Affiliation:
Institute of Molecular Biology and Biotechnology, Bahauddin Zakariya University, Multan 60000, Punjab, Pakistan
Email: nadiyamuzaffar22@gmail.com
ORCID: https://orcid.org/0009-0007-3614-9092
Affiliation:
Institute of Molecular Biology and Biotechnology, Bahauddin Zakariya University, Multan 60000, Punjab, Pakistan
ORCID: https://orcid.org/0000-0002-9123-2792
Affiliation:
Institute of Molecular Biology and Biotechnology, Bahauddin Zakariya University, Multan 60000, Punjab, Pakistan
ORCID: https://orcid.org/0009-0007-3333-5682
Explor Asthma Allergy. 2025;3:100984 DOI: https://doi.org/10.37349/eaa.2025.100984
Received: March 23, 2025 Accepted: May 23, 2025 Published: June 30, 2025
Academic Editor: Garry M. Walsh, University of Aberdeen, UK
Over 330 million people globally have asthma, a chronic disease with multiple endotypes and observable phenotypes. This disease has a substantial socioeconomic impact. A thorough assessment of multiple clinical aspects (such as the presence of atopy, comorbidities, and clinical presentations), characteristics of lung function (such as the degree of bronchial reversibility, the involvement of the airway obstruction, and airway hyperreactivity), and the interaction of sputum and systemic inflammatory factors (such as neutrophilic, eosinophilic, and mixed) are required to determine the specific endotypes of asthma. The precision medicine approach to asthma represents a new paradigm, with improved opportunities for more effective and appropriate personalized therapies and new insights into the immunological aspects of asthma that demand further research. In the world of precision medicine, biomarker-based therapies for individual patients are just the beginning of an exciting and emerging journey in allergy treatment. A collection of biomarkers might be utilized to define and classify the endotypes based on the phenotyping of asthma, which will be more likely with omics information and unbiased clustering. This review will contribute to the development of personalized therapy for asthma, enabling more accurate treatment. It will also serve as a source of novel targets and new treatments for every identified endotype.
Over 330 million people globally have asthma, a chronic, diverse illness that has a substantial socioeconomic impact, particularly when it is severe and uncontrolled [1–3]. It is estimated that managing asthma in the US will cost over $960 billion in the upcoming 20 years [4]. Asthma is typically characterized by chronic airway inflammation and variable expiratory airway constraints, which can become continuous due to airway remodeling, in addition to variable symptoms such as cough, chest tightness, chest pain, wheezing, and shortness of breath [5]. The concept of asthma encompasses a variety of phenotypes (physically observable characteristics) and endotypes (molecular or cellular level subtypes of disease) depending on factors such as age at onset, airflow, atopy status, exacerbations, comorbidities, treatment, prognosis responsiveness, and underlying airway inflammation [6, 7].
A review by Guilleminault et al. (2017) [8] provides details about different phenotypes of asthma and the necessity of personalized medicine, while Chung (2017) [9] focused on the use of mHealth for personalized medicine along with the development of future biomarkers. A study by Papapostolou and Makris (2022) [10] exerts on the use of allergen-specific immunotherapy (AIT), specific biomarkers for disease identification [10]. A systematic review by Cunha et al. (2021) [11] provides a detailed explanation of asthma phenotypes. An overview of the main immune pathways connected to the disease etiology, phenotypic features, and presentation of severe asthma subtypes with specific treatment recommendations is provided in the research of Gonzalez-Uribe et al. (2022) [12] and Chen et al. (2023) [13]. Ranjbar et al. (2022) [14] provide a detailed overview of the pathology and genetic associations of asthma. The current study will provide a thorough explanation of different phenotypes of asthma and an analysis of biological substances and biomarkers that are now readily accessible for the treatment of severe asthma. The study will reveal the pathophysiology and genetic factors related to asthma and the available treatments and clinical manifestations of personalized medicines.
Data for this review was collected from different databases, including PubMed, Google Scholar, Scopus, and Web of Science, using keywords like “asthma phenotypes, asthma endotypes, personalized medicine for asthma, asthma biomarkers, targeted therapy against asthma, clinical studies, biologic therapies in asthma, and ML in asthma”. The research was limited to publications in the English language. All the retrieved publications were imported into “Mendley Reference Manager” for the screening and removal of duplicates. A total of 432 publications were collected, from which 50 were removed based on duplication. The remaining 382 were screened based on title and abstract. There were 196 excluded publications after a complete screening due to the lack of proper explanation about asthma phenotypes and endotypes, result bias, and not involving proper therapeutic targets. A total of 186 publications were included based on preclinical/clinical research on asthma, targeted drug therapies, and ML approaches. Finally, 166 publications are included in writing the current review after proper evaluation of the full text.
Around 80% of asthmatic children and 40–50% of asthmatic elders have allergic mechanisms [15]. Among people with asthma, the allergic phenotype is particularly prevalent [16, 17]. Figure 1 explains the phenotypes of asthma and the related inflammation trigger mechanism.
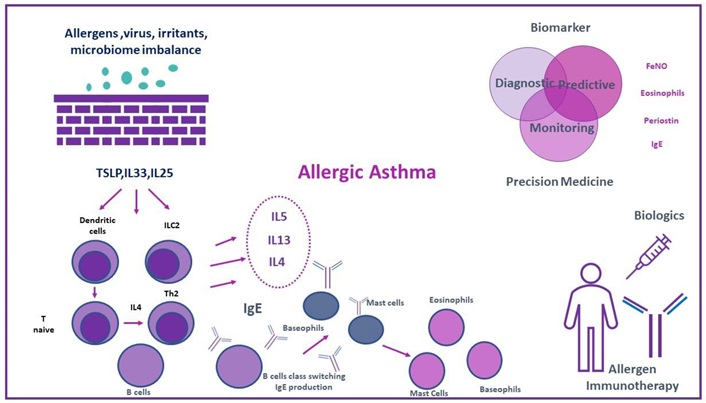
Asthma phenotypes. Reprinted with permission from [10]. © 2022 by Papapostolou N and Makris M
The inflammatory type 2 pathway is activated, which results in the release of type 2 cytokines, namely IL-4, IL-13, and IL-5. Type 2-high inflammation asthma endotypes include allergic asthma. In the eosinophilic asthma endotype, eosinophilic inflammation can exist even in a lack of allergic inflammation. Th1 and Th17 cell activation in type 2-low pathways results in neutrophilic inflammation (mostly non-eosinophillic).
Childhood-onset asthma, also known as early-onset asthma, is usually allergic or atopic. In the case of allergic asthma, the patient is characterized by seasonal variations in exposure to allergens and an allergic mode of action with increased immunoglobulin E (IgE) levels [16]. Non-allergic asthma does not involve IgE, and the patient’s symptoms are triggered by non-allergic factors, like infection, pollution, or cold air [17]. Allergic and non-allergic asthma are associated with blood or sputum eosinophilia [18]. Exposure to allergens raises the fractional exhaled nitric oxide (FeNO) [19].
Cluster analysis has revealed an obesity-related asthma phenotype in adults [20, 21]. According to standard descriptions, it is characterized by late-onset asthma (LOA) [22], low allergy (driven by the inflammation and metabolic factors; cytokinesis from adipose tissues are involved to cause inflammation) [23], frequent episodes [24], noneosinophilic inflammation (obesity-related asthma is more neutrophilic), and heavy drug use [21, 25]. This phenotype may present as atopic with higher IgE levels or a history of allergy, but in obesity-related asthma, the inflammation is driven by non-IgE-mediated pathways related to adipocytes and metabolic factors in obese people. Some patients also have mixed phenotypes of allergic and non-allergic asthma, where they show elevated levels of IgE. Along with markedly reduced airway function, these patients also show considerable airway hyperresponsiveness (AHR) and inadequate asthma control.
According to different studies, obese individuals with severe asthma have higher levels of submucosal eosinophils and sputum interleukin (IL)-5 but not sputum eosinophils [26]. A variety of factors may explain the underlying cause of asthma in obese persons. These comprise oxidative signaling, cytokine disruption, neural signaling pathways, and alterations in lung function and structure (lower lung volume and increased airway reactivity) [27].
Due to its critical importance in the obese population, diet must also be considered in obese asthmatic patients. Research has shown that people with asthma, especially those who are obese, have increased airway inflammation when following a pro-inflammatory meal (such as a high-fat diet) [28]. Obese patients frequently have comorbid conditions like depressive disorders, obstructive sleep apnea, and gastroesophageal reflux syndrome, which might exacerbate the course of asthma [24].
Fungi-related asthma has two phenotypes: allergic bronchopulmonary mycoses and asthma linked to fungal hypersensitivity. These phenotypes have both genetic and ecological predisposing factors. Genes encoding Toll-like receptors, major histocompatibility complex HLA-DR 2 and 5, chitinases (a 24-bp duplication of the CHIT1 gene), and other polymorphisms may be involved [29, 30].
Mold exposure can occur at home, at work, or outside (thunderstorm asthma) and contribute to the development and aggravation of asthma symptoms. Moderate asthma may be linked to fungal hypersensitivity. But if unusually high fungal concentrations surround the patient, it develops into severe asthma [6, 31].
In 2006, the combination of three characteristics was used to characterize severe asthma with fungal sensitization (SAFS):
A total IgE of less than 1,000 IU/mL.
Fungal sensitisation as assessed by prick test or specific IgE testing.
The existence of severe asthma [31].
When SAFS worsens, it can occasionally lead to allergic bronchopulmonary mycosis, which shares many of the same characteristics as SAFS but has total IgE levels of more than 1,000 IU/mL [32].
Severe forms are treated with systemic corticosteroid therapy, long-acting inhaled β2-agonist, and high dosages of inhaled corticosteroids (ICSs). The administration of omalizumab or itraconazole may help control asthma, but there are no official recommendations for it [32, 33].
Certain characteristics, such as exercise-induced bronchospasm, were known before cluster analysis was developed [34]. If treatment is not received, symptoms may last for 30 to 90 minutes after a brief session of exercise [35]. The prevalence varies between 30% and 70%, according to the population under study [35, 36]. Leukotriene receptor antagonists (LTRAs), ICSs, or short-acting β2-agonists administered 15 minutes before exercise are suggested treatments [37].
As per the latest research, there are various subtypes of aspirin-induced asthma that vary in severity, from a kind that mostly affects the upper airways to severe asthma that frequently has exacerbations [38]. Airflow restriction is a defining characteristic of asthma-chronic obstructive pulmonary disease (COPD) overlap syndrome (ACOS), which exhibits characteristics of both COPD and asthma [39]. Adults with asthma have a 20.9% prevalence of ACOS, with a range of 5.2% to 35.4% [40]. Since ACOS is a diverse condition, a number of its phenotypes, including smokers’ asthma and neutrophilic ACOS, have already been discovered [41].
Elderly asthma patients have extremely complicated clinical characteristics that set them apart from young adult asthma patients [42]. Older adults who have asthma have a prevalence of 4.5% to 12.7%, which includes both adult-onset and long-standing early-onset asthma [43, 44]. According to Baan et al. (2022) [45], compared to childhood-onset asthma, LOA may be more likely to cause hospitalizations and acute asthma episodes. Oral corticosteroid courses, which have several adverse effects, are the treatment of choice for exacerbations, which are harmful to lung health and quality of life. Adults with LOA were more likely to have nasal polyposis, overweight/obesity, GERD, and diabetes mellitus than those with earlier-onset asthma. In elderly asthma patients, the clinical syndrome is highly variable and often co-occurs with a variety of phenotypes and comorbidities, including COPD, obesity, depression, atopy (fewer times), and a complicated exposure background [46].
According to Haldar and Pavord (2007) [47] and Piipari et al. (2004) [48], different phenotypes of asthma are seen in older individuals, such as remitting childhood asthma that recurs later in life, LOA, and long-standing asthma (LSA). LSA normally starts before the age of 12, whereas LOA usually occurs beyond the ages of 40 or 60. Studies by Fish and Peters (1999) [49] and de Marco et al. (2006) [50] show that LOA risk factors involve exposure to dust, art materials, and cleaning agents; smoking; rhinitis; weight increase; and respiratory infections. In terms of prognosis, research by ten Brinke et al. (2001) [51] and Mwanga et al. (2021) [52] showed that people with LOA often have stronger bronchodilator responses and greater forced expiratory volume in 1 second (FEV1), whereas those with LSA are more likely to suffer from hyperinflation and permanent airway blockage. Age-adjusted measurements such as forced vital capacity (FVC), forced expiratory volume in 6 second (FEV6), and bronchial hyperresponsiveness (BHR) in the ratios of FEV1/FVC, FEV1/FEV6, BHR, exhaled nitric oxide levels, and CT imaging are necessary for the diagnosis of asthma in older persons, which can be difficult since symptoms including coughing, dyspnea, chest tightness, and wheezing can mimic those of COPD. Bateman et al. (2018) [53] and Yáñez et al. (2014) [54] focused on controlling symptoms, reducing exacerbations, treating comorbidities, and avoiding drug adverse effects, all of which are essential components of effective care. Minimizing smoking exposure is essential, as is managing environmental stressors such as dust mites, pets, and cockroaches. Pharmacologically, controller therapy includes ICSs, LTRAs, and biologics like anti-IL-5 and anti-IgE, while rescue drugs like β2-agonists and short-acting anticholinergics are employed, according to the studies by Pavord et al. (2012) [55] and Pelaia et al. (2015) [56]. Monitoring is essential to avoid adverse effects like cataracts and bone loss, especially while using larger dosages of ICS.
The term “personalized medicine” refers to a method of addressing and preventing disease that considers each patient’s unique genetic makeup, lifestyle, and environment. Since preventive efforts and medicines may be customized for each individual, there is a greater chance of addressing “the right individual with the correct drug at the right time” when using this technique [57]. The term “precision medicine”, which is synonymous with “personalized medicine”, refers to this more exact strategy for medicine since it is supposed that the analysis of proteins, genes, metabolomic profiles, and microbiome profiles will more likely indicate causal pathways and result in an endotype description specific to each patient [46].
Asthma is predominantly treated with conventional anti-inflammatory medications, like bronchodilators, oral corticosteroids (OCSs), and ICSs. However, these methods do not consider the wide variety of asthma characteristics and endotypes [58, 59], as several approaches advised [60–62]. Even with high doses of ICS/long-acting β2-adrenergic agonist (LABA) combinations, along with the addition of other drugs, i.e., leukotriene modifiers and tiotropium OCSs cannot control severe asthmatic conditions. Globally, 5–10% of asthmatic patients have difficult-to-treat asthma and they do not acquire clinical or functional management, even though the above “one size fits all” approach may be successful for the majority of patients [57]. For them, a more targeted and individualised treatment approach is necessary, one that involves identifying unique phenotypes and endotypes to address particular underlying variables [63].
Asthma has been related to more than 60 genetic loci, some of which have been associated with severe forms of the illness. Kids’ and adults’ genome-wide association studies (GWAS) have identified five loci linked to severe exacerbations and associated numerous genes involved in immunological responses, including IL33, IL1RL1, and CDHR3 [64]. Furthermore, 24 loci associated with moderate-to-severe asthma have been discovered by GWAS investigations [65, 66]. Many research studies have explored the role of gene expression in severe and persistent asthma. Numerous cell types have been examined in these studies, comprising whole blood, sputum, bronchoalveolar lavage, and airway epithelial cells [67–70].
Asthma is a diverse, well-defined disease. Different clinical phenotypes are expressed as a result of a complicated interaction between underlying inflammation and environmental and genetic variables. As a result, the underlying pathogenic mechanisms are complex and differ between asthmatics [21]. The identification of asthma type 2 (T2)-high inflammation (T2-high, eosinophilic) and T2-low inflammation (T2-low, non-eosinophilic) endotypes is the result of the current understanding of the classification of immunological dysregulation in asthma (Figure 1).
The hallmarks of T2-high asthma are FeNO levels that are higher and eosinophilic airway inflammation with high blood eosinophil counts [71].
The term “T-helper type 2-high” (Th2-high) inflammation has been replaced by the term “type 2-high” (T2-high) to encompass T2 innate lymphoid cells (ILC2s) and Th2, associated with asthma inflammation. Triggers, such as allergens, can initiate this eosinophilic inflammation through innate or adaptive immunity [64].
On the other hand, paucigranulocytic and neutrophilic asthma are included in T2-low asthma. In the mixed granulocytic endotype, eosinophilic and neutrophilic airway inflammation coexist [72].
The most well-defined asthma model is allergic asthma, which falls within the T2-high endotype and is typified by eosinophilic airway inflammation. It is undeniable that the epithelial barrier has a role in allergic disorders and other chronic illnesses [73].
A subtype of T2-low asthma known as neutrophilic asthma is distinguished by a preponderance of neutrophilic inflammation in the airways as opposed to eosinophilic inflammation. It is usually observed in smokers, obese people, and those who have recurrent respiratory infections. It is also commonly linked to severe LOA. Th1 and Th17 immune system pathways drive this phenotype, which is frequently resistant to corticosteroids and results in elevated levels of cytokines, including IL-8, IL-6, TNF-α, and IL-17. According to Moore et al. (2010) [21], these cytokines encourage the recruitment and activation of neutrophils, which leads to airway remodeling, chronic inflammation, and blockage [59].
Current research is investigating other alternatives like targeted anti-inflammatory medicines and biomarker-driven therapy since neutrophilic asthma is difficult to treat due to its steroid-refractory nature.
In both T2-high and T2-low asthma, activation or dysregulation of epithelial cells is one of the factors that trigger inflammation. Dynamic interactions occur in vulnerable individuals that can induce different inflammatory diseases once rupture of the epithelial barrier represents “leakage”, and multiple immune-regulatory mechanisms arise to reduce the inflammation [73, 74]. Table 1 explains the differences between T2-high and T2-low asthma.
Difference between T2-high and T2-low asthma
| Feature | T2-high asthma | T2-low (non-eosinophilic) asthma |
|---|---|---|
| Type of inflammation | Eosinophilic (allergic/Th2-driven) | Neutrophilic or paucigranulocytic (non-allergic, Th1/Th17-driven) |
| Mechanism | Allergen exposure→activation of dendritic cells→Th2 differentiation→IL-4/5/13 release | Airway epithelial stress (pollutants/infections)→Th1/Th17 activation→IL-17, IL-8 release |
| Key cytokines | IL-4, IL-5, IL-13 | IL-6, IL-8, IL-17, TNF-α |
| Common triggers | Allergens, dust mites, pet dander, pollen | Viral infections, cigarette smoke, air pollution, obesity |
| Steroid response | Good; responsive to inhaled corticosteroids (ICS) | Poor; often steroid-resistant |
| Biologic therapy | Effective (e.g., anti-IL-5, anti-IgE, anti-IL-4R) | Generally ineffective; under investigation |
| FeNO levels | Elevated | Normal or low |
| Sputum eosinophils | High (> 3%) | Low (< 2–3%) or absent |
| Typical patients | Younger, allergic, atopic individuals | Older adults, smokers, obese patients |
| Airway remodeling | Present; often reversible with treatment | More severe and persistent |
Th2: T-helper type 2; IL: interleukin; FeNO: fractional exhaled nitric oxide
Specifically speaking to allergic asthma, exposure to aeroallergens activates the epithelium airways, which in turn causes the release of cytokines derived from the epithelium that are referred to as “alarm signs” or “alarmins”.
According to Mitchell and O’Byrne (2017) [75], these alarmins are:
IL-25.
IL-33.
Thymic stromal lymphopoietin (TSLP).
After exposure to the allergens, the following immune mechanisms start in the body:
In local lymph nodes, antigen-presenting cells expose allergens to naïve T lymphocytes. These cells then differentiate into Th2 cells by combining with co-stimulatory molecules and secreting large amounts of T2 cytokines, such as IL-4, IL-5, and IL-13 [64].
The development of T cells into the Th2 is significantly influenced by IL-4. Furthermore, in response to allergen stimulation, B cells’ IgE isotype changes and the subsequent release of IgE allergen-specific antibodies that are driven by IL-4 and, to a lesser extent, IL-13 [76].
Mast cells (MCs) and basophils (sensitization phase) have a high-affinity IgE receptor (FcεRI) attached to those particular IgE antibodies [77].
Following an allergen exposure, myeloid dendritic cells in patients sensitized to allergens, in conjunction with co-stimulatory molecules, deliver inhaled aeroallergens to tissue-resident memory CD4+ Th2 that express T-cell receptors (TCRs) specific to allergens, causing these cells to respond differently upon activation [78].
Aeroallergens cause a swift and widespread activation by cross-linking IgE receptors on the surface of MCs and basophils [79]. Strong bronchoconstriction, mucus hypersecretion [72, 80], and edema of the airway wall [77] are the results of the degranulation of preformed mediators, such as histamine, tryptase, serotonin, proteases, proteoglycans, carboxypeptidase A, and chymase, combined with the formation of newly synthesized lipid mediators.
A late-phase reaction is brought on by the infiltration of inflammatory cells that support both innate and adaptive immunity, such as eosinophils, basophils, CD4+ T-helper cells, memory cells, monocytes, and neutrophils, in addition to these acute responses and the production of cytokines by Th2 cells, MCs, and basophils [81].
Furthermore, IL-13 has a multifunctional role because it has been linked to increased levels of FeNO due to stimulation of the nitric oxide synthase enzyme in bronchial epithelial cells, mucus hypersecretion, goblet-cell hyperplasia, airway hyperreactivity, and subepithelial fibrosis [81, 82]. The maturation, survival, differentiation, and proliferation of eosinophils are all regulated by IL-5. Additionally, it causes them to release and activate their mediators, which in turn causes inflammation and tissue damage [83]. Figure 2 explains the pathophysiology of asthma phenotypes.
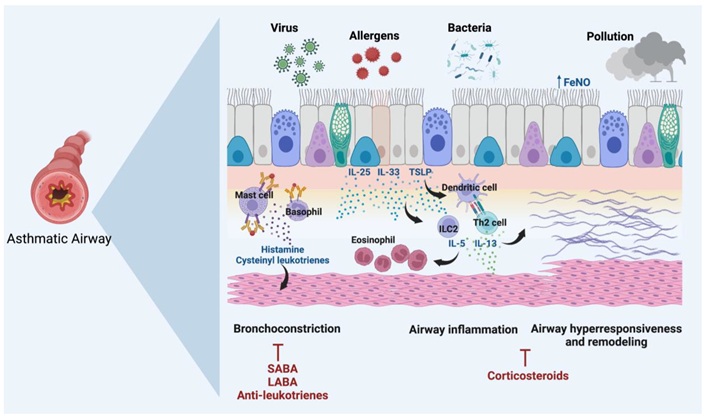
Pathophysiology of asthma patients in response to allergens. SABA: short-acting beta-agonists; LABA: long-acting beta-agonists; AHR: airway hyperresponsiveness; ILC2: type 2 innate lymphoid cell; Th2: T-helper type 2. Reprinted with permission from [14]. © 2022 by Ranjbar M, Whetstone CE, Omer H, Power L, Cusack RP, and Gauvreau GM
This picture depicts the pathophysiology of an asthmatic airway in reaction to pollutants, viruses, allergens, and bacteria, which causes airway remodeling, inflammation, and constriction. Allergens and pollutants that provoke asthma also activate mast cells, basophils, and dendritic cells. These cells then release inflammatory mediators, including histamine and cysteinyl leukotrienes, which narrow the airways. ILC2 and Th2 cells are activated by damaged epithelial cells, which release cytokines such as IL-25, IL-33, and TSLP. These cells then release IL-5, IL-4, and IL-13, which further aggravate airway inflammation and draw in more eosinophils. Over time, the thickening and rigidity of the airway walls are brought on by this chronic inflammation, which also causes AHR and airway remodeling. Treatments for these side effects include anti-leukotrienes, corticosteroids, SABA, and LABA. These aim to ease bronchoconstriction, lower inflammation, and avoid long-term damage to the airways.
The severity of asthma and the risk of exacerbations are closely correlated with eosinophil counts in peripheral blood, bronchoalveolar lavage fluid (BALF), and airway biopsy specimens, particularly in eosinophilic or T2-high asthma [77, 84]. Furthermore, ILC2s play a crucial role in the pathophysiology of asthma through the production of Type 2 cytokines such as IL-5 and IL-13, though they produce little or no IL-4 [85]. These cells lack antigen-specific receptors but express IL-7 receptor alpha (CD127), Th2 cells (CRTh2), and interleukin-1 receptor-like 1 (ST2) and are activated by epithelial-derived cytokines such as IL-25, IL-33, and thymic stromal lymphopoietin (TSLP). Additionally, those cells exhibit high amounts of GATA3, which helps activate Th2, which in turn triggers the release of T2 cytokines [86]. The eosinophilic, non-allergic T2 endotype is linked to the mechanism of ILC2 activation [87].
IgE class flipping is predominantly driven by IL-4, but eosinophilic inflammation is caused by the secretion of IL-5 and IL-13 from ILC2s. Therefore, eosinophils in general contribute to both forms of T2 asthma; IgE-driven mechanisms are linked to allergic asthma, while ILC2-driven mechanisms are linked to eosinophilic asthma [88–90]. Another largely unexplored issue is the persistence of allergic asthma and the natural course of remodeling. As was already noted, there may be differences between the elements that cause or aggravate asthma and those that contribute to the illness’s persistence [91]. Asthma persistence has been associated with allergen sensitivity, particularly in early childhood [92, 93].
Any objectively measured trait that can be utilized for disease monitoring, diagnosis, and treatment response prediction is referred to as a biomarker [94, 95]. A biomarker must be able to identify a phenotype and/or endotype of the disease, forecast and assess treatment response, and track the advancement of the disease in order to be useful in clinical practice [96]. The search for the ideal biomarker with the aforementioned characteristics is critical as we enter the era of personalized medicine. This is especially true for severe asthma endotypes that overlap with other T2 conditions, such as allergies, for which selecting the best course of action for individual patients can be very difficult [87]. Despite the fact that a number of biomarkers have been linked to T2 inflammation, allergic asthma frequently manifests as high levels of periostin, blood and sputum eosinophils, FeNO, and IgE [87, 97].
Patients with allergic asthma have been identified by measuring total IgE levels and sensitization ratios to environmental allergens, either by measuring particular IgEs in serum or by doing skin tests in vivo [98]. Fitzpatrick et al. (2006) [99] also found a positive correlation between IgE levels and the severity of asthma. It is widely acknowledged that not all atopic individuals would exhibit a clinical manifestation of their IgE sensitization and, consequently, the development of allergic asthma, despite the undeniable connection between atopy and asthma [84].
FeNO is a non-invasive biomarker linked to eosinophilic airway inflammation [19, 100, 101]. Nitric oxide synthase in bronchial epithelial cells is stimulated by IL-13, which raises FeNO levels as a result [65, 81]. When evaluating individuals with severe asthma, the Global Initiative for Asthma (GINA) suggests looking for FeNO values greater than 25 parts per billion (ppb) as a sign of T2 inflammation [102]. FeNO concentrations over 50 ppb and 35 ppb in adults and kids, respectively, are linked to ICS response and eosinophilic inflammation [19]. Elevated FeNO levels have been linked to early-onset asthma, atopy, and airway hyperreactivity, according to data from the Severe Asthma Research Cohort [82]. A recent systematic review and meta-analysis shows that measuring FeNO levels provides a good specificity for diagnosing asthma, and the specificity increases with higher cut-off values (> 50 ppb) [97].
One of the most well studied biomarkers for determining a patient’s severity of asthma is sputum eosinophil. This procedure is typically carried out in institutions that specialize in adult populations with severe asthma due to sample acquisition constraints, particularly in juvenile populations [96]. Sputum eosinophilia was found to have a modest correlation with blood eosinophils, FeNO levels, and total serum IgE [65], according to the results of a systematic review and meta-analysis [40, 103].
Patients with allergic asthma have been found to have higher levels of peripheral eosinophils and Th2-polarized responses in comparison to non-allergic asthma patients [104]. Research on blood eosinophil levels in individuals with allergic asthma varies; in RCT and real-world research, approximately 50% of patients had increased levels (> 300 cells/µL) [104–106]. STELLAIR research, a retrospective real-world investigation evaluating the efficacy of omalizumab in patients with allergic asthma has shown that 52.1% of adults and 73.8% of children had a blood eosinophil count of ≥ 300 cells/µL [104]. According to Casale et al. (2019) [105], 711 patients’ baseline blood eosinophil counts ranged from 0 to 2,340 cells/µL, with 35.4% of patients having levels greater than 300 cells/µL.
Periostin is a matricellular protein that has a role in T2 inflammation and airway remodeling in asthma. It is elevated in bronchial epithelial cells by T2 cytokines, IL-4 and IL-13 [107, 108]. Periostin is a valuable biomarker in evaluating T2 inflammation and remodeling since it can be measured in the serum of asthmatic patients [109, 110]. Serum periostin measurement is correlated with various T2 biomarkers, including AHR, serum eosinophils, IgE levels, and eosinophilic cationic protein [111, 112]. Lastly, since children’s baseline periostin levels are higher due to bone growth, the potential use of periostin as a biomarker in pediatric populations has not yet been investigated [113].
Omalizumab was the first biologic medication licensed by the FDA to treat severe asthma. It was also the most extensively used medication in adults, adolescents, and children as young as six years old. This monoclonal antibody (mAb) functions in two ways: it inhibits the production of the mast cell IgE receptor while also preventing IgE from attaching to its FcεRI [114]. Omalizumab has been the first mAb to be included in the GINA suggestions since 2004. It has been demonstrated to be beneficial in enhancing lung function, lowering the risk of exacerbation, easing symptoms, and controlling asthma [115–117].
Exacerbation rates between weeks 16 and 60 were found to be decreased by 26% in the placebo group and 16% in the omalizumab group, according to a review of 25 clinical studies published in 2014 [118]. During weeks 28 and 60, the rate of hospitalization risk dropped from 3% in the placebo group to 0.5% in the omalizumab group. Compared to patients undergoing a placebo, patients taking omalizumab were also more likely to lower or stop taking their ICS dose [32, 117]. However, there was no obvious distinction in OCS withdrawal [118].
Omalizumab dosage is determined by the patient’s age and baseline free total IgE level. Patients 12 years of age and above with total IgE levels between 30 and 700 kU/L and children aged 6 to 11 years old with total IgE levels between 30 and 1,300 kU/L are viable candidates. However, a recent study assessing the evolution of symptoms of asthma and biomarkers in adults administered omalizumab found no correlation between exacerbation rates and elevated FeNO ≥ 25 ppb or eosinophil count ≥ 300 cells/µL, and similar benefits were observed in patients with T2-high and T2-low asthma [116, 119]. Omalizumab seems to have a significant impact on non-specific severe asthma, and IgE levels are not a reliable indicator of how well a patient would respond to treatment.
Omalizumab exhibits a long-term efficacious, safe, and tolerable profile, making it a viable therapeutic treatment choice for severe asthma and some allergic disorders [104, 105].
IL-4 and IL-13 are essential cytokines in the pathophysiology of asthma in T2 inflammation. IL-4 is essential for T-cell differentiation to drive the immunological response that results in allergic inflammation [120–124]. A promising approach to treating asthma has been found to target these cytokines, especially in those with eosinophilic characteristics [121, 125, 126].
IL-4 receptor alpha chain is a desirable target for biological therapy because it is a shared element in the signaling pathways of IL-4 and IL-13. Therapies that block IL-4 receptor alpha can successfully lower T2 inflammation, which is a primary cause of asthma attacks [73].
Dupilumab is a flexible treatment option for people with T2-driven asthma since it can target the IL-4 and IL-13 pathways. This leads to notable enhancements in lung function, quality of life, and asthma management [127]. It inhibits the signaling pathways of both IL-4 and IL-13 by targeting IL-4 receptor alpha. According to clinical trials, patients with moderate-to-severe asthma, especially those with high eosinophil counts or sputum eosinophilia, show a significant 87% reduction in exacerbations [82, 128]. Its effectiveness in treating allergic disorders is not limited to treating pulmonary problems. It also shows promise in the treatment of patients with concomitant atopic dermatitis [129].
Dupilumab has been demonstrated to dramatically lower severe asthma exacerbations and enhance lung function in many people with moderate-to-severe asthma. The advantages of dupilumab have been extensively studied in different age groups. This drug has the capacity to permit glucocorticoid dosage reduction without affecting asthma control [130].
Pitrakinra is another noteworthy treatment that also inhibits IL-13 and IL-4. Although it has shown some promise in treating allergic asthma, it is not as widely used as dupilumab [129, 130].
The IL-13-specific inhibitors lebrikizumab and tralokinumab are available. Despite ICSs, lebrikizumab has been demonstrated to dramatically improve lung function in patients with poorly managed asthma [131], particularly in those with increased periostin levels [105, 132]. Tralokinumab, on the other hand, enhances lung function without having a noticeable effect on the overall management of asthma [126].
The efficacy of these treatments is being evaluated in ongoing Phase III trials. They may eventually be more widely available for mild-to-moderate asthma, but at the moment, they are mostly utilized for severe asthma [131].
The survival, growth, and activation of eosinophils vital to the inflammation associated with eosinophilic asthma depend on IL-5. Managing this particular subtype of asthma now mostly involves targeting IL-5 and its receptor.
Mepolizumab is an anti-IL-5 mAb that successfully lowers eosinophil levels, resulting in fewer exacerbations and better patient outcomes [133]. It is now a crucial medication for individuals with severe eosinophilic asthma who do not respond to traditional treatments. It is administered subcutaneously at intervals of four weeks [133, 134].
Another anti-IL-5 mAb that can be injected is called reslizumab. It gives individuals with eosinophilic asthma another choice and provides advantages comparable to mepolizumab [58, 123].
On the other hand, benralizumab targets IL-5 receptor alpha, causing a significant decrease in basophils and eosinophils by antibody-dependent cellular cytotoxicity [116]. Clinical trials have demonstrated that benralizumab dramatically lowers exacerbations in addition to improving asthma management [135, 136].
These treatments have completely changed how eosinophilic asthma is treated by offering focused methods that minimize the requirement for OCSs and lower the possibility of systemic side effects.
A preclinical research by Lamb et al. (2021) [137] shows that retinoic acid receptor-related orphan receptor-gamma t (RORγt), a transcription factor crucial for the generation of Th17 cytokines (particularly IL-17 and IL-22) in severe T2-low asthma, has become a prospective pharmaceutical target. IL-17-positive cells and a unique IL-17-associated gene expression profile were found in the bronchial biopsies of individuals with severe asthma [138]. Experimental models demonstrated that IL-17 and IL-22 together, but not separately, caused neutrophilic inflammation, pro-inflammatory cytokine production, and AHR. Notably, the RORγt inhibitor BIX119 outperformed anti-IL-17 therapy alone in reducing IL-17 and IL-22 levels, AHR, and airway neutrophilia. These findings implicated corticosteroid-resistant γδ-T cells as important mediators, since they closely matched gene expression patterns in patient tissues with severe asthma. Therefore, RORγt inhibition-based dual blocking of IL-17 and IL-22 may have better therapeutic promise for treating the Th2-low neutrophilic asthma phenotype [137].
TSLP is a cytokine synthesized by epithelial cells that is essential for starting the inflammatory cascade in asthma. Targeting TSLP offers a novel treatment approach by suppressing T2 inflammation at an early stage [78, 85].
A first-in-class biopharmaceutical called tezepelumab inhibits TSLP and stops it from interacting with the TSLP receptor [78, 85]. This blockage inhibits antigen-presenting cells and leads to the induction of adaptive immunological responses. Tezepelumab has been shown in clinical trials to lower blood eosinophil count, FeNO levels, and exacerbations—all important indicators of inflammation [139, 140]. Furthermore, tezepelumab promotes lung function, decreasing the requirement for OCSs and making it a viable treatment for people with severe asthma [141].
Tezelumab provides a broad-spectrum strategy by targeting TSLP, which may be beneficial for a variety of asthma phenotypes, particularly those that are not caused by eosinophilic inflammation [131]. Table 2 provides a summary of different biological agents for asthma treatment.
Overview of biologic therapies for severe asthma
| Biologic therapy | Approval year | Targets | Indication | Mechanism of action | Age range | Clinical outcomes | Trials/Studies | Dosage | References |
|---|---|---|---|---|---|---|---|---|---|
| Omalizumab | 2003 | IgE | Severe asthma, allergic asthma | - Prevents IgE binding to FcεRI- Inhibits mast cell IgE receptor expression | ≥ 6 years old | - Reduces exacerbations- Improves asthma control- Reduces hospitalization risk- Allows ICS dose reduction | - Clinical trials: GAIN, EXTRA, and others- Review of 25 trials (2014) | - Based on age and total IgE levels | [32, 115, 116, 119] |
| Mepolizumab | 2015 | IL-5 | Severe asthma with eosinophilic phenotype | - Prevents IL-5 from binding to IL-5Rα | ≥ 6 years old | - Reduces exacerbations by 53%- Improves quality of life and symptom control- Increases FEV1 | - Trials: DREAM, SIRIUS, MUSCA, MENSA | - 100 mg every 4 weeks | [58, 133, 134] |
| Benralizumab | 2017 | IL-5Rα | Severe asthma with eosinophilic phenotype | - Prevents IL-5 from acting on eosinophils- Induces eosinophil apoptosis | ≥ 12 years old | - Decreases exacerbation rates- Reduces OCS use- Improves asthma symptoms- Enhances quality of life and lung function | - Trials: ZONDA, SIROCCO, ANDHI, CALIMA | - 30 mg every 4 weeks for 3 doses- Then every 8 weeks | [135, 136] |
| Reslizumab | 2016 | IL-5 | Severe eosinophilic asthma | - Prevents IL-5 from binding to IL-5Rα | ≥ 18 years old | - Improves FEV1- Reduces exacerbations- Decreases eosinophil count- Improves quality of life | - Trials: RESPIRE 1 and 2, and others | - 3 mg/kg every 4 weeks | [58, 123] |
| Dupilumab | 2018 | IL-4Rα | Severe asthma, eosinophilic phenotype or OCS-dependent asthma | - Blocks IL-4 and IL-13 from binding to IL-4Rα | ≥ 6 years old | - Improves asthma control- Enhances lung function- Reduces exacerbations and OCS use | - Trials: LIBERTY ASTHMA QUEST, VENTURE, and others | - Dosage not specified | [120, 122, 124, 129, 130] |
| Tezepelumab | 2021 | TSLP | Severe asthma | - Blocks TSLP from interacting with its receptor | ≥ 12 years old | - Decreases exacerbation rates- Increases FEV1- Improves quality of life- Reduces OCS dose | - Trials: PATHWAY, NAVIGATOR, SOURCE, DESTINATION | - 210 mg every 4 weeks | [78, 85, 139–141] |
IgE: immunoglobulin E; FcεRI: high-affinity immunoglobulin E receptor; ICS: inhaled corticosteroids; IL: IL: interleukin; FEV1: forced expiratory volume in 1 second; OCS: oral corticosteroid; TSLP: thymic stromal lymphopoietin
Several alternative therapies are becoming more popular in the management of asthma, in addition to biological therapies that target cytokines, especially for individuals with severe or refractory types of asthma.
A non-pharmacological treatment called bronchial thermoplasty includes carefully applying heat to the bronchial walls to decrease the portion of smooth muscle tissue in the airways [142, 143]. Asthma’s defining feature, decreased AHR, results from this decline. Bronchial thermoplasty fails to significantly enhance lung function, but it may improve life quality while lowering the likelihood of exacerbations, according to clinical trials such as the anti-inflammatory reliever (AIR) and AIR2 investigations [144, 145]. Individuals with severe asthma who continue to have symptoms after receiving the most effective medical care are given special consideration for this therapy [146].
It has been demonstrated that losing weight via diet, exercise, or bariatric surgery can improve asthma management and decrease airway inflammation in people with obesity-complicated asthma. Comparably, continuous positive airway pressure (CPAP) may help people with both sleep apnea and asthma control, as well as enhance sleep quality by reducing inflammation in the airways [12, 119].
The potential of C-X-C motif chemokine receptor 2 (CXCR2) receptor antagonists, such as SCH527123, to lessen neutrophilic inflammation in asthma is also being studied. Not all treatments have proven effective. For instance, the anti-IL-17 antibody brodalumab did not demonstrate overall effectiveness but might help a small group of patients with high bronchodilator reversibility [93, 147].
Along with clinical data, extensive multi-omics datasets including genomic/epigenomic, transcriptomic, proteomic, metabolomic, and lipidomic profiles are now publicly accessible. A deeper exploration of molecular characteristics and their associations with asthmatic features is made possible by the abundance of data available [148]. When treatment options that target specific pathways are implemented, these genetic features may eventually develop into endotypes based on their known relationship with different disease outcomes. But the shift from genetic characteristics to endotypes requires extensive testing, which is still a step away.
With differing levels of clinical preparedness, machine learning (ML) has become a formidable instrument in asthma research, demonstrating considerable promise in forecasting exacerbations, hospitalizations, and treatment failures. As per the “no free lunch theorem”, algorithm selection is crucial since no single method works for all datasets. Predictive power was low in early ML research on asthma, which was frequently based on tiny genetic datasets or clinician comparisons. However, performance has significantly improved with new large-scale deployments that use electronic health records (EHRs), home monitoring systems (like ProAir Digihaler®), and environmental data (including weather reports and air quality). High accuracy (AUCs > 0.80) has been shown by algorithms including gradient boosting machines (GBM), random forest, logistic regression, light GBM, XGBoost, support vector machines (SVM), and deep neural networks (DNN) in forecasting outcomes like hospital readmissions and asthma flare-ups [149–151].
For example, Zein et al. (2021) [149] used light GBM to reach AUCs of up to 0.88, and Finkelstein and Jeong claimed 100% accuracy with adaptive Bayesian networks. Disease control scores, peak expiratory flow (PEF), demography, weather, and medication usage were all predictive factors. Notably, in terms of stability and interpretability, XGBoost and logistic regression continuously performed better than the others. Despite these developments, integration into clinical workflows, low incidence rates, and data unpredictability pose obstacles to real-world adoption [151, 152].
Combining the multi-omics traits in one person can reveal important information that would be hidden if each type of data were examined separately. It is also necessary to think about adding and combining clinical data so that a thorough analysis may be performed while taking important variables into account. The combination of ML and multi-omics approaches opens up a variety of research directions [148]. Research on asthma has seen an increase in the use of omics technologies, which include proteomics, metabolomics, transcriptomics, genomics, and other cutting-edge approaches. They are also used for objectively identifying putative biomarkers [94, 96].
Through the use of these techniques, asthma may be examined from a variety of angles, allowing for a more comprehensive understanding of the complex and multidimensional nature of the illness [94, 153]. As an example, genomics is essential for determining genetic risk factors and possible targets for asthma treatment [153]. On the other hand, transcriptomics provides important information about asthma-related gene expression patterns, improving our understanding of the pathophysiology of the illness [154]. Moreover, changes in protein and metabolite levels are shown by proteomics and metabolomics, respectively, and they could be useful as asthma biomarkers [153]. Furthermore, combining multi-omics data with thorough phenotyping and clinical results offers a path toward gaining significant functional understanding of complex diseases like asthma [154].
Deciphering the clinical nuances and etiology of asthma requires a deliberate incorporation of omics information that makes use of data from many sources and patient groups [154, 155]. However, it is important to recognize that even while omics technologies have reduced the distance between clinical research and patient treatment, a number of methodological and design challenges need to be overcome before omics can be fully incorporated into asthma patient management. Notwithstanding these obstacles, omics has a bright future in the treatment of asthma and has the ability to improve the accuracy of care for those who have asthma [154, 156]. Moreover, deep learning techniques that were formerly complex and opaque are now easier to understand thanks to recent developments. They are now more accessible thanks to techniques like backpropagation, which allow for conclusions that go beyond simple predictions. These methods can combine several multi-omics datasets to uncover trends in the molecular characteristics of asthma. To identify different endotypes, these patterns can then be exposed to hypothesis-driven research and associated perturbation systems [94].
Thoroughly choosing a suitable approach that takes these factors into account opens the door to reliable scientific results that are repeatable and useful. Furthermore, transcriptomics improves our understanding of the pathophysiology of asthma by providing insights into the gene expression patterns underlying the illness [154]. On the other hand, epigenomics can reveal changes in histone modification, DNA methylation, and other epigenetic markers that may impact gene expression and aid in the development and progression of asthma [148, 154]. Accordingly, metabolomics can highlight changes in metabolite concentrations and may serve as important asthma biomarkers. By means of the well-coordinated integration of information derived from these many omics approaches, researchers can cultivate a more comprehensive understanding of the complex molecular mechanisms underlying asthma [154]. This comprehensive understanding, in turn, makes it easier to identify different asthma endotypes, which are subtypes of asthma distinguished by certain pathophysiological mechanisms. Understanding these endotypes is essential for determining the best route for individualized treatment plans that are specifically designed to meet the needs of each patient [154, 156].
Moreover, integrating multi-omics data with detailed phenotyping and clinical results offers a way to get a deeper functional understanding of complex diseases like asthma. The systematic combination of omics data, which utilizes data from several sources and a range of ethnic backgrounds, might provide essential insights into the complex clinical features and etiology of asthma [153, 154, 156]. Omics data can be difficult to comprehend because they can be large and complicated. Advanced computational techniques are needed to integrate various omics information and find significant patterns; the outcomes may not always be obvious. Even while omics research produces insightful information, it might be difficult to turn these discoveries into treatments that work. Pharmacological development aimed at specific biological pathways found by omics techniques necessitates substantial validation and clinical studies. However, ML models have great potential to improve individualized asthma treatment, especially when they integrate behavioral, environmental, and clinical data.
Mesenchymal stromal cells (MSCs) have received interest as a possible alternative for addressing respiratory disorders due to their therapeutic qualities, including anti-inflammatory, antiapoptotic, antibacterial, and antifibrotic attributes [157]. However, the initial outcomes of clinical trials looking into MSCs for respiratory illnesses have not lived up to expectations, even with the theoretical possibility for these cells [143, 158].
One way to further optimize the beneficial effects of MSCs is through genetic engineering. This tactic includes the manipulation of genes involved in immunomodulation and cell survival pathways, which can be accomplished by plasmid transfection, viral vector transduction, or miRNA and small interfering RNA use [159].
For example, in a lung disease model, decreased lung injury histopathological indices, reduced pulmonary edema, fewer neutrophil counts, lowered TNF-α levels, decreased protein concentration in BALF, and decreased myeloperoxidase activity in lung homogenates were the outcomes of increased expression of the Developmental Endothelial Locus-1 gene in MSCs derived from murine bone marrow [132, 159, 160].
However, turning MSC transplantation into a successful process is a significant hurdle. Even though there is a significant amount of experimental data supporting various approaches to improving MSC functionality—including genetic modification—there is still a long way to go before these approaches are applied in a clinical environment [132]. Approved for use, genetically engineered cells are currently being used in early-stage clinical trials for patients with pulmonary hypertension, such as the Pulmonary Hypertension and Angiogenic Cell Therapy (PHACeT) experiment [161]. However, additional investigation is necessary to fully understand the advantages and disadvantages of using genetically engineered MSCs to treat respiratory disorders.
The development of more personalized treatment strategies is needed to control asthma care. Combining biological molecules or drugs that target several pathways is an exciting field of study. It could lead to greater benefits by concurrently tackling many aspects of the disease. Furthermore, the ongoing discovery of trustworthy biomarkers will be crucial for informing treatment choices and guaranteeing that patients get the best medications based on the unique features of their diseases [162].
As research advances, people with milder types of asthma as well as those with severe cases would have easier access to these tailored and targeted medications. The ultimate objective is to personalize the therapy regimens to the distinct immunopathology of each patient, which will enhance their prognosis and quality of life [60, 102].
The prognosis for asthma and general quality of life might be improved by using ML to support clinical judgment and prescribe the right medication. Directly applying ML to smaller and/or less specified datasets might be challenging. In these situations, a preliminary natural language processing (NLP) phase might assist in supplying a carefully selected dataset for the ML model’s input. In addition to other elements in the patient chronology that may be used as ML inputs, clinical NLP can be used to extract clinical features from clinician records, including clinic visits, hospital treatments, and pulmonary function tests (PFTs) [163].
The interpretability of ML algorithms is challenging, and medical professionals can be enhanced by employing simpler algorithms that provide plain language explanations of their predictions or by displaying the algorithm’s decision-making process, which can then be examined by humans [163].
Because inflammatory pathways involving the innate and adaptive immune systems express themselves in different ways in different patient groups, asthma is a complicated and multifaceted illness. It is no longer reasonable to see asthma as a single, monolithic illness with symptoms including airway obstruction, bronchial hyperreactivity, persistent inflammation, and structural changes to the airways. For both T2-high and T2-low asthma, some patients still have acute exacerbations even after using bronchodilators, ICSs, leukotriene modifiers, and biologic drugs.
Numerous factors, such as the medical history of a patient, asthma endotypes, pulmonary function, biomarker phenotype, degree of support from the healthcare system, adherence to prescribed therapy, comorbidities, individual behaviors, and environmental conditions, all have an impact on the course of their asthma and the severity of their condition. Given this knowledge, treating asthmatic patients with a more customized approach is essential. This means incorporating precision medicine, which makes it easier to distinguish patients more precisely.
Asthma-related morbidity and medical expenses can be considerably decreased by employing disease management strategies to more accurately predict asthma exacerbations. In order to enhance the prognosis for asthma and the patient’s quality of life, ML may be used to examine these patient experience characteristics and support clinical decision-making.
ACOS: asthma-chronic obstructive pulmonary disease overlap syndrome
AHR: airway hyperresponsiveness
BALF: bronchoalveolar lavage fluid
COPD: chronic obstructive pulmonary disease
FcεRI: high-affinity immunoglobulin E receptor
FeNO: fractional exhaled nitric oxide
FEV1: forced expiratory volume in 1 second
GBM: gradient boosting machines
GINA: Global Initiative for Asthma
GWAS: genome-wide association studies
ICSs: inhaled corticosteroids
IgE: immunoglobulin E
IL: interleukin
ILC2s: type 2 innate lymphoid cells
LOA: late-onset asthma
LSA: long-standing asthma
LTRAs: leukotriene receptor antagonists
mAb: monoclonal antibody
MCs: mast cells
ML: machine learning
MSCs: mesenchymal stromal cells
NLP: natural language processing
OCSs: oral corticosteroids
ppb: parts per billion
SAFS: severe asthma with fungal sensitization
T2: type 2
Th2: T-helper type 2
TSLP: thymic stromal lymphopoietin
NM: Conceptualization, Data curation, Investigation, Writing—original draft, Writing—review and editing. MB: Data curation, Validation, Writing—review and editing. HIM: Conceptualization, Data curation, Investigation, Writing—review and editing. All authors read and approved the submitted version.
The author declares that there are no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
