Affiliation:
1División de Ciencias e Ingenierías, Universidad de Guanajuato, León, Guanajuato C.P. 37150, México
2Vicerrectoría de Investigación, Universidad La Salle México, Ciudad de México C.P. 06140, México
Email: pu.munozgonzalez@ugto.mx
ORCID: https://orcid.org/0000-0003-4072-3783
Affiliation:
1División de Ciencias e Ingenierías, Universidad de Guanajuato, León, Guanajuato C.P. 37150, México
ORCID: https://orcid.org/0009-0009-3303-0509
Affiliation:
1División de Ciencias e Ingenierías, Universidad de Guanajuato, León, Guanajuato C.P. 37150, México
Affiliation:
1División de Ciencias e Ingenierías, Universidad de Guanajuato, León, Guanajuato C.P. 37150, México
Affiliation:
1División de Ciencias e Ingenierías, Universidad de Guanajuato, León, Guanajuato C.P. 37150, México
Affiliation:
1División de Ciencias e Ingenierías, Universidad de Guanajuato, León, Guanajuato C.P. 37150, México
Affiliation:
1División de Ciencias e Ingenierías, Universidad de Guanajuato, León, Guanajuato C.P. 37150, México
ORCID: https://orcid.org/0000-0001-6604-395X
Explor BioMat-X. 2025;2:101344 DOI: https://doi.org/10.37349/ebmx.2025.101344
Received: May 17, 2025 Accepted: August 07, 2025 Published: August 31, 2025
Academic Editor: Juan Mareque-Rivas, Swansea University, UK
This review presents key molecular biology techniques used to investigate interactions between biomaterials and biological systems, emphasizing their role in evaluating biocompatibility and cellular responses. We focus on methodologies such as recombinant DNA technology, polymerase chain reaction (PCR), in situ hybridization, immunocytochemistry (ICC), and immunohistochemistry (IHC). These tools enable the detection and quantification of gene and protein expression, particularly those involved in inflammation and tissue regeneration, providing molecular-level insights into how cells respond to biomaterial cues. We discuss the relevance of these techniques in identifying inflammatory markers, tracking cell differentiation, and understanding tissue integration processes, as well as how their implementation faces technical challenges, including interference from the physicochemical properties of biomaterials, difficulties in sample preparation, and the standardization of protocols across different platforms. Addressing these limitations is vital to ensure data reliability and reproducibility. Looking ahead, we highlight emerging opportunities involving the integration of 3D imaging technologies and artificial intelligence to manage and interpret high-dimensional biological data. This article also serves as a practical tool for emerging investigators who are entering the field of biomaterials, offering accessible guidance on the selection and application of essential molecular biology techniques. These innovations promise to accelerate the rational design of biomaterials tailored to specific clinical applications and patient needs. In conclusion, molecular biology techniques provide a foundational toolkit for characterizing biological responses to biomaterials, supporting the development of safer and more effective therapeutic materials and empowering emerging investigators to contribute meaningfully to the next generation of biomedical solutions.
Biomaterials represent one of the most important fields in biomedical engineering and technology, with applications ranging from the development of prosthetics that serve solely structural and mechanical support functions to tissue engineering scaffolds capable of providing matrices that promote tissue regeneration [1]. According to the National Institute of Biomedical Imaging and Engineering, a biomaterial is defined as any material designed to interact with biological systems to evaluate, treat, augment, or replace any tissue, organ, or function of the body, emphasizing that biomaterials not only must avoid causing an adverse immune response but can also have active functions, such as promoting tissue regeneration or delivering drugs [2]. These materials, which can be natural or synthetic, have evolved significantly over the years from simple inert structures to bioactive platforms capable of directly interacting with surrounding cells and tissues [3]. Biomaterials provide not only physical support but also trigger specific biological responses through controlled release of growth factors or modulation of inflammatory processes. Biomaterials play a key role in various medical fields, including regenerative medicine, implantology, and drug delivery [4]. Biomaterial-based solutions to unmet medical and clinical needs require addressing scientific issues not only of materials synthesis and manufacturing methods but also of biological responses to the material. Among the wide variety of biomaterials, collagen-based matrices are widely used for soft tissue repair, synthetic polymers offer tunable mechanical and degradation properties suitable for drug delivery and bone regeneration, nanoparticles enable targeted therapeutic delivery and immune modulation, and graphene-based scaffolds show promise in neural tissue engineering due to their electrical conductivity and biocompatibility [5–8]. Graphene-based scaffolds, due to their electrical conductivity and biocompatibility, have also gained increasing attention, particularly in neural tissue engineering and regenerative applications, where three-dimensional architectures and interactions with stem cells show promising potential [9, 10].
Biocompatibility as a biomaterial feature refers to their ability to function safely in medical treatments without causing harmful effects while promoting appropriate cellular or tissue responses and ensuring optimal clinical performance [11]. Determining biocompatibility requires analyzing various biological parameters through consecutive strategies, including in vitro, ex vivo, and in vivo, preclinical and clinical studies, which are often standardized by international organizations such as the International Standards Organization (ISO) and the American Society for Testing and Materials (ASTM), overseeing proper cell culture techniques for cytotoxicity evaluation [12]. Biocompatibility is one of the key challenges in biomaterials research for the development of new products and applications, as illustrated in Figure 1. The first aspect is that the material must not cause toxicity or adverse reactions and must be accepted by the body without triggering a negative immune response. Tissue integration is another crucial challenge, as biomaterials must be safe and effective in their intended function, whether promoting tissue regeneration, facilitating healing, enhancing organ functionality, or merely being an inert interaction during biomaterial implantation [13, 14]. Successful integration is related to factors such as the material’s structure, mechanical and biochemical properties, and its interactions with surrounding cells and tissues, depending on the target tissues treated (Figure 1a). Biological variability further complicates biomaterial development, as each patient has a unique biological profile that can influence the body’s response to a given material. For example, at best a hydrogel can accomplish its commitment to inducing wound healing (Figure 1b) [15], while there are complications of breast implant rejection due to fibrotic encapsulation of the implant, having the risk of causing carcinoma (Figure 1c) [16], or periimplantitis in bone implants that occur over time, which can be prevented by coating metal implants with antimicrobial additives (Figure 1d) [17–19]. This highlights the need for biomaterial technology that considers biological variability along with manufacturing and production issues for commercial medical devices. Additionally, regulatory approval is a significant hurdle, as biomaterials must meet strict safety and efficacy standards before clinical use. This process is often lengthy and costly, potentially limiting innovation and delaying the introduction of new products to the market.
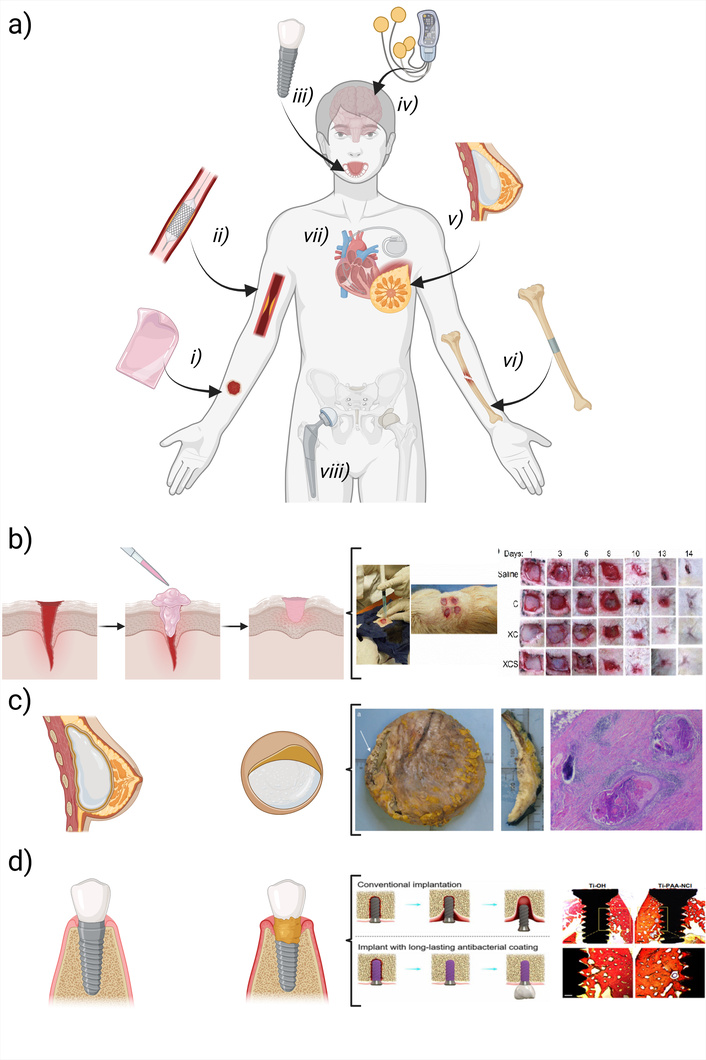
Diverse biomaterial-host interactions depend on technology and function. a) Examples include (i) hydrogel dressings for wound healing, (ii) thrombosis-resistant vascular stents, (iii) dental implants integrating with tissues, (iv) neural stimulators without inflammation, (v) fibrosis-free breast implants, (vi) bone plates reinforcing fractures, (vii) rejection-free cardiac pacemakers, and (viii) durable hip prostheses. A schematic illustrates b) gel state materials made of crosslinked collagen that are designed to accelerate wound healing and promote tissue regeneration instead of scar tissue formation [15], c) the role of the surface of polyurethanes when this causes an undesirable response such as breast implant fibrotic encapsulation where histological analysis suggests that the implant may cause carcinoma [16], or d) the role of drug-eluding dental implants to prevent peri-implantitis over time, where antibacterial coatings made of polyacrylic acid-chlorine allow osteointegration while this is capable of preventing bacterial infections [19]. Created in BioRender. Muñoz, P. (2025) https://BioRender.com/99c3z4c. b) Partly adapted with permission from [15]. © 2022 IOP Publishing Ltd. c) partly adapted from [16]. © The Author(s) 2023. Licensed under a CC-BY 4.0. d) Partly adapted from [19]. © The Author(s) 2021. Licensed under a CC-BY 4.0
Molecular biology techniques play a fundamental role in this process by enabling us to evidence the influence of biomaterials on gene expression, protein synthesis, and cellular behaviors such as proliferation, differentiation, and apoptosis. These results are inputs to establish biocompatibility and biofunctionality for biomaterials in different applications, including modulation of immune cells to regulate wound healing in response to an implant, or to promote tissue regeneration rather than a fibrotic outcome [12, 20, 21]. Given these challenges, training researchers in these techniques is essential to advance biomaterials science and technology. Molecular biology provides powerful tools to study the interactions between biomaterials and living tissues at the cellular and molecular levels. Techniques such as PCR, electrophoresis, and DNA sequencing enable researchers to analyze and manipulate genetic material, facilitating the development of biomaterials that integrate effectively with biological tissue. For instance, genetic modifications can enhance cell receptivity to a specific biomaterial, improving the success of biomedical treatments. Additionally, cell culture techniques play a crucial role in assessing biocompatibility by simulating biological environments and evaluating cellular responses to different materials, helping predict their behavior in vivo. Furthermore, proteomic and metabolomic analyses allow researchers to examine how biomaterials influence metabolic pathways and gene expression, which is essential for designing materials that are not only safe to repair defects but also biointeractive to promote regenerative mechanics [22, 23]. By applying molecular biology techniques to biomaterials research, crucial insights are gained into the biocompatibility and long-term behavior of a material, so training research staff in these techniques is strongly suggested [15, 24–27].
Biomaterials research and development is a growing field where distinct disciplines converge to drive innovation, including biotechnology, medicine, chemistry, and engineering. According to Precedence Research, the global biomaterials market was valued at USD 171.35 billion in 2024 and is projected to reach approximately USD 523.75 billion by 2034, growing at a compound annual rate of 11.82% during this period. North America leads the market, supported by organizations such as the National Institute of Standards and Technology and the National Science Foundation, while the Asia-Pacific region is expected to experience rapid growth due to expansion strategies [28]. Additionally, initiatives to strengthen this field are emerging worldwide through the training of new generations of researchers in the roadmap to support the biomaterials’ translation [29]. Collaboration with experts in molecular biology and related areas remains essential to ensure continued progress and strengthen the growth of the field in the different regions of the world, which requires the expertise to interact in an inter- and transdisciplinary manner.
This article presents five fundamental molecular biology techniques commonly used in biomaterials research, outlining their background, relevance, and applications. It provides a simplified overview of basic protocols along with examples of how researchers have utilized these techniques for biomaterial characterization. By detailing these methodologies, the article serves as a resource for scientists entering the field while also emphasizing the close relationship between molecular biology and biomaterials. Additionally, it offers guidance to both early-career and experienced researchers interested in exploring this branch of biomedical engineering. The goal is to promote interdisciplinary collaboration across scientific fields. Rather than replacing the expertise of molecular biologists, this work aims to provide a foundation for assessing the biomedical potential of newly developed biomaterials.
Nucleic acid hybridization is a key molecular biology technique for detecting and analyzing specific DNA or RNA sequences based on base complementarity. Among its methods, the Southern blot stands out, involving DNA extraction, restriction enzyme digestion, electrophoretic separation, and transfer to a membrane for hybridization with a labeled DNA probe [30–32]. The Northern blot follows a similar approach but targets RNA [33, 34]. A significant variant is in situ hybridization (ISH), performed directly on tissues, cells, or chromosomes, first applied in 1969 to detect DNA in Xenopus. This technique has since advanced into a crucial tool for studying gene expression, cell proliferation, apoptosis, and disease-related genes, including cancer-related ones [35]. Its RNA application focuses on detecting specific mRNAs. Hybridization relies on the renaturation of single-stranded DNA or RNA following denaturation, which disrupts hydrogen bonds and hydrophobic interactions through heat or chemical agents. Controlled cooling allows complementary strands to reanneal, forming hybrids if the pairing strand differs from the original. This property enables hybridization-based gene and nucleic acid identification [36]. When talking of ISH, two key components are required: A target sequence generated by proteolytic enzymes and a labeled probe complementary to it. The probe, marked with a radioactive, fluorescent, or enzymatic tag, enables visualization of hybridization sites within tissues or cells [36]. In biomaterials research, ISH serves as a crucial tool for assessing biocompatibility, offering molecular insights into material-tissue interactions, and validating biomedical applications.
Below is a simplified protocol for implementing ISH, which is graphically summarized in Figure 2. For the complete process, refer to [37, 38]. In Fluorescent in situ hybridization (FISH), the preparation and application of probes are critical for accurate detection of chromosomal abnormalities. Three main types of probes are commonly used: chromosome painting probes, which label entire chromosomes and enable the identification of structural and numerical rearrangements using techniques like Multicolor FISH (M-FISH) and Spectral Karyotyping (SKY); repetitive sequence probes, which target repetitive chromosomal regions such as telomeric or centromeric sequences, aiding in the detection of aneuploidies like monosomies or trisomies; and locus-specific probes, which are derived from genomic clones in PAC or BAC vectors and are used to detect specific chromosomal aberrations such as translocations, deletions, or inversions.
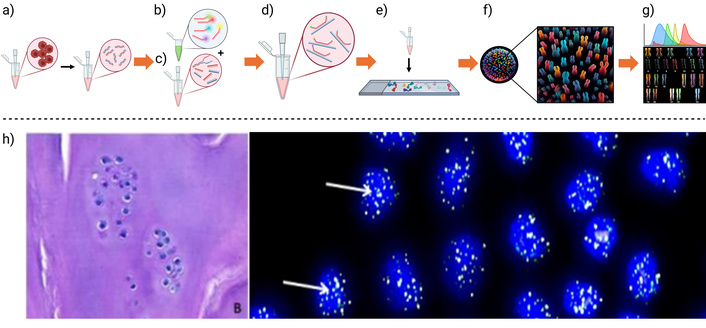
A standard FISH-SKY method is graphically summarized, describing the main steps. a) cytological preparation where cells are fixed, and then squashed to expose the DNA, b) probe fluorescent labeling, attaching different fluorescent markers for each probe, c) DNA denaturation, d) probe application to the denatured DNA, e) labeled DNA hybridization, showing a schematic representation of f) chromosome disposition and g) rearrangement after fluorescent signal detection. h) Application example: FISH analysis of telomere length in cartilage tissue after autologous chondrocyte implantation (left). Telomeres were labeled with green fluorescence, allowing their size to be monitored as an indicator of cellular senescence over time (right). Created in BioRender. Muñoz, P. (2025) https://BioRender.com/400p3en. h) Partly adapted from [16]. © The Author(s) 2023. Licensed under a CC-BY 4.0
Cytological preparation is essential for clear hybridization signals (Figure 2a). For chromosomal studies, cells—such as root tips in the mitotic phase—are fixed in an ethanol:acetic acid solution (3:1), stained with 1% acetocarmine, squashed in 45% acetic acid, and can be stored at –80°C for up to a year. DNA probes are labeled (Figure 2b) using non-radioactive molecules like biotin, digoxigenin, fluorescein, or rhodamine through techniques such as nick translation, random priming, or PCR. Detection is then performed using antibodies conjugated to fluorochromes or enzymes.
ISH methods (Figure 2c–g) vary depending on the approach. In non-radioactive ISH, chromosomal DNA is denatured with formamide and SSC at 68–70°C, the probe is applied and incubated, and hybridization sites are detected with streptavidin-peroxidase and developed with DAB (to be defined) and H2O2, followed by Giemsa counterstaining. Genomic in situ hybridization (GISH) involves fragmenting genomic DNA, labeling it with biotin-16-dUTP, and hybridizing similarly, using the same detection steps. In FISH, the DNA is denatured, the fluorescently labeled probe is hybridized, and the slides are blocked with PBS and Triton-100, then incubated with fluorescent antibodies (e.g., FITC). A counterstain with propidium iodide allows the observation of fluorescent signals under a microscope, enabling precise chromosomal analysis.
ISH can be used to investigate how biomaterials interact with cells in a living environment. This tool is valuable for understanding cellular responses to implanted materials and how they interact with the genetic material of surrounding cells. In addition to reflecting the genetic interaction between biomaterials and host cells, ISH also allows for the evaluation of gene expression, immune response, and other aspects related to material biocompatibility. Some of the earliest applications of this technique in this field date back to 1996, when the Institute of Pathology and the Department of Trauma and Reconstructive Surgery in Germany analyzed osteoblast activity at the biomaterial-bone interface using ISH. In this study, the amount of mRNA expressed by osteoblasts was evaluated, confirming RNA loss during the decalcification process [39].
Additionally, ISH has a direct application in biomaterial development. One example is a study that proposed a new method for the biosynthesis of a three-dimensional scaffold using the in-situ hybridization of carbon nanotubes (CNT) with bacterial cellulose (BC). This method achieved homogeneous and effective hybridization between CNT and BC, resulting in hybrid scaffolds with excellent osteoinductive and osteoconductive properties. This, in turn, promoted higher bone density, improving regeneration efficiency. This innovation highlights the potential of ISH of three-dimensional scaffolds with nanomaterials as a new approach for regenerative medicine [40].
To provide a clearer and more structured overview of ISH applications in biomaterials research, Table 1 summarizes key case studies from the literature. These examples demonstrate how ISH has been used to assess biocompatibility, immune responses, and scaffold performance by targeting specific genes or cellular processes.
Applications of in situ hybridization (ISH) in biomaterials
| Application | Type of biomaterial or system | Purpose of ISH | Benefits | Limitations | Ref. |
|---|---|---|---|---|---|
| Evaluation of osteoblasts at the bone–biomaterial interface | β-tricalcium phosphate hybridized with procollagen | Assess the temporal expression of osteogenic genes during new bone formation | High spatial and temporal resolution detects gene activity associated with bone remodeling | RNA degradation due to decalcification; no signal at early stages | [40] |
| Quantification of mRNA expression in cancer tissue samples | Formalin-fixed, paraffin-embedded (FFPE) clinical samples | Quantitative detection of nucleic acid biomarkers | High signal-to-noise ratio; suitable for diagnostic pathology; works with FFPE tissues; cost-effective and sensitive | Complex procedures, possible uncertainty in results; image quality can be compromised in conventional single-molecule FISH (smFISH) | [41] |
| Non-invasive detection of endometrial cancer (EC) | Vaginal swabs from patients with suspected EC | Identification of genomic imbalances in exfoliated tumor cells | Non-invasive; high sensitivity and negative predictive value | Requires validation; false positives are possible; relies on the quality of brushing and cell preservation | [42] |
However, the ISH technique faces limitations, such as difficulty in identifying targets with low DNA or RNA copy numbers. Despite this, strategies continue to be developed to improve its sensitivity, either by amplifying nucleic acid sequences before hybridization or by enhancing signal detection after completion [43]. In a study evaluating gene expression changes on biomaterial surfaces, the combination of ISH for mRNA extraction with real-time quantitative PCR allowed researchers to identify expressed genes. However, this technique did not provide information on the abundance of different cell types, which is problematic since inflammatory processes mainly involve neutrophils and macrophages in wound healing. Understanding the balance between these cells and the presence of specific macrophage subpopulations is crucial for a better comprehension of these processes [44].
Although there are areas requiring further development, the information obtained so far is fundamental to ensuring the safety and advancement of any material intended for biological applications. ISH is used to study the effects of biomaterials on the gene expression of exposed cells, allowing researchers to observe how materials influence the activation or repression of key genes for biocompatibility. For example, ISH can assess the expression of genes related to inflammation or cell proliferation in tissues interacting with implants or scaffolds.
Genetic engineering has led to the development of recombinant DNA, a groundbreaking tool in modern molecular biology. By combining DNA sequences from different organisms, recombinant DNA allows for precise genetic manipulation, enabling the modification of traits and the introduction of new biological functions. This technology has revolutionized fields like medicine, agriculture, and biomaterials, facilitating advances in biotechnology with high precision and control [45]. Central to recombinant DNA technology are restriction enzymes, which cut DNA at specific sites, and DNA ligase, which joins the fragments to form new molecules. These recombinant molecules are introduced into host organisms, such as bacteria, yeast, or mammalian cells, where they can replicate and express proteins or molecules. This technique has enabled the large-scale production of human proteins, including insulin, growth factors, and monoclonal antibodies [45, 46].
In biomaterials, recombinant DNA technology offers new possibilities for designing functional materials. It allows for the modification of biological and physical properties, optimizing interactions with cellular and tissue environments. By introducing specific genes into cells interacting with biomaterials, researchers can control protein production to promote tissue regeneration, wound healing, or implant integration [47, 48]. A key application of recombinant DNA in biomaterials is creating platforms for controlled expression of bioactive molecules. For example, tissue engineering scaffolds can be designed to contain genetically modified cells that secrete growth factors, enhancing tissue formation. This is especially important in bone and cartilage regeneration, where proteins like BMP (bone morphogenetic protein) are essential for cellular differentiation and bone formation [49].
Recombinant DNA technology also plays a vital role in improving biomaterial biocompatibility and reducing immune system rejection. By incorporating genes that encode immunomodulatory molecules, biomaterials can be designed to regulate immune responses, minimizing inflammation and rejection in applications like stents, prostheses, and other medical implants [47, 48]. Additionally, recombinant DNA enables the development of responsive biomaterials. By incorporating genes that react to external stimuli like temperature, pH, or mechanical signals, biomaterials can adjust their properties in real-time to meet the needs of the biological environment. For example, biomaterials that release therapeutic proteins in response to inflammation could be used to treat conditions like rheumatoid arthritis or cancer, expanding the potential of recombinant DNA in personalized medicine [49].
Ultimately, recombinant DNA technology is not only advancing biomaterials but also enabling the creation of more accurate models for human diseases, aiding the development of new therapies. In summary, recombinant DNA has ushered in a new era in biomaterials science, blending biology and engineering to create materials that are not only compatible but actively involved in healing and regeneration processes [47].
This section outlines a generic protocol for applying recombinant DNA technology to biomaterials, which is graphically summarized in Figure 3. For a more detailed overview of the protocol, refer to references [50, 51]. The process of creating recombinant DNA begins with the selection of appropriate vectors and host organisms (Figure 3a), where the host is chosen based on the intended biomedical application, and the cloning vector is a DNA molecule capable of replicating within the host while carrying the target gene. Next, both the vector and the target DNA are prepared (Figure 3b, c) by isolating them and using restriction enzymes to cut at specific sites, allowing for precise ligation. The recombinant DNA is then formed (Figure 3d) by combining the prepared vector and target DNA using DNA ligase, resulting in a single construct containing the desired gene. This recombinant DNA is introduced into the host organism through transformation methods such as electroporation or the use of viral vectors (Figure 3e), followed by the selection of host cells that successfully incorporate the recombinant DNA. Finally, detection and identification are carried out (Figure 3f) using techniques like PCR, restriction analysis, or sequencing to confirm successful incorporation, after which the recombinant DNA is expressed.
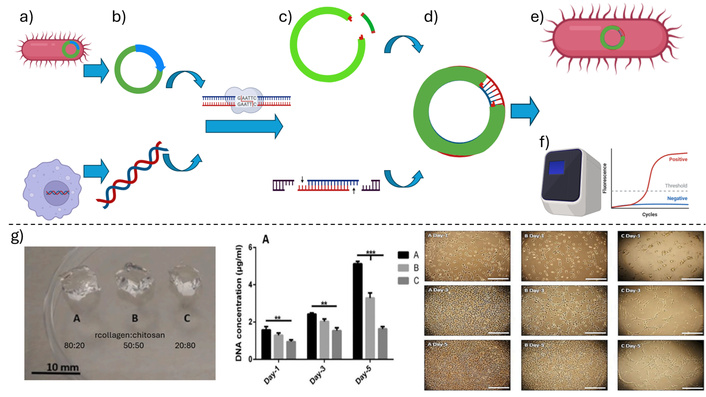
A schematic representation of the main steps of the recombinant DNA technology protocol, showing. a) The selection of the host (up) and cloning (down) cells, b) the isolation of the host (up) and cloning (down) DNA, c) the preparation of the host plasmid (up) and cloning (down) DNA, where the restriction enzyme has cut these molecules exposing the sticky ends, d) the cloning plasmid after host DNA incorporation, e) the plasmid inserted into the host cell, and f) the quantification of the interest gene expression via polymerase chain reaction. g) Application example: recombinant collagen–chitosan hydrogels prepared at three different collagen ratios (left). Hydrogels with higher recombinant collagen content promoted greater fibroblast DNA content (center) and increased cell proliferation over five days (right). Created in BioRender. Muñoz, P. (2025) https://BioRender.com/lk1rhie. g) Adapted with permission from [52]. © 2021 Elsevier.
Recombinant DNA technology is a fundamental tool in modern biotechnology, used to produce key biomolecules, such as therapeutic proteins, which are utilized in the creation of biomaterials for various medical interventions. Through genetic engineering, it is possible to insert genes of interest into host organisms such as bacteria, yeasts, or mammalian cells, enabling them to produce proteins that would otherwise be difficult or costly to obtain. Clear examples include the production of human insulin, growth hormones, and clotting factors, which have revolutionized the treatment of diseases such as diabetes and hemophilia. These recombinant proteins can be integrated into biomaterials to enhance their functionality in medical applications, such as scaffolds for tissue regeneration or matrices for controlled drug delivery [51].
The first breakthrough in medical biotechnology using recombinant DNA technology was the production of human insulin by Genentech in the 1980s. By inserting the human insulin gene into Escherichia coli bacteria, a safe and efficient source of insulin for diabetes treatment was achieved. This recombinant insulin eliminated the dependency on animal-derived insulin, which carried the risk of adverse immune reactions [53]. In hemophilia A, a deficiency in coagulation factor VIII causes severe blood clotting issues. Recombinant DNA technology enabled the production of factor VIII in mammalian cells, offering a safer and purer alternative to human plasma concentrates, which carried the risk of transmitting viruses such as HIV and hepatitis [54]. This treatment revolutionized the lives of hemophilia patients by providing reliable therapy with no risk of infection.
Collagen is a structural protein essential in tissues like skin, tendons, and bones. The production of recombinant human collagen has been achieved by inserting collagen genes into yeast or mammalian cells. A notable example is the company FibroGen, which produces recombinant collagen for use in manufacturing dermal patches used in the healing of chronic wounds such as diabetic ulcers, offering a safer alternative to animal-derived collagen [55, 56]. In the field of regenerative medicine, genetically modified stem cells have been used to enhance tissue regeneration. An example is the use of mesenchymal stem cells genetically modified to express specific growth factors, such as transforming growth factor beta (TGF-β), which is crucial in bone tissue repair. These cells, combined with biodegradable biomaterial scaffolds, have demonstrated accelerated bone fracture healing in clinical trials [57, 58]. Recombinant collagen has been applied as hydrogels in combination with other biomolecules, such as chitosan (Figure 3g), obtaining that the recombinant collagen presence improves several aspects of this biomaterial, such as the biocompatibility of these hydrogels, cell infiltration, and accelerates wound healing [52].
Recombinant DNA technology has also benefited the development of monoclonal antibodies [59]. Monoclonal antibodies, such as trastuzumab (Herceptin), are recombinant proteins produced using recombinant DNA technology and are used to treat cancers like HER2-positive breast cancer. These antibodies specifically target cancer cells expressing the HER2 receptor, blocking their growth and signaling. Recombinant DNA technology has enabled the large-scale production of these antibodies, improving the treatment of aggressive cancers [60]. An emerging application is the controlled release of recombinant proteins through biomaterial matrices. A notable case is the development of a controlled release system for recombinant VEGF (vascular endothelial growth factor) to promote angiogenesis in tissue regeneration. This system, based on biodegradable polymers, has shown effectiveness in treating ischemic wounds by promoting the formation of new blood vessels [61, 62]. These key examples of recombinant DNA applications in regenerative and therapeutic medicine (where therapeutic medicine refers to medical approaches aimed at treating or curing diseases) are summarized in Table 2, highlighting the host systems, medical uses, and associated benefits of each strategy.
Applications of recombinant DNA technology in biomaterials and regenerative medicine
| Application | Product or strategy | Host system | Medical purpose | Benefits | Ref. |
|---|---|---|---|---|---|
| Recombinant hydrogel fabrication | Recombinant human collagen (RHC)/carboxylated chitosan (CHI) hydrogels | RHC from engineered cells | Soft tissue engineering and wound healing | Avoids viral contamination and immunogenicity from animal collagen; tunable mechanical/degradation properties; promotes cell adhesion, proliferation, and in vivo wound healing | [52] |
| Improving the silk mechanical properties | Transgenic silkworm silk expressing MaSp1-type repeats with polyalanine motifs | Transgenic Bombyx mori via the piggyBac system | Production of bionic silk for biomedical materials | Increased β-sheet content leads to higher strength and stiffness; potential for scalable production of high-performance silk-based biomaterials | [63] |
| Repair of skull defect | TGF-β3/recombinant human-like collagen/chitosan (TRFS) scaffold loaded with human periodontal ligament stem cells | Recombinant human-like collagen (produced via engineered microbial or eukaryotic systems) | Cranial bone regeneration in traumatic skull defects | Enhances osteogenic differentiation, biocompatibility, and scaffold biodegradability; promotes bone regeneration via stem cell delivery | [64] |
| Osteogenic differentiation of stem cells | Graphene nanogrids fabricated from oxidatively unzipped multi-walled carbon nanotubes (rGONR grid) | Human mesenchymal stem cells (hMSCs) from umbilical cord blood | Bone tissue engineering/regeneration | Highly accelerated and patterned osteogenic differentiation (~2.2× vs rGO); enhanced cytoskeletal organization and biocompatibility | [65] |
| Tendon regeneration | Recombinant periostin (rPOSTN) integrated into an aligned collagen scaffold | Tendon stem/progenitor cells (TSPCs), rat model | Achilles tendon repair | rPOSTN promotes TSPC proliferation and tenogenic potential; the scaffold supports hierarchical collagen formation; functional recovery in vivo is demonstrated | [66] |
PCR is a fundamental technique in molecular biology that enables the amplification of specific DNA sequences from minimal samples. Developed by Kary Mullis in the 1980s, PCR has transformed genetic analysis, disease diagnostics, and biotechnology by facilitating the exponential replication of target DNA regions [67]. The process involves cyclic thermocycling steps: denaturation (separating DNA strands), primer annealing (binding of short DNA primers), and extension (DNA polymerase-driven synthesis of new strands). Essential reaction components include template DNA, primers, nucleotides (dNTPs), DNA polymerase, buffer, and Mg2+ ions. The amplified DNA can be analyzed using agarose gel electrophoresis to confirm successful amplification [68–70].
PCR plays a crucial role in biomaterial research by enabling the characterization of genetic material extracted from biological samples. It is used to identify genetic variants, analyze gene expressions, and investigate critical molecular interactions in fields such as tissue engineering and biomedical diagnostics [71–73]. Additionally, real-time PCR (RT-PCR) allows the study of RNA by converting it into complementary DNA (cDNA) via reverse transcription [67, 68].
This method, which is both rapid and highly efficient, has revolutionized areas like genetic research, molecular diagnostics, and the development of targeted therapies, among other biotechnological applications. PCR is a method that allows for the exponential amplification of DNA sequences. It uses a heat-stable DNA polymerase to replicate specific DNA segments through controlled temperature cycles. This technique can also be adapted to RT-PCR to work with RNA, converting it to cDNA using the enzyme reverse transcriptase.
This section outlines a generic PCR protocol with specific applications in biomaterials research, which is graphically summarized in Figure 4. PCR follows a series of essential steps to ensure successful DNA amplification. These steps include primer design, reaction mixture preparation, PCR execution, and thermal cycling adjustments. For a more exhaustive description of the procedure, it is recommended to consult references [74, 75].
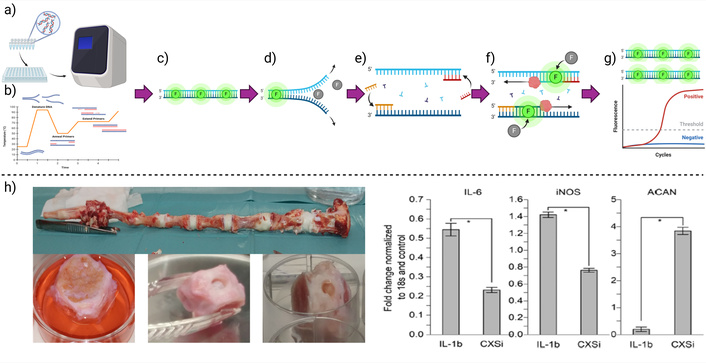
A standard quantitative-PCR method is graphically summarized, describing the main steps. a) Reaction mixture preparation, addition to the well plate, and introduction into the thermocycler, b) representation of a basic thermal cycle program, where each thermal stage: c) initiation, d) DNA denaturation, e) primer annealing, f) elongation, g) having the final product and a representative replication quantitative result curve. h) Application example: quantification of gene expression (IL-6, iNOS, and ACAN) in an ex vivo model of intervertebral disc rupture treated with a collagen hydrogel (left). Results (right) show downregulation of the proinflammatory genes IL-6 and iNOS, and upregulation of ACAN, which encodes aggrecan, a key extracellular matrix protein. Created in BioRender. Muñoz, P. (2025) https://BioRender.com/jm8irwg. h) Adapted with permission from [24]. © 2019 Royal Society of Chemistry
Accurate primer design is essential for PCR efficiency and specificity. Primers must bind to complementary DNA strands: one to the sense strand (5'→3') and the other to the antisense strand (3'→5'). Common issues include the formation of secondary structures such as hairpins (self-annealing), primer-dimer formation, and melting temperature (Tm) mismatches that interfere with proper annealing during thermal cycling. Optimal primers are typically 15–30 nucleotides long, have a GC content between 40–60%, and a Tm between 52–58°C, with no more than a 5°C difference between the forward and reverse primers. The 3' end should contain a G or C base to enhance stability, and complementarity between 3' ends should be avoided to prevent dimer formation. Repeats and homopolymeric stretches should also be minimized.
A precise reaction mix is crucial for consistent PCR results. The mix includes sterile water, 10× buffer (with or without MgCl2), dNTPs, primers, template DNA, and DNA polymerase. Each reaction should be set up in clean, labeled 0.2 mL PCR tubes or a 96-well plate placed on ice. It is important to include a negative control (no DNA) to check for contamination, and a positive control (known DNA template and primers) to validate the reaction. Table 3 shows a typical concentration of the reagents that can be used during a reaction mixture preparation.
Typical concentrations of the reagents are added to the reaction mixture
| Component | Concentration | Volume |
|---|---|---|
| Buffer (MgCl2) | 10× | 5 μL |
| dNTPs | 10 mM | 1 μL |
| Mg2+ | 1.5 mM | - |
| Each primer | 20–50 pmol | - |
| Template DNA | 1–1,000 ng | 0.5 μL |
| DNA polymerase | 0.5–2.5 units | - |
| Water | - | Complete 50 μL |
The basic PCR protocol begins by placing PCR tubes or a 96-well plate on ice to maintain reagent stability. Each tube is clearly labeled using ethanol-resistant markers to prevent misidentification. The reaction mixture is assembled by sequentially adding sterile water, PCR buffer, dNTPs, MgCl2 (if not already included in the buffer), forward and reverse primers, and the DNA template. DNA polymerase is added last to minimize nonspecific amplification before thermal cycling. The components are mixed gently using a micropipette, ensuring no bubbles are formed. After sealing the tubes, they are loaded into a thermocycler programmed with the appropriate cycling conditions. Once the amplification process is complete, the PCR products are stored at 4°C. These products are subsequently analyzed using agarose gel electrophoresis, typically stained with ethidium bromide or a safer alternative like SYBR Safe, to visualize the amplified DNA fragments.
PCR relies on precise and rapid temperature changes to enable DNA denaturation, primer annealing, and DNA strand elongation. A typical thermal program includes an initial denaturation step, followed by 25–35 cycles of denaturation, annealing, and extension, with a final extension and cooling step (Table 4). During this final extension, Taq polymerase typically adds a 3' A-overhang to the amplified DNA, a feature that is particularly useful for TA cloning strategies.
Typical thermal cycling conditions
| Step | Temperature | Time | Notes |
|---|---|---|---|
| Initial denaturation | 94–98°C | 1–3 min | Longer for hot-start enzymes |
| Denaturation (per cycle) | 94°C | 10–60 s | Melts DNA strands |
| Annealing | ~Tm –5°C | 30 s | Depending on the primer Tm |
| Extension | 70–75°C | ~1 min per 1–2 kb | 72°C is typical for Taq, 75°C for Pfu |
| Final extension | 72°C | 5 min | Ensures complete product synthesis |
| Hold | 4°C | Indefinite | Preserves samples until retrieval |
PCR is a key tool for assessing gene expression in response to biomaterial interactions with living tissues. It is widely used to study genes involved in inflammatory responses, wound healing, and tissue regeneration, providing insights into the biocompatibility of biomaterials.
Biomaterials, whether natural or synthetic, are designed for medical applications to restore or replace biological functions. Despite their promising physical, chemical, and biological properties, they do not always integrate seamlessly with living tissues. A common issue is the immune system recognizing the material as foreign, triggering an inflammatory response. PCR allows for the identification of gene expression changes, helping to evaluate tissue reactions at the molecular level (Table 5 summarizes representative examples of these applications in various tissues and biomaterials).
Applications of PCR in the evaluation of biological responses to biomaterials
| Application | Target genes | Model or system | Biological purpose | Benefits | Ref. |
|---|---|---|---|---|---|
| Quantification of the expression of inflammation/regeneration | IL6, iNOS, ACAN | Ex vivo model of intervertebral disc degeneration | Evaluate the effectiveness of the treatment with collagen hydrogels to promote immunomodulation | Real-time tracking of the effectiveness of the treatment on the inflammatory pathology | [24] |
| Plasmonic thermocycling RT-qPCR for rapid diagnostics | SARS-CoV-2 RNA (including variants) | Human saliva and nasal specimens with a plasmonic nanoparticle-enhanced PCR system | Real-time detection and quantification of viral RNA in clinical samples | High sensitivity and specificity, reduced thermocycling time, compact and portable for point-of-care diagnostics | [76] |
| Evaluation of cardiac tissue response to conductive hydrogel | Cardiac-specific markers | Mouse intracardial injection model; 3D culture of human iPSC-derived cardiomyocytes | To assess the biocompatibility and functional integration of reverse thermal hydrogel in cardiac tissue engineering | Enables minimally invasive delivery, in situ gelation, and evaluation of cardiac gene expression profiles | [77] |
| Improvement of decellularized extracellular matrix scaffolds for liver bioengineering | Nano-graphene oxide crosslinked decellularized liver scaffold | Mouse liver failure model | Liver tissue regeneration: an alternative to donor organ transplantation | Inhibits enzymatic degradation (MMPs); promotes M2 macrophage polarization; reduces inflammation; improves scaffold longevity and function | [78] |
MMPs: matrix metalloproteinases; M2 macrophage: immune cells associated with a prohealing phenotype
PCR has been applied to study biomaterial-tissue interactions in various contexts. For instance, biomaterial patches in the gastrointestinal tract were analyzed, revealing increased expression of the vitamin D binding protein in gastric tissue following implantation [79]. Similarly, in studies of bioactive polymer scaffolds for bone regeneration, PCR has been used to assess osteogenesis-related genes, demonstrating how biomaterials influence stem cell differentiation into osteoblasts [80]. In cardiac tissue engineering, PCR has helped monitor the gene expression in biomaterial grafts that simulate myocardium, identifying materials that promote integration without chronic inflammation [81, 82]. Even this technique has been helpful to determine if modified collagen hydrogels can modulate the inflammatory response, promote regeneration in ex vivo intervertebral discs rupture models, by quantifying the expression of genes that code inflammatory cytokines IL-6 and iNOS, and the extracellular matrix protein aggrecan (Figure 4h) [24].
In research on biocompatibility, PCR has been used to evaluate inflammatory markers such as IL-6 and TNF-α in macrophages interacting with biomaterials, providing critical data for optimizing material design [83, 84]. PCR has also been employed to study angiogenesis by analyzing VEGF expression in tissues implanted with bioactive scaffolds, which is essential for applications requiring vascularization, such as chronic wound healing [85]. Additionally, in nerve regeneration studies, PCR has been used to measure the expression of NGF and BDNF, which are crucial for neuronal survival and repair, helping identify biomaterials that support nerve growth [86].
In bone regeneration, PCR has been applied to analyze gene expressions in response to materials like hydroxyapatite and bioglass, focusing on markers such as RUNX2 and ALP, which are essential for osteointegration [87]. Understanding the molecular response to biomaterials is fundamental for improving their design and ensuring their functionality without triggering adverse biological reactions.
Despite its advantages, PCR has limitations. Sample preparation is critical for accurate results, and conventional PCR requires complementary techniques like gel electrophoresis for interpretation. RT-PCR enables precise quantification but requires specialized equipment and strict sample handling. Additionally, the technique is limited in amplifying long DNA sequences, which can be a constraint in complex biomaterial studies [88, 89].
PCR remains an essential technique for evaluating biomaterial tissue interactions, providing valuable information for the development of more effective and biocompatible medical materials.
ICC is a powerful technique for visualizing cell proteins using labeled antibodies. Developed in the 1940s by A.H. Coons, it has since evolved into a crucial method in histopathology, cytopathology, and biomedical research. ICC can be applied to both cultured cells and tissue sections using fluorescence or visible light microscopy, depending on the marker [90, 91]. A key feature of ICC is its ability to be used in both animal and plant cells, provided a specific antibody is available. This technique relies on antigen-antibody interactions, allowing precise protein localization. The specificity of antibodies enables the detection of minimal antigen amounts, making ICC a highly sensitive method. Advances in antibody generation continue to expand its applications [92, 93]. Initially, ICC was limited to cultured cells, while IHC was reserved for paraffin-embedded tissues. Today, ICC is widely used for formaldehyde-fixed animal tissues, which can be cryosectioned and incubated with antibodies [94].
The core principle of ICC is the antigen-antibody reaction. Antibodies, produced by the immune system in response to antigens, can be purified and conjugated with fluorescent dyes for detection. ICC employs two types: polyclonal antibodies, which recognize multiple epitopes of an antigen, and monoclonal antibodies, which bind to a single epitope [95, 96]. For instance, a polyclonal antibody against actin can be generated by injecting purified actin into a rabbit, triggering an immune response. The resulting antibodies are then isolated and labeled for use in ICC [97]. ICC techniques include: i) Direct Immunocytochemistry: Uses a primary antibody directly conjugated to a fluorophore or enzyme. While simple, it has limited sensitivity [98]. ii) Indirect Immunocytochemistry: Enhances sensitivity by using an unlabeled primary antibody, followed by a labeled secondary antibody that binds to the primary. This amplification improves detection and reduces the need for labeling multiple primary antibodies individually [99].
Detection markers vary depending on the application. Fluorescent dyes allow imaging with fluorescence or confocal microscopy, enzymatic markers like peroxidase enable colorimetric detection under a light microscope, and colloidal gold nanoparticles are used in electron microscopy [100]. ICC is extensively used in oncology for detecting tumor biomarkers, as well as in basic research to study protein distribution in cells and tissues. It has been instrumental in understanding processes such as the cell cycle, apoptosis, and intracellular signaling [101–103].
This section outlines a standardized protocol for ICC with specific applications in biomaterials research, which is graphically summarized in Figure 5. For a more detailed procedure, refer to references [104–106].
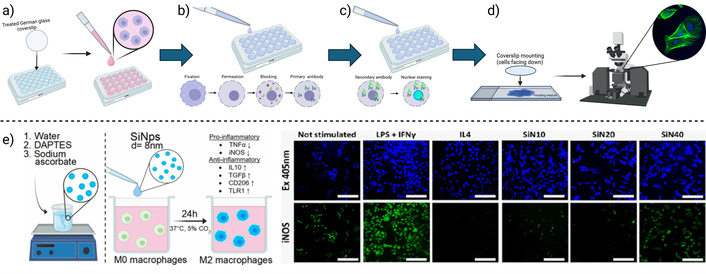
A basic immunocytochemistry method is graphically summarized, describing the main steps. a) Cell culture, b) cell fixation, membrane permeation, protein blocking, and primary antibody incubation, c) secondary antibody incubation, and nuclear staining, d) coverslip mounting and visualization. e) Example application: Immunocytofluorescence was used to evaluate the inflammatory response of macrophages to silicon nanoparticles. The expression of the proinflammatory molecule iNOS was detected (right) after stimulation with different nanoparticle doses (left). Notably, only the 40 μg/mL dose induced green-fluorescent labeling of iNOS. Created in BioRender. Muñoz, P. (2025) https://BioRender.com/15xojll. e) Adapted with permission from [107]. © 2023 American Chemical Society
To prepare coverslips (Figure 5a), German glass coverslips are first placed in a holder to allow solution access to both sides. They are incubated in a 1% Liquinox solution for 10 minutes, followed by three washes with deionized water to remove detergent residue. Coverslips are then incubated in 1 M HCl for 30 minutes, rinsed again three times with deionized water, and dried overnight at 65°C. Once dry, they are transferred to a clean glass plate and sterilized by autoclaving. For optimal cell adhesion, sterile coverslips are placed in a multiwell plate under a laminar flow hood. A coating step begins with the application of 10 µg/mL poly-L-lysine D in sterile water, incubated for 5 minutes at room temperature. After removing the poly-L-lysine, the coverslips are incubated with 20 µg/mL laminin in EMEM medium for 4 hours or overnight at 37°C. Finally, the coverslips are rinsed with PBS before use. Cells are seeded onto the prepared coverslips at a density of 100,000 to 300,000 cells/mL per well and incubated at 37°C to allow adherence for at least 4 hours. At this stage, cells can optionally be stimulated with experimental biomaterials, depending on the specific design of the study.
Fixation begins with the preparation of a paraformaldehyde-based fixative (Figure 5b), carefully pH-adjusted to dissolve without heat. After removing the culture medium, pre-warmed fixative is added to the cells and incubated for 5 to 15 minutes at room temperature (typically 10 minutes). Fixative is then properly discarded as hazardous waste, and cells are washed three times with PBS (5 minutes each wash), avoiding any drying between steps. For intracellular antigens, permeabilization is performed by adding 0.3% Triton X-100 in PBS. Due to its viscosity, Triton X-100 should be added slowly to ensure complete dissolution. Cells are incubated with this solution for 5 minutes, followed by three PBS washes. Blocking is conducted with 5% bovine serum albumin (BSA) in PBS, filtered through a 0.45 µm syringe filter. Cells are incubated with the blocking solution for 1 hour at room temperature. Subsequently, the primary antibody is diluted in 1% BSA in PBS and applied to a parafilm-covered glass plate (30 µL for 25 mm coverslips, 20 µL for 18 mm, or 15 µL for 12 mm). Coverslips are placed cell-side down onto the antibody droplets, covered with another layer of parafilm, and incubated overnight at 4°C.
After primary antibody incubation (Figure 5c, d), the parafilm is carefully removed, and coverslips are inverted into a plate with PBS, ensuring cells face up. Coverslips are washed three times with PBS for 5 minutes each. Secondary antibodies specific to the species of the primary antibody are diluted in 1% BSA in PBS. If multiple primary antibodies are used, they must originate from different species or isotypes to allow selective detection. Coverslips are incubated with the secondary antibody for 2 hours at room temperature in the dark to protect fluorescent signals. For nuclear staining, cells are washed twice with PBS, then incubated for 1 minute in Hoechst stain (2 µg/mL in PBS). If signal intensity is insufficient, incubation time or concentration may be increased. A final PBS wash is performed before mounting. To mount coverslips, a drop of fluorescence-compatible mounting medium is applied to the slide: 12 µL for 25 mm coverslips, 6 µL for 18 mm, or 3 µL for 12 mm. The back of the coverslip is rinsed with deionized water to remove salt residues and gently dried. Coverslips are then placed cell-side down onto the mounting medium, avoiding air bubbles. Slides are labeled with the date and sample details and dried in the dark. Excess mounting medium can be removed with gentle suction, and edges may be sealed with nail polish for long-term storage. Prepared slides should be stored at –20°C.
ICC enables the analysis of cellular interactions with biomaterials by identifying key proteins involved in adhesion, proliferation, and differentiation. These techniques have revolutionized biomaterials research by providing detailed insights into the physiological responses triggered by implants [108–110]. Initially, biomaterials were designed to minimize immune responses and avoid rejection or chronic inflammation. However, advances in regenerative medicine have shifted the focus toward bioactive biomaterials capable of modulating cellular responses to promote tissue regeneration [1–3].
For instance, in bone regeneration, biomaterials should not only be biocompatible but also stimulate osteoblast activity. Here, immunochemical techniques play a crucial role by enabling the analysis of these complex interactions, providing detailed information on how cells respond to different implanted materials [111, 112]. Immunocytochemical studies have evaluated titanium surfaces for cell adhesion, revealing that differentiated osteoblasts respond favorably to surface roughness while stem cells proliferate independently of topography [108]. This type of analysis is essential for developing optimized surfaces for medical implants, as cell adhesion and proliferation are determinants of implant success. Similarly, collagen matrices for urethral reconstruction in rabbits demonstrated proper integration and muscle fiber organization six months post-implantation [113]. Hydroxyapatite implants have been analyzed using IHC, revealing increased expression of osteopontin and TGF-β, key markers of osteogenesis [114]. In vascular engineering, electrospun polymer grafts assessed with endothelial markers (CD31, vWF) demonstrated new blood vessel formation and cellular integration in rat models, while recent advances in microvascular tissue engineering explore biofabrication strategies such as extrusion-based and droplet-based bioprinting, Kenzan, and biogripper techniques to create nano- and micro-sized aggregates and microspheres that mimic capillary networks through the inclusion of specific cell types and proangiogenic factors [115, 116].
Collagen scaffolds modified with silver nanoparticles (AgNPs) have been shown to reduce proinflammatory macrophages (CD68) and increase reparative macrophages (CD206), creating a favorable environment for wound healing [117]. Additionally, cross-linked collagen hydrogels either containing colloidal silica or modified with epoxyeicosatrienoic acids promoted an anti-inflammatory macrophage response, identified via CD206 immunostaining [14, 24, 25, 118]. The immunomodulatory effect of plain silicon nanoparticles has been evaluated, where immunocytofluorescence has been used to prove that the inflammatory response of macrophages to these particles can be dose-regulated, proving that doses over 40 μg/mL promote the expression of the proinflammatory cytokine iNOS (Figure 5e), while lower doses promote the expression of the antiinflammatory molecules CD206 and TLR1 [107].
Myocardial patches using induced pluripotent stem cells (iPSCs) on fibrin gel scaffolds have been evaluated through immunohistochemical techniques, confirming differentiation into cardiomyocytes via troponin I and α-actinin expression [119, 120]. Controlled drug release systems using biodegradable PLGA polymers have been studied in animal models, with immunohistochemical analysis revealing an initial inflammatory response that was mitigated over time by integrated anti-inflammatory drugs, demonstrating their potential for chronic inflammatory conditions [121, 122].
Despite their advantages, immunochemical techniques have limitations, including potential errors in sample preparation, antibody concentration optimization, and cross-reactivity. Fluorescence-based methods require careful selection of filters and lasers to prevent signal interference [123, 124]. Future developments in 3D bioprinting and tissue engineering will increasingly rely on ICC to evaluate cellular responses to novel biomaterials. The integration of these techniques with super-resolution microscopy and single-cell protein analysis will deepen our understanding of biomaterial-cell interactions, opening new frontiers in regenerative medicine and personalized therapies [104].
IHC is a technique that enables the localization of proteins in tissue sections, allowing for the visualization of molecular structures using enzymatic or fluorescent detection methods [124, 125]. Combining principles from immunology, histology, and chemistry, IHC plays a crucial role in biomaterials research by identifying biological markers related to inflammation, wound healing, angiogenesis, apoptosis, and tissue regeneration [126–133]. Scientific literature provides guides, protocols, and textbooks that are essential for those seeking to deepen their use of IHC and achieve proper preparation for biomaterial evaluations. Biocompatibility is fundamental for medical applications, as it defines a material’s ability to interact with biological systems without adverse effects. IHC is widely used to evaluate biomaterials in both in vivo and in vitro studies, providing insights into immune responses and tissue integration [6, 126].
The technique relies on antibodies, primarily from the IgG class, which specifically bind to target antigens [125, 134]. Detection can be direct, where the primary antibody is conjugated with a marker, or indirect, using a secondary antibody to enhance signal sensitivity [135]. Fluorescent markers are preferred for frozen tissues, while enzymatic labels are used in formalin-fixed, paraffin-embedded samples [136]. Proper tissue fixation is essential for preserving molecular structures. Cross-linking fixatives, such as formaldehyde, stabilize proteins, whereas precipitating fixatives, like alcohols, remove water to preserve tissue morphology [125]. Both types are selective for proteins, while lipids and carbohydrates remain trapped within them.
Fixation methods include perfusion, distributing the fixative through the vasculature, immersion, and submerging tissue in a fixative solution [125]. Samples are then sectioned into thin slices (5–15 µm) for microscopic analysis. Frozen tissue preparation requires precision to prevent ice crystal formation, which can damage cellular structures. A sucrose solution is often used to protect tissue integrity before embedding in a suitable compound for sectioning and analysis [137]. These controlled procedures ensure optimal conditions for IHC, enhancing the accuracy of biomaterial evaluations.
This section describes a general IHC protocol with specific applications in biomaterials research, which is graphically summarized in Figure 6. Refer to references [138, 139] for a more comprehensive description. The IHC process is summarized below.
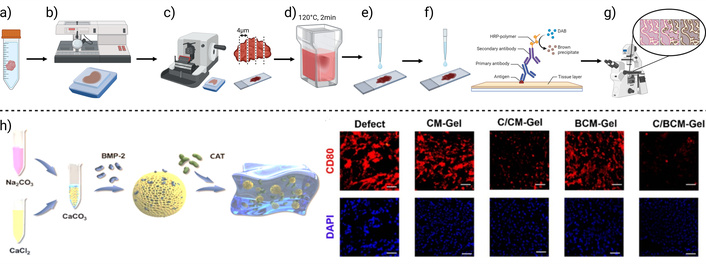
An immunohistochemistry method is graphically summarized, describing the main steps. a) Tissue fixation, b) tissue processing and block formation, c) tissue sectioning, d) epitope recuperation, e) protein and endogenous enzyme blocking, f) detection system and counterstaining, and g) visualization. h) Example application: Immunohistofluorescence was used to evaluate osteoimmunomodulatory effects of composites (left). Expression of the M1 macrophage marker CD80 was detected using red-fluorescent labeling (right). Results show that composites containing catalase did not promote this undesired inflammatory response. Created in BioRender. Muñoz, P. (2025) https://BioRender.com/1hs3m5j. h) Adapted from [140]. © The Author(s) 2024. Licensed under a CC-BY 4.0
Proper tissue fixation is critical for IHC reproducibility. Surgical specimens are typically fixed in 10% buffered formalin for 24 hours at room temperature, maintaining a tissue-to-fixative volume ratio between 1:1 and 1:20. Alternatively, frozen sections may be used, requiring specific protocols [124]. After fixation, tissues undergo dehydration through a graded ethanol series (50% to 98%) using a tissue processor, followed by paraffin infiltration. Each step generally lasts one hour. The processed tissues are then embedded into paraffin blocks for subsequent sectioning.
Paraffin-embedded tissues are sectioned at approximately 4 µm thickness. Prolonged storage, particularly beyond two months, may cause epitope degradation and affect staining quality. Since fixation can mask epitopes, a retrieval step is necessary. Heat-induced epitope retrieval (HIER), commonly performed at 120°C for 2 minutes, is used to enhance antibody binding and signal sensitivity.
To reduce background staining and improve specificity, two types of blocking are implemented. First, protein blocking prevents the nonspecific binding of antibody Fc regions. Common reagents include 5–10% normal serum, 0.1–0.5% BSA, gelatin, or skim milk. The incubation ranges from 30 minutes to overnight at room temperature or 4°C. Second, endogenous enzyme blocking prevents false signals: 3% hydrogen peroxide is used to inhibit peroxidases, 10 mM levamisole for alkaline phosphatases, and avidin/biotin steps for endogenous biotin.
Primary antibodies are applied after blocking, typically diluted in 1% BSA in PBS, and incubated overnight at 4°C. Detection is performed using species-specific secondary antibodies conjugated to enzymes or fluorophores. Systems such as avidin-biotin complex (ABC), streptavidin-biotin, polymer-based detection, and tyramide signal amplification are commonly used. The latter two enhance detection sensitivity significantly, up to 50-fold. Automated IHC platforms offer high reproducibility, though they may limit flexibility in reagent or protocol selection.
Following secondary antibody incubation, tissue sections are counterstained to provide contrast. Hematoxylin is commonly used, though multiplex IHC may involve diverse counterstains for target discrimination. Finally, stained slides are visualized using light or fluorescence microscopy, depending on the labeling method.
IHC is a powerful tool for evaluating how biomaterials influence biocompatibility by analyzing inflammatory, angiogenic, and apoptotic markers in surrounding tissues. This technique enables the investigation of tissue-biomaterial interactions, providing insights into specific proteins associated with cellular responses such as adhesion, proliferation, and differentiation within the tissue context. Understanding these molecular-level interactions is crucial for assessing how implanted biomaterials modulate cellular behavior within tissues. By allowing the precise localization of key proteins, IHC has revolutionized biomaterials research, offering a detailed perspective on physiological processes triggered in response to implants [141].
In a study on hyaluronic acid-based hydrogels for IL-10 delivery in idiopathic pulmonary fibrosis (IPF), IHC played a critical role in identifying and quantifying fibrosis-related markers and cellular activity in lung tissue treated with IL-10 hydrogels. Using specific antibodies, IHC detected proteins associated with fibrosis, such as type I collagen and TGF-β, in tissue sections obtained after intratracheal administration of the treatment in a murine model. This allowed for an assessment of IL-10’s antifibrotic effects, demonstrating a reduction in collagen expression and decreased activation of fibroblasts and myofibroblasts in lung tissue. IHC provided detailed histological and molecular insights into treatment-induced changes, confirming the potential of this IL-10 delivery system in mitigating fibrosis progression and preserving lung structure [142].
In a study on calcium carbonate (CaCO3)-based composite hydrogels for bone regeneration, IHC played a critical role in identifying and quantifying osteoimmunomodulatory markers and cellular activity in bone defects treated with BMP-2 and catalase (CAT)-loaded hydrogels. Using specific antibodies, IHC detected proteins associated with inflammation and bone healing, such as CD80 and CD206, in tissue sections obtained after implantation in an animal model. This allowed for an assessment of the hydrogel’s ability to regulate macrophage polarization, demonstrating that CAT presence in the composites downregulated the expression of CD80 (Figure 6h), and upregulated the expression of CD206, associated with proinflammatory M1, and anti-inflammatory macrophage phenotypes, respectively. Additionally, IHC provided detailed histological and molecular insights into treatment-induced changes, confirming the potential of this biomaterial to modulate the bone microenvironment, enhance osteogenesis, and promote bone defect healing [140].
Similarly, in tissue regeneration studies, IHC was used to evaluate the efficacy of bioactive polypeptide hydrogels (QK-SF) in modulating macrophage polarization and promoting skin regeneration in a murine wound model. IHC enabled the visualization and quantification of macrophage subpopulations in wound tissue, highlighting the shift from proinflammatory M1 macrophages to pro-healing M2 phenotypes. This was evidenced by an increase in M2-specific markers and a reduction in M1 markers in skin sections treated with the hydrogel. Additionally, IHC facilitated the assessment of keratinocyte differentiation and collagen deposition, providing a comprehensive view of tissue healing and regeneration. The findings, supported by in vitro data, reinforced the conclusion that QK-SF hydrogel not only promotes angiogenesis but also regulates immune responses, making it a promising candidate for tissue engineering and wound healing applications [143].
In the context of coronary artery disease treatment, IHC was utilized to assess the effectiveness of an immunomodulatory biomaterial in macrophage polarization and cardiac tissue regeneration following myocardial infarction. IHC analyses of heart tissue sections from rats treated with alginate gels loaded with immunomodulatory factors, such as colony-stimulating factor (CSF-1) and anti-inflammatory interleukins (IL-4/6/13), revealed the expression of markers associated with prohealing macrophages. This indicated a cellular shift toward phenotypes that support tissue repair. Furthermore, IHC was complemented by echocardiography to evaluate overall cardiac function, providing a comprehensive assessment of the treatment’s impact on myocardial recovery. The results underscored the therapeutic potential of the proposed biomaterial, demonstrating its ability to modulate immune responses and enhance cardiac function, suggesting a novel approach for ischemic tissue revascularization [144].
In a study on pelvic organ prolapse (POP), IHC was employed to evaluate the effectiveness of a biologically functionalized hydrogel in promoting connective tissue repair in abdominal wounds in rabbits. IHC analysis of fibroblast regeneration markers in tissue sections obtained after injection of polyisocyanide (PIC) hydrogel provided insights into new tissue growth and immune activity at the hydrogel-tissue interface, indicating a favorable therapeutic response. Additionally, IHC identified cellular composition changes and extracellular matrix remodeling, crucial for understanding how hydrogel injection enhances collagen deposition and tissue stiffness over time compared to controls. These findings support the hypothesis that PIC hydrogel induces a regenerative fibroblast response, offering an innovative solution for improving surgical outcomes in POP patients and motivating further translational research [145].
Decellularized bronchial grafts have been investigated as promising biomaterials for airway reconstruction, addressing a critical clinical need. These grafts, derived from porcine lungs, demonstrated effective integration with native tissue and epithelial layer formation, as evidenced by histological and electron microscopy analyses. IHC revealed proper revascularization, with positive CD31 staining indicating a favorable healing environment. Although a slight reduction in tensile strength was observed after one month, it was restored within two months. The immune response to the grafts was comparable to that of autografts, suggesting that these biomaterials are safe and could be applied clinically for tracheal reconstruction, offering innovative solutions for patients with airway defects [146].
Overall, IHC is an essential tool in biomaterials research, allowing for the assessment of graft-host tissue interactions. This technique provides critical insights into processes such as revascularization, re-epithelialization, and immune responses, offering valuable information on biomaterial efficacy and safety in tissue regeneration. Highlighting its significance underscores the crucial role of IHC in advancing regenerative therapies and its potential to improve clinical outcomes across various biomedical applications.
The biological evaluation of biomaterials requires selecting techniques that provide specific and detailed information about their impact at the cellular and molecular levels. ISH enables the identification of spatial gene expression patterns, such as those associated with inflammatory or regenerative responses, providing localized analysis of gene expression in cells and tissues in contact with the biomaterial. PCR is essential for measuring gene expression in cells exposed to biomaterials, allowing the detection of inflammatory genes and markers of different cellular responses. ICC and IHC techniques complement this perspective by offering visualization of protein distribution in cells and tissues. While ICC is ideal for observing changes in cells directly interacting with the material, IHC allows the assessment of tissue responses, detecting indicators of biocompatibility or inflammation at the implantation site [50, 68, 124, 141, 147]. Recombinant DNA technology facilitates the genetic modification of cells to induce the production of specific proteins, enhancing material biocompatibility or promoting cell adhesion and proliferation. However, significant challenges exist in applying these techniques to biomaterials. Material-tissue interactions can affect result accuracy, as certain materials, such as synthetic polymers or metals, interfere with detection probes used in ICC and IHC studies. Additionally, the structural and compositional properties of biomaterials may require specific adjustments in sample fixation and preparation protocols to preserve their integrity and obtain reliable data. Overcoming these challenges involves establishing specialized preparation procedures, such as microtomy techniques, and optimizing fixatives according to material type [148].
One of the biggest obstacles is the complex interaction between the material and tissue: Designed to integrate into biological systems, biomaterials can modify sample quality, particularly in techniques like ICC and IHC. Since these techniques rely on antibody interactions with specific proteins, autofluorescence or the physical properties of many materials can interfere with the signal. This issue is particularly problematic in structurally complex materials or those requiring multiple fixation layers to preserve tissue integrity. To address these issues, protocol adjustments are necessary, such as modifying fixation methods and using additional blocking steps to minimize unwanted signals and autofluorescence. Furthermore, the hardness or flexibility of some biomaterials poses a challenge in obtaining uniform sections for analysis, requiring specialized microtomes [24, 149]. Sample preparation is also critical for bioactive and porous materials, such as scaffolds for tissue regeneration. The structural complexity and porosity of these materials can hinder the penetration of fixatives and reagents, affecting protein preservation and the cellular architecture of interest. Therefore, pilot tests are necessary to optimize each step of the protocol, from fixative concentrations to specific incubation times. The lack of standardized procedures adds another layer of difficulty, as each new biomaterial may require a different protocol, increasing costs and reducing reproducibility across laboratories [150, 151].
Traditional biotechnologies such as ISH, recombinant DNA technology, PCR, and ICC or histochemistry play a pivotal role in advancing biomaterials science by providing detailed molecular and cellular insights into biomaterial–tissue interactions. ISH provides spatial resolution of gene expression patterns, facilitating the elucidation of localized cellular responses to biomaterials. Recombinant DNA technology enables the engineering of bioactive molecules that are incorporated into materials to enhance their functionality and biocompatibility. PCR techniques allow rapid and sensitive quantification of gene expression changes associated with inflammation, regeneration, and cellular differentiation, accelerating biomaterial evaluation. Immunocytochemical and histochemical methods reveal protein localization and cellular phenotypes critical for understanding tissue integration and immune modulation. While each technique has limitations, their combined application offers a powerful toolkit that drives the design of smarter, more effective biomaterials tailored for regenerative medicine and personalized therapies.
Regarding future perspectives, advances in analytical techniques are opening new opportunities to characterize biomaterials more comprehensively and accurately. Omics technologies, such as transcriptomics and proteomics, allow researchers to analyze how biomaterials affect gene expression and the cellular proteomic profile, offering an integrated view of the biomaterial’s impact. These technologies can help identify specific biocompatibility or inflammation biomarkers for each material type, promoting the design of personalized biomaterials for regenerative medicine and implants. In parallel, electrically conductive scaffolds, including carbon-based biomaterials that are designed for cardiac tissue regeneration, require detailed evaluation of gene and protein expression associated with electrical integration, contractility, and inflammatory modulation [152]. Additionally, omics analyses can reveal metabolic pathways altered by biomaterials, providing insights into their systemic effects [153, 154]. Another relevant advancement is the integration of 3D imaging, such as fluorescence tomography, confocal microscopy, and second harmonic generation microscopy, which enables visualization of biomaterial-tissue interactions in a 3D context. This approach is particularly useful for complex biomaterial designs, such as those used in bioprinting and three-dimensional scaffolds, where spatial arrangement is crucial for material functionality. 3D imaging allows researchers to observe cell distribution around the biomaterial, assess cell penetration depth into the scaffold, and detect inflammatory or regenerative signals in different regions. Additionally, 3D analysis provides the advantage of real-time dynamic tracking, enabling the observation of material-tissue interaction evolution, which is essential for evaluating implant durability and biocompatibility [155, 156]. Finally, automation and artificial intelligence (AI) are becoming increasingly relevant in biomaterial characterization. With AI, it is possible to analyze large volumes of gene expression data, proteomic profiles, or 3D images quickly and accurately, facilitating the identification of complex patterns in cellular and tissue responses to biomaterials. AI and machine learning can also be applied to predict how different cell types will respond to new biomaterials, optimizing their design before preclinical or clinical studies. Together, these innovations not only help overcome current challenges but also pave the way for the development of more effective and specific biomaterials capable of optimal integration into tissues and with high potential for applications in personalized and regenerative medicine [157].
The molecular biology techniques presented in this article are fundamental tools for analyzing and improving the interaction between biomaterials and biological systems. These methodologies enable a detailed and quantitative study of critical aspects of biocompatibility and cellular response, contributing to a deeper understanding of the molecular processes activated in the presence of these materials. Gene expression, in cells and tissues in contact with biomaterials, is assessed by PCR, which allows us to quantify the expression of specific genes related to processes or indicators that reveal whether the material is well tolerated or induces an adverse response. IHC and ICC complement this approach by allowing visualization of the distribution and localization of key proteins, either in surrounding tissues responding to the material or in individual cells encountering the material. The integration of these methodologies into biomaterials research not only strengthens the development of safe and effective medical devices but also accelerates the creation of advanced treatments, particularly using the principles of regenerative medicine and tissue engineering. These techniques allow biomaterials to be adjusted or enriched to better adapt to the biological environment, facilitating the development of customized devices and materials. The exploration of biomaterial-based innovative strategies worldwide will be promoted by incorporating molecular biology techniques. This article thus serves as a valuable guide for those interested in molecular biology applied to biomaterials, as it provides an overview of tools and their applications. With knowledge of these techniques, new researchers are better prepared to contribute to the design of next-generation biomaterials, which will positively impact health and the future of medicine.
AI: artificial intelligence
BC: bacterial cellulose
BMP: bone morphogenetic protein
BSA: bovine serum albumin
CAT: catalase
cDNA: complementary DNA
CNT: carbon nanotubes
FISH: Fluorescent in situ hybridization
ICC: immunocytochemistry
IHC: immunohistochemistry
ISH: in situ hybridization
PCR: polymerase chain reaction
POP: pelvic organ prolapse
RT-PCR: real-time polymerase chain reaction
TGF-β: transforming growth factor beta
Tm: melting temperature
VEGF: vascular endothelial growth factor
The authors thank Daniela P. Gárnica-Robledo and Karla P. Martínez-Velázquez for their valuable collaboration during the conception of the idea of this article.
PUMG: Conceptualization, Writing—original draft. LOEM: Conceptualization, Writing—original draft. DSR, JMM, JSL, USA: Writing—original draft. BMN: Writing—original draft, Writing—review & editing.
The authors declare that they have no competing interests.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
LOEM thanks Mexico’s Secretary of Sciences, Humanities, Technology and Innovation (SECIHTI) and the University of Guanajuato for the given scholarships. We thank SECIHTI for project CF-2023-I-2285. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 4715
Download: 147
Times Cited: 0
