodium falciparum merozoite invasion, paving the way for future advancements in malaria control and vaccine development.
MUM: Conceptualization, Investigation, Project administration, Validation, Writing—original draft. DBR: Investigation, Validation, Visualization, Writing—review & editing. FGQ: Conceptualization, Validation, Writing—review & editing. All authors read and approved the submitted version.
Fanny Guzmán-Quimbayo who is the Guest Editor of Exploration of Drug Science had no involvement in the decision-making or the review process of this manuscript. The other authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
The dataset analyzed for this study can be found on the PDB Homepage (https://www.rcsb.org/) and AlphaFold Protein Database (https://alphafold.com/). Additionally, the sequences of the proteins that were constructed by AlphaFold Server were taken from UniProt (https://www.uniprot.org/). The database used to search for articles was PubMed (https://pubmed.ncbi.nlm.nih.gov/) where the keywords used were “Plasmodium falciparum + peptides + (Name of the protein)”, for the articles done in Instituto de Inmunología “Patarroyo ME” was added to the search. The software Protein BLAST by NCBI was used to determine the conserved regions of the peptides in different strains, while the sequences of the strains were obtained from UniProt.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 4292
Download: 115
Times Cited: 0
