Affiliation:
1Key To Business, R&D, 00144 Roma, Italy
Email: m.mameli@key2.it
ORCID: https://orcid.org/0000-0002-5269-9939
Affiliation:
2Department of Neurosciences, Catholic University of the Sacred Heart, 00168 Rome, Italy
3IRCCS “A. Gemelli” University Polyclinic Foundation, 00168 Rome, Italy
Explor Digit Health Technol. 2025;3:101158 DOI: https://doi.org/10.37349/edht.2025.101158
Received: February 27, 2025 Accepted: May 27, 2025 Published: August 11, 2025
Academic Editor: Anastasios Koulaouzidis, University of Southern Denmark (SDU), Denmark
The article belongs to the special issue Deep Learning Methods and Applications for Biomedical Imaging
Aim: Colorectal cancer is a leading cause of cancer-related mortality, emphasising the need for accurate polyp segmentation during colonoscopy for early detection. Existing methods often struggle to generalize effectively across diverse clinical scenarios. This study introduces DeepPolyp, an artificial intelligence framework designed for comprehensive benchmarking and real-time clinical deployment of polyp segmentation models.
Methods: Transformer-based segmentation models, SegFormer and SSFormer, were trained from scratch using an extensive dataset comprising public collections (CVC-ClinicDB, ETIS-LaribPolypDB, Kvasir) and recently augmented datasets (PolypDataset-TCNoEndo, PolypGen). Training involved standardized data augmentation, learning rate schedules, and early stopping. Models were evaluated using Dice and Intersection over Union (IoU) metrics. Real-time inference performance was assessed on an NVIDIA Jetson Orin device with ONNX and TensorRT optimizations.
Results: SegFormer-B4 achieved the highest accuracy (Dice: 0.9843, IoU: 0.9694), but was not selected for clinical deployment due to computational constraints. SegFormer-B2 provided comparable accuracy (Dice: 0.9787, IoU: 0.9588) with significantly faster inference (94 ms per frame), offering an optimal balance suitable for real-time clinical use. SSFormer showed lower accuracy and slower inference, limiting its practical deployment.
Conclusions: DeepPolyp enables systematic evaluation of polyp segmentation models, assisting in selecting models based on both performance and computational efficiency. Despite superior accuracy from SegFormer-B4, SegFormer-B2 was selected for clinical deployment due to its advantageous balance between accuracy and real-time execution efficiency.
Colorectal cancer remains one of the leading causes of cancer-related mortality worldwide, with more than 1.9 million new cases diagnosed annually [1]. Early detection and diagnosis are essential to increase patient survival by enabling timely medical intervention and treatment planning [2, 3]. Polyp segmentation, defined as the task of identifying and delineating polyps in endoscopic images, plays a crucial role in the screening and diagnosis of colorectal cancer [4]. Accurate segmentation assists gastroenterologists in differentiating between benign and malignant lesions, guiding decisions during procedures such as polypectomy [5–7]. However, polyp segmentation remains a difficult task due to high variability in polyp size, shape, texture, and contrast with surrounding tissue [8, 9].
Anatomical differences, varying camera angles, and inconsistent lighting across endoscopic equipment create complex visual patterns, making reliable polyp segmentation especially challenging. Traditional segmentation approaches often rely on hand-crafted features, thresholding, and region-based techniques [10]. These methods usually apply filters to extract texture, edges, or color features and use post-processing techniques such as morphological operations to refine segmentation masks. While effective in controlled environments, these techniques struggle to generalize under varying imaging conditions and are unable to capture complex spatial dependencies [11]. As a result, segmentation masks generated using traditional methods are often incomplete or inaccurate [9].
The introduction of deep learning, particularly convolutional neural networks (CNNs), has brought significant improvements in polyp segmentation by enabling automatic learning of hierarchical features from image data. Encoder-decoder architectures such as U-Net and its variants have become standard in medical image analysis due to their ability to localize features and refine object boundaries [12]. However, CNNs have limitations in modelling long-range dependencies because of their inherently local receptive fields [13].
Transformer-based architectures have recently emerged as powerful alternatives by using self-attention mechanisms to capture global dependencies. These models have demonstrated strong performance across many vision tasks, including segmentation. Hybrid architectures, such as SegFormer and SSFormer, combine the strengths of convolutional layers for local feature extraction with transformer blocks for global context modelling, achieving state-of-the-art results in several segmentation benchmarks [14]. Lightweight transformer models, including Enhanced Nanonet, have also been developed to reduce computational cost while maintaining high segmentation accuracy, supporting deployment on low-power devices [15].
Despite the progress achieved by deep learning, several challenges remain. Many current segmentation models are designed with highly specialized architectures and require extensive hyperparameter tuning to reach optimal performance. Such specialization limits model adaptability to different imaging conditions or datasets. Additionally, most models are trained on relatively small and homogeneous datasets, which fail to capture the diversity of polyp appearances encountered in clinical practice. Variations in polyp morphology and imaging modalities across patients and devices further reduce generalization performance.
Another critical issue is the computational complexity of advanced models, which restricts their deployment on embedded or portable systems that require real-time operation. This limitation is particularly relevant in clinical settings where fast and reliable analysis is essential. Consequently, there is a growing demand for general-purpose segmentation networks that are robust, adaptable to various scenarios, and efficient enough for use on edge devices.
Recent advances in transformer-based segmentation models offer promising solutions, yet their application in medical imaging—and specifically in polyp segmentation—remains limited. Many existing studies rely on standard benchmark datasets, which may not adequately represent the complexity of real clinical scenarios. New datasets, such as PolypDataset-TCNoEndo [16] and PolypGen [17], provide additional variability in polyp appearance and imaging modalities, and therefore represent more realistic testbeds for evaluating model generalization.
In addition, the deployment of segmentation models on resource-constrained hardware for real-time use has not been thoroughly investigated. Although interest in edge AI is increasing, few works evaluate segmentation models in terms of latency, segmentation quality, and computational efficiency under real hardware constraints.
To address these gaps, a modular AI framework named DeepPolyp is introduced. This framework is designed to benchmark and evaluate the performance of general-purpose transformer-based segmentation models, including SSFormer [18] and SegFormer [19], when trained on a diverse and extended set of datasets. In addition to widely used public datasets such as CVC-ClinicDB [20], CVC-ColonDB [21], ETIS-LaribPolypDB [22], and Kvasir [23], two newer datasets—PolypDataset-TCNoEndo [16] and PolypGen [17, 24, 25]—are included to ensure higher variability in the evaluation.
While recent transformer-based architectures and hybrid models have shown promising results in medical image segmentation, several gaps persist in current research. First, most existing models are designed as specialized solutions tailored to specific datasets, which limits their generalizability across diverse imaging conditions. Second, research efforts often rely on benchmark datasets that do not fully reflect the variability present in real-world clinical environments. Third, there is limited investigation into the practical feasibility of deploying such models on edge devices for real-time clinical use. These limitations are addressed in this work through the following specific contributions:
Introduction of DeepPolyp, a novel AI framework specifically designed for the comprehensive evaluation of polyp segmentation models in terms of accuracy, generalization, and deployment feasibility.
A systematic assessment of state-of-the-art general-purpose segmentation architectures, namely SSFormer [18] and SegFormer [19], retrained and evaluated on a large and diverse collection of polyp datasets.
Expansion of existing evaluation settings beyond commonly used datasets (CVC-ClinicDB [20], CVC-ColonDB [21], ETIS-LaribPolypDB [22], and Kvasir [23]) by including two additional recent datasets: PolypDataset-TCNoEndo [16] and PolypGen [17, 24, 25], providing a more realistic and challenging evaluation setting.
Evaluation of model performance under computational constraints, including inference time and resource usage, to explore deployment feasibility in resource-limited clinical environments using edge hardware.
The remainder of the paper is structured systematically. State-of-the-art section provides a structured comparison and detailed critique of existing literature, highlighting the strengths and limitations of current methodologies relative to the proposed DeepPolyp framework. Materials and methods section outlines the methodology, including dataset preparation, model training protocols, and evaluation metrics. Results section presents experimental results, emphasizing model comparison and generalization capabilities. Discussion section discusses implications for clinical deployment, particularly focusing on computational efficiency and real-time capabilities.
Polyp segmentation has advanced significantly with deep learning techniques, which have overcome limitations of traditional methods [26]. Traditional approaches struggle with the variability in polyp size, shape, texture, and contrast, resulting in inconsistent segmentation. In contrast, deep learning models, particularly CNNs and transformer-based architectures, demonstrate superior accuracy and robustness. Recent research has focused on developing novel architectures and optimization strategies, including hybrid models that combine CNNs with transformers to capture both local features and global context. These advances have improved segmentation performance, enabling more accurate and reliable clinical applications.
CNN-based models have established strong baseline performance for polyp segmentation due to their ability to extract hierarchical features. However, these models often struggle with capturing long-range dependencies and maintaining consistent performance across varied polyp morphologies.
Fan et al. [27] pioneered a parallel reverse attention network that combines global and local features to improve boundary detection and segmentation accuracy. While effective for well-defined polyps, this approach may underperform with flat or sessile polyps that lack clear boundaries.
ResUNet variants have shown promising results. Jha et al. [28] enhanced ResUNet with squeeze-and-excitation blocks, attention gates, and residual connections to boost feature extraction and segmentation performance. Though effective, these models require significant computational resources, limiting their deployment on resource-constrained devices.
DilatedSegNet [29] employs a ResNet50 backbone with a Dilated Convolution Pooling (DCP) block, achieving reliable segmentation at 33.68 FPS. While computationally efficient, it may struggle with very small polyps due to information loss during pooling operations.
MSRF-Net [30] uses Dual-Scale Dense Fusion (DSDF) blocks to preserve high-resolution features, addressing the detail loss common in CNN architectures. However, it requires careful parameter tuning to maintain optimal performance across datasets.
HarDNet-MSEG [31] achieves over 0.9 mean Dice score with an 86 FPS inference speed using a low-memory CNN backbone, making it suitable for clinical applications. Its focus on efficiency may occasionally compromise performance on challenging cases.
Transformer models excel at capturing global contextual information but often require significant computational resources and may lose fine local details critical for accurate boundary delineation.
Dong et al. [32] present a transformer-based approach with attention mechanisms in both encoder and decoder, refining outputs while preserving the UNet-like decoder structure. This approach effectively captures global dependencies but may struggle with real-time applications due to computational overhead.
SSFormer [18] integrates a transformer-based pyramid encoder with a Progressive Locality Decoder (PLD) and Stepwise Feature Aggregation (SFA), mitigating attention dispersion issues. While effective for capturing global context, it faces challenges with very small polyps and has increased latency compared to lightweight CNN models.
FCBFormer [33] combines convolutional and transformer-based methods through a dual-branch architecture, enhancing robustness. This approach balances global and local feature extraction but requires careful optimization to manage computational complexity.
Polyp-PVT [32] leverages pyramid vision transformers, integrating a cascaded fusion module, camouflage identification module, and similarity aggregation module. Though powerful, it requires substantial GPU resources that may not be available in all clinical settings.
Hybrid models aim to combine the strengths of CNNs and transformers, addressing the limitations of individual approaches. These models typically offer better performance but often at the cost of increased complexity and computational requirements.
Zhang et al. [34] combine CNN and transformers, where the transformer encoder captures global dependencies, and a cascaded CNN upsampler refines local features. This approach effectively balances global context with local detail but introduces additional complexity in training and deployment.
The authors [35] introduce a fusion of Meta-Former with UNet, incorporating a multi-scale upsampling block and level-up augmentation to enhance texture representation. While this approach improves texture delineation, it requires careful balancing of the two architectural components.
FeDNet [36] introduces a Feature Decoupled Module (FDM) leveraging Laplacian pyramid decomposition for targeted optimization. Integrated with a vision transformer-based Feature Pyramid Network (FPN), FeDNet demonstrates strong accuracy and generalization but at increased computational cost.
LDNet [37] introduces a lesion-aware dynamic kernel, Lesion-aware Cross-Attention (LCA), and Efficient Self-Attention (ESA) to improve contrast between polyps and the background. This approach excels with challenging cases but requires careful implementation to maintain efficiency.
Zhou et al. [38] propose a cross-level feature aggregation and boundary prediction network, utilizing a two-stream structure to capture hierarchical semantic information. The model integrates a Cross-level Feature Fusion module to handle scale variations but may struggle with very small or flat polyps.
BDG-Net [39] employs a Boundary Distribution Map (BDM) for segmentation precision, addressing the challenge of accurate boundary delineation. However, it requires additional computational steps that may impact real-time performance.
DCRNet [40] captures contextual relations within and across images using an episodic memory mechanism. While effective for maintaining consistency across video frames, this approach requires sequential processing that increases latency.
ColonFormer [41] employs a hierarchical transformer encoder and a CNN-based decoder with multiscale feature representation. This approach effectively handles scale variations but faces challenges with real-time deployment due to its complexity.
PolypSeg+ [5] integrates an Adaptive Scale Context module and an Efficient Global Context module for real-time segmentation. It balances performance and efficiency but may still underperform on datasets with significant domain shifts.
HarDNet-DFUS [42] optimizes the HarDNet-MSEG model with ShuffleNetV2 concepts and a Lawin Transformer decoder, enhancing computational efficiency while maintaining accuracy. This approach represents a promising direction for clinical deployment.
DuAT [43] balances local and global representations with Global-to-Local Spatial Aggregation (GLSA) and Selective Boundary Aggregation (SBA). This comprehensive approach addresses multiple challenges but increases model complexity.
FuzzyNet [27] employs a Fuzzy Attention module to refine segmentation near polyp boundaries, addressing a critical challenge in clinical applications. However, it requires careful parameter tuning to achieve optimal results.
HSNet [44] combines Transformer-CNN frameworks, integrating a Cross-Semantic Attention module and Multi-Scale Prediction module for high performance. While effective, it introduces additional complexity that may challenge deployment in resource-constrained environments.
UACANet [45] enhances segmentation with Uncertainty Augmented Context Attention, improving robustness to ambiguous boundaries. This approach addresses a key clinical challenge but at the cost of increased computational overhead.
M2SNet [46] applies subtraction-based feature fusion to improve edge preservation, addressing a common limitation in polyp segmentation. This approach effectively captures boundaries but may struggle with flat or sessile polyps.
MSNet [47] employs a subtraction-based extraction mechanism for boundary delineation. While effective for well-defined polyps, it may underperform with polyps that have gradual transitions to surrounding tissue.
SANet [48] introduces a color exchange operation and probability correction strategy for small polyp segmentation. This approach specifically addresses the challenge of small polyps but may not generalize well to larger, more complex cases.
TransFuse [34] integrates CNN and Transformer models with a BiFusion module for precise segmentation. This balanced approach effectively combines global and local features but requires careful implementation to manage computational demands.
CaraNet [49] enhances small object segmentation through a Context Axial Reverse Attention Network. While effective for small polyps, it may introduce unnecessary complexity for larger, more obvious cases.
FANet [50] refines segmentation iteratively with a Feedback Attention Network. This approach improves accuracy through multiple refinement steps but increases inference time, potentially limiting real-time applications.
Enhanced U-Net [51] improves robustness with a Semantic Feature Enhancement Module (SFEM) and Adaptive Global Context Module (AGCM). This approach effectively balances performance and efficiency but still faces challenges with very small or flat polyps.
Recent works have focused on addressing specific challenges in polyp segmentation, such as boundary delineation, small polyp detection, and domain generalization.
The authors [28] introduce an advanced ResUNet-based architecture with residual units, squeeze-and-excitation blocks, and attention mechanisms, achieving strong results on Kvasir-SEG. However, complex attention mechanisms increase computational demands.
Tomar et al. [29] propose a dual decoder attention network, with one decoder acting as an autoencoder, enhancing feature maps through attention mechanisms. This approach improves feature representation but at the cost of model complexity.
The authors [28] develop a multi-scale residual fusion network with cross multi-scale attention, improving generalizability. While effective for handling domain shifts, this approach introduces additional parameters that increase memory requirements.
Guo et al. [52] address threshold selection by learning adaptive threshold maps through a confidence-guided manifold mixup approach, achieving a Dice coefficient of 87.307% on EndoScene. This approach improves segmentation consistency but requires careful implementation to avoid overfitting.
Despite significant advances, challenges remain in polyp segmentation, including:
Balancing computational efficiency with segmentation accuracy for clinical deployment
Addressing performance variations across different polyp morphologies
Enabling reliable deployment on resource-constrained edge devices
Providing systematic comparison frameworks for evaluating model performance
These challenges highlight the need for a comprehensive framework to evaluate and compare different segmentation models under uniform conditions, particularly for edge deployment scenarios. This study focuses on analyzing the DUCK-Net, SSFormer, and SegFormer models for potential deployment on edge devices, addressing a critical gap in current research.
This section introduces DeepPolyp, an advanced AI framework for polyp segmentation and detection, designed to systematically evaluate the effectiveness of specialized and general-purpose segmentation models. The workflow, illustrated in Figure 1, is organized into four main stages: Data preparation, SOTA model selection, model comparison, and edge porting. This structured approach ensures a rigorous evaluation of segmentation models under different imaging conditions, providing comprehensive information on their performance and feasibility. Further details are given in the following subsections.
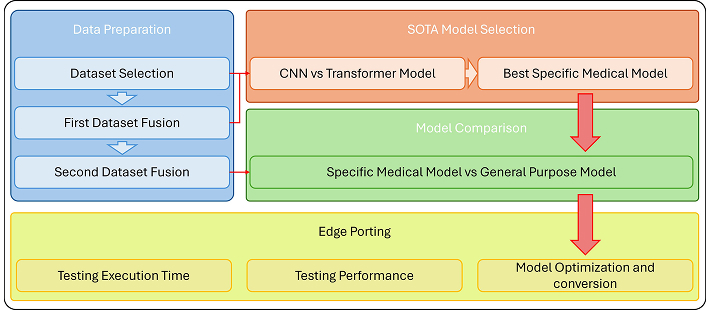
Workflow of DeepPolyp. The framework consists of four main stages: (1) data preparation, including dataset selection and fusion to improve model generalisation; (2) SOTA model selection, comparing CNNs and transformer-based architectures to identify the best specialised medical model; (3) model comparison, evaluating the robustness of the specialised medical model against a general-purpose segmentation model; and (4) edge porting, optimising and deploying the model on edge devices for real-time clinical applications. This systematic approach ensures comprehensive evaluation, high segmentation accuracy and efficient real-time performance
Data preparation is key to robust model training and evaluation. In DeepPolyp, this phase is divided into three key steps: dataset selection, first dataset fusion, and second dataset fusion, to ensure comprehensive and diverse training data for effective model generalisation.
Public datasets have significantly advanced automatic polyp segmentation research by providing standardized benchmarks for deep learning models. These datasets offer diverse polyp images with detailed annotations, enabling reproducible research and fair model comparisons. The DeepPolyp framework leverages several key datasets to address the limitation of existing models being overly specialized to specific datasets.
Kvasir-SEG [23]: This widely used dataset contains 1,000 polyp images with corresponding segmentation masks. The images have varying resolutions (332 × 487 to 1,920 × 1,072 pixels) stored in JPEG format with bounding box information in JSON format. This resolution diversity helps models become more robust by learning from different input sizes.
CVC-ClinicDB [20]: Consisting of 612 frames extracted from colonoscopy videos, this dataset includes polyps with ground truth segmentation masks. It effectively represents real clinical scenarios, making it valuable for evaluating segmentation algorithms.
ETIS-LaribPolypDB [22]: This dataset provides a comprehensive collection of polyp images with detailed annotations. Its diverse polyp appearances contribute to better model generalisation across different clinical settings.
PolypGen [17]: One of the most comprehensive datasets available, containing 1,537 polyp images, 2,225 positive polyp video sequences, and 4,275 negative frames. Data collected from six medical centers across Europe and Africa ensures significant imaging variability, enhancing model adaptability to real-world clinical settings.
CVC-ColonDB [21]: This dataset offers 300 colonoscopy images with corresponding polyp segmentation annotations, supporting the development of accurate machine learning models.
CVC-300 [53]: Comprising 912 images from 44 colonoscopy sequences with ground truth segmentation masks. It is frequently used as a test set alongside other datasets to evaluate model generalisation capabilities. The inclusion of multiple sequences from different procedures provides a comprehensive evaluation of segmentation algorithms.
PolypDataset-TCNoEndo [16]: This dataset is an augmented version of Kvasir-SEG, not a new dataset. It contains approximately 19,000 images generated through various data augmentation techniques, including color modification, lighting adjustment, and contrast alteration. These augmentations introduce greater variability in imaging conditions, crucial for training models that generalise well across different clinical scenarios.
To address the generalisation limitations of existing models, DeepPolyp employs a systematic dataset fusion approach:
Preprocessing: All images undergo standardized preprocessing before fusion:
Normalization to a common intensity range [0, 1]
Resizing to a uniform dimension (512 × 512 pixels)
Color space standardization (RGB)
Contrast enhancement using adaptive histogram equalization
To improve model generalisation, individual datasets are combined to create a mixed dataset, which is designed to simulate real-world scenarios by including a wide range of polyp appearances, lighting conditions and imaging devices. The mixed dataset contains images from all selected datasets, ensuring better model generalisation and robustness.
The datasets are divided into training, validation and test sets in a ratio of 80-10-10. Table 1 summarises the distribution.
Number of images for each dataset split
| Dataset | Training | Validation | Test |
|---|---|---|---|
| CVC-300 | 43 | 11 | 6 |
| CVC-ClinicDB | 440 | 110 | 62 |
| CVC-ColonDB | 273 | 69 | 38 |
| ETIS-LaribPolypDB | 140 | 36 | 20 |
| Kvasir | 720 | 180 | 100 |
| Mixed dataset | 1,077 | 381 | 704 |
To further improve model generalisation, a second dataset fusion is performed. This stage includes additional datasets, particularly images without polyps, to reduce false positives. This enriched dataset consists of:
Training set: 19,657 images
Validation set: 5,027 images
Test set: 10,660 images
This comprehensive dataset allows the model to learn from a wide range of scenarios, improving segmentation accuracy and robustness.
The DeepPolyp framework’s unique contribution lies in this structured fusion approach, which addresses the key challenges in polyp segmentation: limited generalisation across datasets, insufficient diversity in training data, and the gap between laboratory performance and clinical deployment. By integrating diverse datasets through a systematic methodology, DeepPolyp enables the training of more robust segmentation models that perform consistently across different clinical settings and imaging conditions.
This section evaluates existing segmentation models to establish a benchmark for comparison. The comparison systematically assesses CNN-based models against transformer-based architectures and identifies the best-performing specialized medical model for further evaluation.
DUCK-Net [35] and SSFormer [18] are trained on the mixed dataset. DUCK-Net was selected as a representative CNN-based model with established performance in medical image segmentation, while SSFormer represents the newer transformer-based approaches specifically designed for medical applications.
The evaluation uses two standard metrics: Dice coefficient and mean Intersection over Union (mIoU) [28]. Each model’s performance is tested on individual datasets to assess their ability to generalize across different imaging conditions.
DUCK-Net shows acceptable performance on larger datasets such as Kvasir and CVC-ClinicDB but performs poorly on smaller datasets like CVC-ColonDB. In contrast, SSFormer consistently outperforms DUCK-Net across all datasets, achieving higher Dice and mIoU scores. This superior performance stems from the transformer-based architecture’s ability to capture global dependencies in the images.
This phase aims to improve the model’s ability to generalize for polyp segmentation from RGB images. A key goal is to ensure the network can correctly handle images where no polyp is present, thus reducing false positives. To achieve this, additional datasets were incorporated to diversify the training data.
By combining these new datasets with those previously selected in Dataset selection section, the final dataset for the second verification step includes 19,657 images for training, 5,027 images for validation, and 10,660 images for testing.
This comprehensive dataset enables a direct comparison between a specific medical model (SSFormer) and a general-purpose segmentation model (SegFormer). SegFormer was selected as a state-of-the-art general-purpose segmentation model to benchmark against the specialized medical approach of SSFormer. The main objective is to evaluate how robust these models are under varied imaging conditions, especially in scenarios with no polyp present, thereby assessing their false-positive rates and overall segmentation accuracy.
To ensure fair comparison, both models are trained using identical hyperparameter settings, data augmentation strategies, and evaluation metrics. This standardization allows for an unbiased assessment of each model’s ability to generalize, highlighting their strengths and limitations across diverse datasets.
The results of this comparative analysis are discussed in detail in the following sections, focusing on performance differences between specific medical models and general-purpose models in terms of segmentation accuracy, generalization capability, and clinical applicability.
Model Training Settings: The model comparison stage evaluates the best specialized medical model (SSFormer) against a general-purpose segmentation model (SegFormer) to assess their robustness in handling different image variations. This comparison includes:
Training from scratch: Both models are trained without pre-trained weights to ensure unbiased learning.
Data augmentation: Standard techniques, including normalization, color jitter, and contrast adjustment, are applied consistently.
Learning rate scheduling and early stopping: These techniques optimize convergence and prevent overfitting.
Training parameters for both models are detailed in Table 2.
Training parameters used for SSFormer and SegFormer
| Parameter | SSFormer | SegFormer |
|---|---|---|
| Learning rate | 1e−4 | 1e−5 |
| Epochs | 200 | 50 |
| Optimizer | AdamW | SGD |
| Learning rate scheduler | Activated | Activated |
| Early stopping | Not activated | Activated |
Both SegFormer and SSFormer models were trained from scratch to ensure fair learning from the newly incorporated datasets, PolypDataset-TCNoEndo and PolypGen dataset. Standard data augmentation techniques were randomly applied to the input data. Early stopping terminated training when evaluation metrics showed no improvement for five consecutive epochs, preventing overfitting and optimizing computational resources.
The SegFormer model was trained in two variants, B2 and B4, both showing robust learning behavior. As shown in Figure 2 and Figure 3, SegFormer-B4 achieved superior evaluation metrics. The validation phase at each epoch confirmed that both variants approached their maximum metric values asymptotically, demonstrating their effectiveness for polyp segmentation.
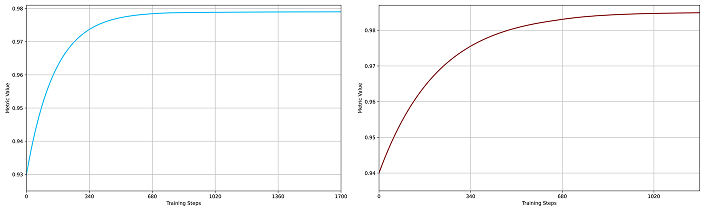
Dice metrics for SegFormer models. (a) SegFormer-B2 performance (blue curve) and (b) SegFormer-B4 performance (red curve) during training. In both plots, the x-axis represents training steps, and the y-axis shows the metric value (maximum is 1). The SegFormer-B4 variant achieves higher overall performance and better generalization, indicated by a smaller gap between training and validation curves
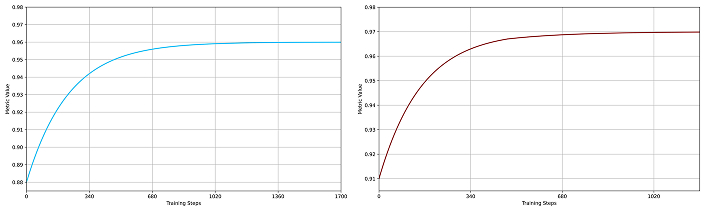
IoU metrics for SegFormer models. (a) SegFormer-B2 training performance (blue curve); (b) SegFormer-B4 training performance (red curve). In both plots, the x-axis represents training steps, and the y-axis shows the IoU metric value (maximum is 1). The convergence patterns illustrate steady improvement, with the SegFormer-B4 variant demonstrating more stable learning and superior final performance compared to the B2 variant. IoU: Intersection over Union
The SSFormer model was trained in two variants, small and large, both demonstrating effective learning dynamics. As depicted in Figure 4 and Figure 5, SSFormer-Large consistently achieved higher evaluation metrics, despite experiencing more fluctuations during training. Training curves for both variants illustrate rapid initial improvements followed by gradual optimization towards their peak performance, highlighting the robustness and suitability of SSFormer for polyp segmentation tasks.
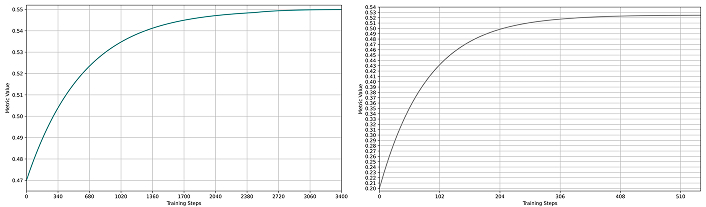
Dice metrics for SSFormer models. (a) SSFormer-Small training performance (green curve); (b) SSFormer-Large training performance (gray curve). In both plots, the x-axis represents training steps, and the y-axis shows the metric value (maximum is 1). Training curves exhibit rapid initial improvement followed by gradual optimization, with the large variant achieving higher final performance but displaying more pronounced fluctuations during training
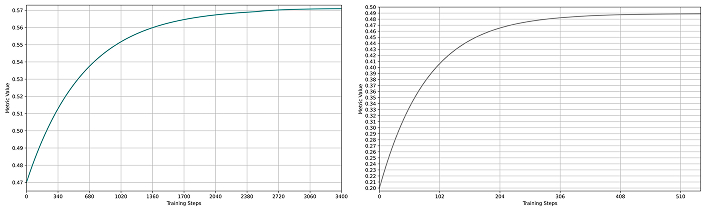
IoU metrics for SSFormer models. (a) SSFormer-Small training performance (green curve); (b) SSFormer-Large training performance (gray curve). In both plots, the x-axis represents training steps, and the y-axis shows the IoU metric value (maximum is 1). The convergence behavior resembles the Dice metrics, with both variants achieving strong performance; however, the large variant demonstrates superior final results despite exhibiting greater oscillations during training. IoU: Intersection over Union
This section presents the main experimental results obtained through the DeepPolyp framework. DeepPolyp is designed as a modular benchmarking platform to evaluate segmentation models on diverse datasets, with the possibility to extend it to additional architectures in future studies. The framework enables consistent comparison of both specialised medical models and general-purpose segmentation models using standard metrics and reproducible conditions. It also supports testing models in edge deployment settings.
The first set of experiments involved comparing DUCK-Net, a CNN-based model, and SSFormer, a Transformer-based model designed for medical imaging. Both models were trained on a mixed dataset composed of CVC-300, CVC-ClinicDB, CVC-ColonDB, ETIS-LaribPolypDB, and Kvasir. Evaluation was performed using the Dice coefficient and mean mIoU.
As shown in Table 3, DUCK-Net achieved good segmentation accuracy on larger datasets such as Kvasir (Dice: 0.9042) and CVC-ClinicDB (Dice: 0.8847). However, its performance dropped significantly on smaller datasets like CVC-ColonDB (Dice: 0.7169), revealing its limited generalisation. Additional experiments training DUCK-Net on single datasets (Tables 4, 5, 6, 7) confirmed this dependency on dataset size and distribution.
DUCK-Net results with training on the mixed dataset
| Dataset | Dice 17 | mIoU 17 | Dice 34 | mIoU 34 |
|---|---|---|---|---|
| CVC-300 | 0.8711 | 0.7717 | 0.8608 | 0.7556 |
| CVC-ClinicDB | 0.8583 | 0.7517 | 0.8847 | 0.7932 |
| CVC-ColonDB | 0.5331 | 0.3634 | 0.7169 | 0.5587 |
| ETIS-LaribPolypDB | 0.8268 | 0.7048 | 0.8957 | 0.8111 |
| Kvasir | 0.8423 | 0.7275 | 0.9042 | 0.8251 |
17 and 34 refer to the number of filters incorporated in the models: A model with 17 filters is identified as an optimal smaller model, whereas a model with 34 filters effectively represents a larger model. mIoU: mean Intersection over Union
DUCK-Net results with training on the CVC-300 dataset
| Dataset | Dice 17 | mIoU 17 | Dice 34 | mIoU 34 |
|---|---|---|---|---|
| CVC-ClinicDB | 0.1564 | 0.0848 | 0.0299 | 0.0152 |
| CVC-ColonDB | 0.02091 | 0.1167 | 0.2097 | 0.1171 |
| ETIS-LaribPolypDB | 0.2750 | 0.1594 | 0.0572 | 0.0294 |
| Kvasir | 0.0679 | 0.0352 | 0.0118 | 0.0059 |
17 and 34 refer to the number of filters incorporated in the models: A model with 17 filters is identified as an optimal smaller model, whereas a model with 34 filters effectively represents a larger model. mIoU: mean Intersection over Union
DUCK-Net results with training on the CVC-ClinicDB dataset
| Dataset | Dice 17 | mIoU 17 | Dice 34 | mIoU 34 |
|---|---|---|---|---|
| CVC-300 | 0.5055 | 0.3382 | 0.7348 | 0.5808 |
| CVC-ColonDB | 0.5751 | 0.4037 | 0.6032 | 0.4318 |
| ETIS-LaribPolypDB | 0.2319 | 0.1311 | 0.2207 | 0.1240 |
| Kvasir | 0.5909 | 0.4194 | 0.5896 | 0.4181 |
17 and 34 refer to the number of filters incorporated in the models: A model with 17 filters is identified as an optimal smaller model, whereas a model with 34 filters effectively represents a larger model. mIoU: mean Intersection over Union
DUCK-Net results with training on the CVC-ColonDB dataset
| Dataset | Dice 17 | mIoU 17 | Dice 34 | mIoU 34 |
|---|---|---|---|---|
| CVC-300 | 0.8935 | 0.8074 | 0.9200 | 0.8519 |
| CVC-ClinicDB | 0.5310 | 0.3615 | 0.6773 | 0.5121 |
| ETIS-LaribPolypDB | 0.6063 | 0.4350 | 0.6587 | 0.4911 |
| Kvasir | 0.4310 | 0.2747 | 0.6626 | 0.4954 |
17 and 34 refer to the number of filters incorporated in the models: A model with 17 filters is identified as an optimal smaller model, whereas a model with 34 filters effectively represents a larger model. mIoU: mean Intersection over Union
DUCK-Net results with training on the ETIS-LaribPolypDB dataset
| Dataset | Dice 17 | mIoU 17 | Dice 34 | mIoU 34 |
|---|---|---|---|---|
| CVC-300 | 0.1246 | 0.0665 | 0.2995 | 0.1761 |
| CVC-ClinicDB | 0.3518 | 0.2134 | 0.4310 | 0.2747 |
| CVC-ColonDB | 0.2517 | 0.1440 | 0.2902 | 0.1698 |
| Kvasir | 0.5261 | 0.3570 | 0.6537 | 0.4855 |
17 and 34 refer to the number of filters incorporated in the models: A model with 17 filters is identified as an optimal smaller model, whereas a model with 34 filters effectively represents a larger model. mIoU: mean Intersection over Union
In contrast, SSFormer achieved consistently higher accuracy across all datasets. For instance, Table 8 reports a Dice score of 0.9295 on CVC-300 using SSFormer-Large, surpassing DUCK-Net’s best result. This superior performance is attributed to the transformer-based attention mechanism, which captures global dependencies more effectively than convolutional filters. Further evidence is provided in Tables 9, 10, and 11, where SSFormer demonstrates strong generalization capabilities across various training scenarios. Notably, when trained on CVC-300 (Table 9), SSFormer achieves a Dice Small of 0.7265 on the Kvasir dataset, and when trained on CVC-ClinicDB (Table 10), it reaches a Dice Small of 0.9122 on CVC-ColonDB. The best performance is observed when trained on CVC-ColonDB (Table 11), where it obtains a Dice Large of 0.9490 on CVC-300, highlighting its robustness and cross-dataset generalizability.
SSFormer results trained on the mixed dataset
| Dataset | Dice Small | mIoU Small | Dice Large | mIoU Large |
|---|---|---|---|---|
| CVC-300 | 0.8064 | 0.7204 | 0.9295 | 0.8734 |
| CVC-ColonDB | 0.5703 | 0.4845 | 0.9069 | 0.8539 |
| CVC-ClinicDB | 0.6869 | 0.5678 | 0.9212 | 0.8757 |
| ETIS-LaribPolypDB | 0.6027 | 0.5164 | 0.8857 | 0.8349 |
| Kvasir | 0.7534 | 0.6365 | 0.9386 | 0.8970 |
mIoU: mean Intersection over Union
SSFormer results trained on the CVC-300 dataset
| Dataset | Dice Small | mIoU Small | Dice Large | mIoU Large |
|---|---|---|---|---|
| CVC-ClinicDB | 0.5716 | 0.4842 | 0.5465 | 0.4759 |
| CVC-ColonDB | 0.6708 | 0.5515 | 0.6476 | 0.5337 |
| ETIS-LaribPolypDB | 0.5826 | 0.4991 | 0.6147 | 0.5296 |
| Kvasir | 0.7265 | 0.6131 | 0.7143 | 0.5982 |
mIoU: mean Intersection over Union
SSFormer results trained on the CVC-ClinicDB dataset
| Dataset | Dice Small | mIoU Small | Dice Large | mIoU Large |
|---|---|---|---|---|
| CVC-300 | 0.8485 | 0.7779 | 0.8453 | 0.7790 |
| CVC-ColonDB | 0.9122 | 0.8575 | 0.9211 | 0.8689 |
| ETIS-LaribPolypDB | 0.8068 | 0.7222 | 0.8012 | 0.7332 |
| Kvasir | 0.8693 | 0.7898 | 0.8691 | 0.7962 |
mIoU: mean Intersection over Union
SSFormer results with training on the CVC-ColonDB dataset
| Dataset | Dice Small | mIoU Small | Dice Large | mIoU Large |
|---|---|---|---|---|
| CVC-300 | 0.9442 | 0.8979 | 0.9490 | 0.9073 |
| CVC-ClinicDB | 0.8708 | 0.7945 | 0.9191 | 0.8573 |
| ETIS-LaribPolypDB | 0.7825 | 0.6911 | 0.7893 | 0.7138 |
| Kvasir | 0.8010 | 0.6999 | 0.7978 | 0.7064 |
mIoU: mean Intersection over Union
Qualitative examples support these findings. Figure 6 shows that DUCK-Net struggles with polyps in low contrast or irregular shapes, often producing discontinuous or oversmoothed masks. On the other hand, SSFormer masks (Figure 7) preserve anatomical boundaries and fine details. Despite this, SSFormer also exhibits some failure cases under complex visual conditions, as illustrated in Figure 8 and Figure 9. These results suggest room for architectural improvement.
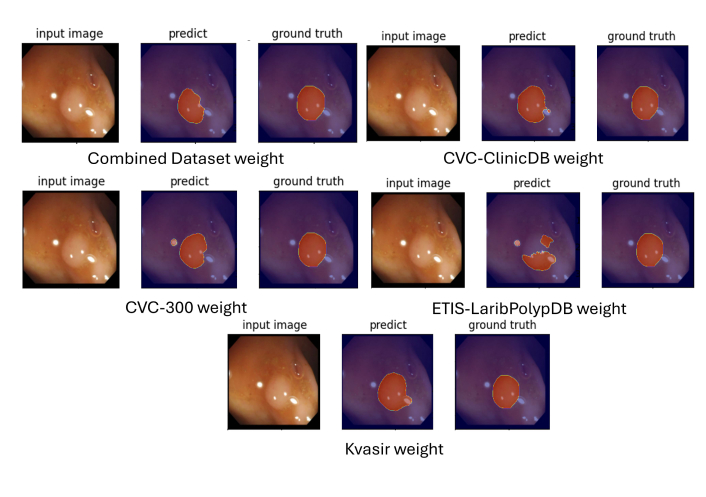
Example segmentation masks generated by DUCK-Net. The masks reveal challenges in accurately segmenting polyps, particularly in complex scenarios such as low-contrast regions or irregular polyp shapes
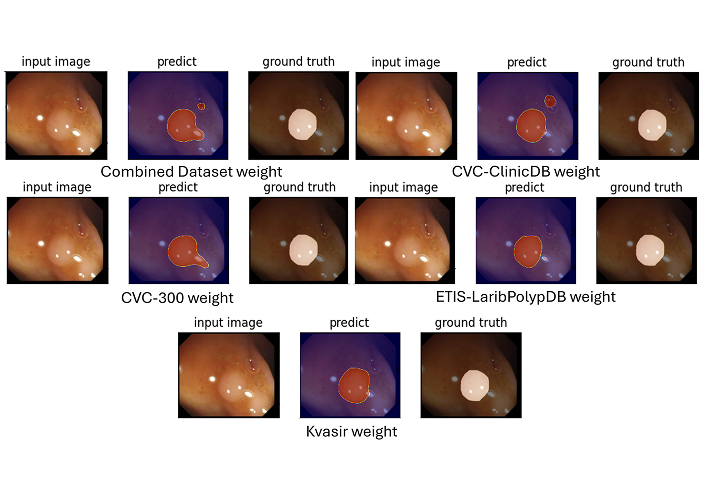
Example segmentation masks generated by SSFormer. SSFormer demonstrates significantly higher precision in polyp delineation, preserving fine-grained structures and achieving superior discrimination between polyps and the background
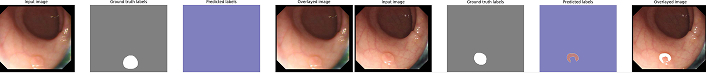
SSFormer small variant error for mask generation. In complex conditions such as low contrast or occlusion, SSFormer fails to produce accurate masks, highlighting the need for architectural optimizations
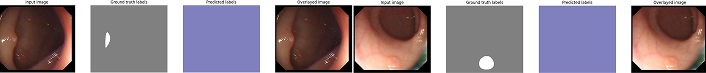
SSFormer large variant error for mask generation. Similar to the small variant, the large variant struggles with complex scenarios, indicating areas for future improvement
To further assess model robustness, the best-performing medical model (SSFormer) was compared to SegFormer, a general-purpose transformer segmentation model. Both SegFormer-B2 and SegFormer-B4 variants were evaluated using the same training and test conditions.
As shown in Table 12, SegFormer-B4 achieved the highest accuracy, with a Dice score of 0.9843 and IoU of 0.9694. These scores are significantly higher than those of SSFormer, whose performance remained below 0.18 in all configurations. Figure 10 and Figure 11 demonstrate SegFormer’s ability to produce accurate segmentation masks across variable polyp appearances and sizes.
Performance metrics (Dice and IoU) for SegFormer and SSFormer on the test set
| Metric | SegFormer-B2 | SegFormer-B4 | SSFormer-Small | SSFormer-Large |
|---|---|---|---|---|
| Dice | 0.9787 | 0.9843 | 0.1659 | 0.1780 |
| IoU | 0.9588 | 0.9694 | 0.1590 | 0.1616 |
IoU: Intersection over Union
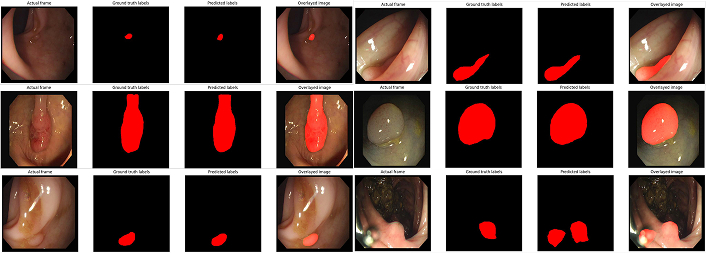
Segmentation masks generated by SegFormer-B2. The model accurately identifies polyp boundaries, showing high reliability and precision even in challenging cases with varying polyp sizes, shapes, and contrast levels
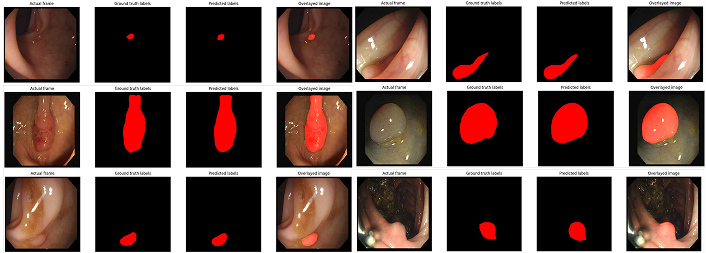
Segmentation masks generated by SegFormer-B4. The B4 variant demonstrates superior reliability and precision compared to B2, highlighting the impact of its enhanced architectural features
The strong performance of SegFormer, especially the B4 variant, is likely due to its larger capacity, improved architecture, and pretraining on diverse datasets. This suggests that general-purpose models, when fine-tuned on domain-specific data, can outperform models designed specifically for medical segmentation.
To evaluate whether the observed differences in segmentation performance between models were statistically significant, a two-tailed paired t-test was conducted on the Dice and IoU scores obtained from the test datasets. The test compared the performance of SegFormer-B4 with SSFormer-Large, as these two models represented the best-performing variants within their respective categories.
The results of the statistical analysis (Table 13) indicated that the difference in Dice scores between SegFormerB4 and SSFormer-Large was statistically significant, with a p-value < 0.001. Similarly, the IoU scores also showed a significant difference with p-value < 0.001. These results confirm that SegFormer-B4 consistently outperformed SSFormer-Large across all test datasets, and the difference in performance was not due to random variation.
Statistical comparison of segmentation performance between SegFormer and SSFormer variants
| Comparison | Metric | Mean ± Std (SegFormer) | Mean ± Std (SSFormer) | p-value (t-test) |
|---|---|---|---|---|
| SegFormer-B4 vs SSFormer-Large | Dice | 0.9843 ± 0.0052 | 0.1780 ± 0.0417 | < 0.001 |
| SegFormer-B4 vs SSFormer-Large | IoU | 0.9694 ± 0.0063 | 0.1616 ± 0.0389 | < 0.001 |
| SegFormer-B2 vs SSFormer-Small | Dice | 0.9787 ± 0.0061 | 0.1659 ± 0.0452 | < 0.01 |
| SegFormer-B2 vs SSFormer-Small | IoU | 0.9588 ± 0.0074 | 0.1590 ± 0.0428 | < 0.01 |
IoU: Intersection over Union
A similar test was performed between SegFormer-B2 and SSFormer-Small, also resulting in statistically significant differences (p-value < 0.01 for both Dice and IoU scores).
These findings reinforce the conclusions drawn from the quantitative and qualitative analyses and support the claim that SegFormer, despite being a general-purpose model, offers superior performance in the context of polyp segmentation. The inclusion of statistical validation adds reliability to the benchmarking procedure conducted through the DeepPolyp framework.
Overall, the experiments suggest that general-purpose models like SegFormer, especially variant B4, are effective for polyp segmentation when properly trained. SSFormer shows promising results as a medical-specific alternative, but requires architectural optimisation for deployment efficiency. DUCK-Net is outperformed in most scenarios, particularly on smaller datasets.
DeepPolyp enables fair and reproducible comparisons, integrates deployment analysis, and supports future extensions to novel models. Its modular design makes it useful for researchers and practitioners seeking to evaluate both accuracy and real-world applicability in medical image segmentation.
In addition to segmentation accuracy, the DeepPolyp framework allows for evaluating model performance in real-time conditions on edge devices. This functionality is essential for clinical settings where low-latency feedback is needed, such as during endoscopic procedures.
The edge deployment process was carried out on an NVIDIA Jetson Orin device and involved three main steps: (1) model conversion and optimization using ONNX and TensorRT for efficient inference, (2) evaluation of segmentation accuracy to ensure consistency with PyTorch-based results, and (3) performance measurement of the full inference pipeline.
Table 14 reports the segmentation metrics before and after deployment. SegFormer-B2 and B4 maintained high accuracy after optimization, with only a minor drop in Dice and IoU scores. On the other hand, SSFormer variants experienced minimal changes, although their overall performance remained low.
Metrics comparison between PyTorch and TensorRT models
| Model | PyTorch results | TensorRT results | ||
|---|---|---|---|---|
| Dice | IoU | Dice | IoU | |
| SegFormer-B2 | 0.9787 | 0.9588 | 0.9231 | 0.8684 |
| SegFormer-B4 | 0.9843 | 0.9694 | 0.9433 | 0.9025 |
| SSFormer-Small | 0.1659 | 0.1590 | 0.1606 | 0.1449 |
| SSFormer-Large | 0.1780 | 0.1616 | 0.1667 | 0.1487 |
IoU: Intersection over Union
After validating segmentation accuracy, a performance analysis of the full execution pipeline was conducted. The analysis considered a 20-second video sequence and measured execution times for each component, including preprocessing, inference, post-processing, and image rendering. Among these, inference remained the most computationally intensive step.
As reported in Table 15, execution times for the inference pipeline, detailed for each component, edge deployment significantly reduced inference time compared to GPU execution. SegFormer-B2 achieved the lowest latency, completing the full pipeline in approximately 94 ms per frame, making it suitable for real-time operation. SegFormer-B4 maintained acceptable latency (135 ms per frame), while SSFormer models were slower but remained within operational limits for semi-real-time tasks.
Execution times for the inference pipeline, detailed for each component
| Function | GPU execution time (ms) | Edge execution time (ms) | ||||||
|---|---|---|---|---|---|---|---|---|
| SegFormer-B2 | SegFormer-B4 | SSFormer-Small | SSFormer-Large | SegFormer-B2 | SegFormer-B4 | SSFormer-Small | SSFormer-Large | |
| Inference | 431.71 | 527.20 | 427.65 | 520.96 | 74.18 | 115.45 | 64.38 | 98.76 |
| Sigmoid | 0.662 | 4.529 | ||||||
| Interpolate | 0.107 | 0.537 | ||||||
| Mask processing | 0.237 | 0.657 | ||||||
| Add weighted | 0.357 | 3.772 | ||||||
| Image encode | 0.112 | 5.052 | ||||||
| Display handle update | 0.35 | 5.508 | ||||||
| Full pipeline | 433.535 | 529.025 | 429.475 | 522.785 | 94.235 | 135.505 | 84.435 | 118.815 |
The results confirm that the DeepPolyp framework supports the deployment of segmentation models in real-time clinical scenarios. The integration of model optimisation (ONNX and TensorRT) and pipeline measurement enables a complete evaluation of both segmentation performance and execution speed.
The large performance gap between SegFormer and SSFormer in edge execution can be explained by their architectural differences. SegFormer is based on standard transformer blocks and convolutional layers that are compatible with ONNX export and TensorRT inference. These standard layers allow efficient graph optimisation, fusion, and quantisation during the conversion process. In contrast, SSFormer includes custom attention blocks and non-standard operations that limit the effectiveness of TensorRT’s optimisation strategies. As a result, SegFormer models benefit from faster execution and better utilisation of hardware resources on edge devices, while SSFormer models require further re-engineering or custom plugin development to match the same level of optimisation.
These findings highlight the importance of architectural compatibility when targeting edge deployment. While SSFormer may offer advantages in feature learning, SegFormer remains more suitable for real-time clinical applications due to its streamlined conversion and execution process.
This study introduced DeepPolyp, a modular framework for evaluating and deploying segmentation models for polyp detection. Among the tested models, SegFormer achieved the highest generalisation performance, consistently delivering accurate results across diverse datasets with different imaging conditions and polyp morphologies. This suggests that SegFormer is able to extract complex visual features relevant to real-world clinical applications. Its cross-scale attention mechanism and efficient feature extraction contributed to its robustness, especially in detecting small, flat, or occluded polyps.
In contrast, SSFormer, although transformer-based, showed lower performance, particularly on smaller or more complex datasets. The lack of multi-scale context integration limited its ability to generalise. However, its architecture, based on self-attention, still proved effective in modelling global dependencies, making it a promising baseline for future optimisation. DUCK-Net, a CNN-based architecture, performed well only with larger datasets, revealing its limitations in generalisation.
The performance differences between these models align with their design choices. SegFormer combines the benefits of lightweight design with multi-scale contextual reasoning, making it ideal for real-time inference and deployment. SegFormer-B2 was chosen for edge deployment using NVIDIA Jetson Orin with TensorRT optimisation, where it achieved low latency and high segmentation accuracy, confirming its suitability for clinical integration.
To assess the real-world utility of the DeepPolyp framework, a questionnaire was administered as part of the ENDO-AI project to both specialised and general medical personnel. The results confirmed strong interest in key features such as automatic polyp detection and data historization.
Feedback from specialised personnel:
Universal agreement on the usefulness of AI-assisted diagnostic tools.
Strong interest in automatic polyp detection (82% rated it “Very” or “Extremely” useful).
A preference for quick workflows: Only 75% saw preliminary visualisations as “Very” useful.
Mixed opinions on 3D reconstruction and measurements: Most found them only “Somewhat” useful.
Broad consensus (91%) that this technology can improve diagnostic accuracy.
Feedback from non-specialist personnel:
94% rated the system as “Very” or “Extremely” useful for diagnostics.
Automatic detection was rated as useful by 94%.
3D reconstruction and measurements were seen as “Extremely” useful by over 75%.
Preliminary result viewing received strong support (84%), though some warned about over-reliance.
Clear endorsement of the system’s role in real-world clinical diagnostics.
This analysis confirms that DeepPolyp addresses clinical needs effectively. Automatic detection, consistent accuracy, and reliable deployment on embedded systems make the framework highly suitable for modern diagnostic workflows. However, the feedback also emphasises areas for future improvement. For example, while 3D visualisation is appreciated, its integration must consider clinical workflow constraints. Training programs should be implemented to ensure balanced use of automation, preserving clinical judgement.
Future work should explore optimising SSFormer with hybrid architectures that combine convolutional and transformer-based layers. Lightweight transformer designs will be evaluated for better efficiency on edge devices. Domain adaptation, generative data augmentation, and semi-supervised learning can enhance generalisation and reduce reliance on manual annotations.
Additionally, the framework’s flexibility makes it adaptable to other medical imaging tasks, such as tumour segmentation in radiology and histopathology. DeepPolyp offers a reliable and extensible platform for advancing AI-driven diagnostics in diverse clinical contexts.
CNNs: convolutional neural networks
mIoU: mean Intersection over Union
We extend our gratitude to all partners and collaborators who contributed to the successful implementation and validation of the proposed system, including Key To Business, Department of Neurosciences – Catholic University of the Sacred Heart, and Studio5T.
MM: Conceptualization, Methodology, Data curation, Investigation, Formal analysis, Writing—original draft. SS: Software, Investigation, Formal analysis. MP: Formal analysis, Writing—review & editing. IGC: Project administration, Formal analysis, Supervision, Conceptualization. All authors read and approved the final version of the manuscript.
The authors declare that they have no conflicts of interest.
The datasets used in this manuscript were sourced from Kvasir-SEG, CVC-ClinicDB, ETIS-LaribPolypDB, PolypGen, CVC-ColonDB, CVC-300, and PolypDataset-TCNoEndo. All datasets and information presented in this article are fully anonymized and do not contain any personally identifiable information. Therefore, ethical approval, consent to participate, and consent to publication are not required.
Not required.
Not required.
The dataset used in this study is derived from publicly available sources cited in the manuscript. The details of each dataset are as follows: (1) Kvasir-SEG: Simula Datasets - Kvasir SEG; (2) CVC-ClinicDB: Simula Datasets - Kvasir SEG; (3) ETIS-LaribPolypDB: ETIS-LaribPolypDB; (4) PolypGen: Simula Datasets - Kvasir SEG; (5) CVC-ColonDB: CVC colon DB | Visual Interaction Group; (6) CVC-300: CVC-300; (7) PolypDataset-TCNoEndo: An augmented version of Kvasir-SEG. These datasets were selected to ensure a diverse and representative collection of polyp appearances, enhancing model generalization and robustness for real-world clinical applications. However, due to current project constraints, the integrated dataset cannot be openly released at this time. Access may be granted upon reasonable request and will be evaluated on a case-by-case basis.
This project has been co-financed by the European Union through the PR FESR 2021–2027 RSI program of Regione Lazio, managed by LazioInnova [CUP F89J23001090007], and approved with the publication of the rankings related to the public notice “Riposizionamento competitivo RSI PR FESR 2021–2027 Regione Lazio” in the BUR on 21/11/2023. The authors would like to thank the European Union and Regione Lazio for their support in enabling this research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Paolo Dell’Aversana
Leyla Ebrahimi ... Sara Hashemi
