Affiliation:
Department of Medical Laboratory Sciences, Islamic University of Gaza (IUG), Gaza P840, Palestine
Affiliation:
Department of Medical Laboratory Sciences, Islamic University of Gaza (IUG), Gaza P840, Palestine
Affiliation:
Department of Medical Laboratory Sciences, Islamic University of Gaza (IUG), Gaza P840, Palestine
Email: hadeerabuwarda@gmail.com; 2hwarda@students.iugaza.edu.ps
ORCID: https://orcid.org/0000-0001-7281-5784
Affiliation:
Department of Medical Laboratory Sciences, Islamic University of Gaza (IUG), Gaza P840, Palestine
ORCID: https://orcid.org/0000-0002-1111-958X
Affiliation:
Department of Medical Laboratory Sciences, Islamic University of Gaza (IUG), Gaza P840, Palestine
ORCID: https://orcid.org/0000-0003-3849-0909
Explor Med. 2025;6:1001360 DOI: https://doi.org/10.37349/emed.2025.1001360
Received: May 26, 2025 Accepted: August 11, 2025 Published: September 21, 2025
Academic Editor: Derek M. Dykxhoorn, University of Miami Miller School of Medicine, USA
Aim: Abnormalities in sperm parameters can result from genetic variations in DNA repair genes. The base excision repair (BER) pathway is responsible for maintaining DNA integrity. Single-nucleotide polymorphisms (SNPs) in BER genes may influence sperm DNA fragmentation (SDF) and other seminal fluid parameters. Therefore, we conducted a study to investigate the impact of SNPs in BER genes, specifically XRCC1, OGG1, MUTYH, and APEX1, on SDF and seminal fluid parameters in a selected male population from the Gaza Strip.
Methods: The case-control study included 75 men with elevated SDF and 74 men with normal SDF. Semen samples underwent conventional semen analysis and the SDF test. DNA extracted from the samples was genotyped for the selected polymorphisms using the allele-specific polymerase chain reaction (AS-PCR) technique. Genotypes and allele frequencies were compared between the case and control groups using standard statistical methods.
Results: In terms of SDF, the XRCC1 rs25487 polymorphism showed a significant difference between cases and controls, with the C allele and the CC genotype being more prevalent (P-value = 0.004) in the control group. Additionally, the MUTYH rs3219489 polymorphism also had a significant difference, with the GC genotype being more frequent (P-value = 0.025) in the control group. However, OGG1 and APEX1 polymorphisms did not show a significant difference between the two groups. The examined polymorphisms were not significantly related to conventional semen parameters.
Conclusions: This study highlights the effects of genetic variations in DNA repair genes, specifically XRCC1 and MUTYH, on SDF. Further studies with a larger sample size are necessary to confirm these findings and explore the impact of these SNPs on reproductive potential.
The World Health Organization (WHO) has recognized infertility as a global health issue. Approximately 8–12% of couples worldwide are affected by infertility, leading to emotional distress, psychological instability, and significant mental strain among those experiencing it [1]. About 40–50% of infertility cases are attributed to “male factor” infertility, indicating that these issues are primarily related to male reproductive health. Additionally, up to 2% of all men exhibit sperm parameters that fall below the standard quality, potentially contributing to fertility challenges [2].
Male infertility is directly linked to the quality and quantity of sperm in the seminal fluid. Successful fertilization relies on the transmission of high-quality and sufficient numbers of sperm to the partner, as well as the ability of the sperm to reach the fertilization site and successfully complete the fertilization process. Abnormalities in the quantity and quality of sperm can be caused by various factors, ranging from inherent birth defects and genetic disorders to lifestyle choices and environmental conditions [1].
The analysis of semen plays a crucial role in evaluating male reproductive function, enabling appropriate treatment for male subfertility. A comprehensive assessment of seminal fluid can aid clinicians in making informed decisions about further examinations and managing couples facing fertility challenges.
Sperm DNA fragmentation (SDF) has emerged as a new and valuable biomarker for identifying infertile men and providing valuable insights into the outcomes of assisted reproductive technology (ART). Sperm DNA carries half of the genomic material responsible for the offspringʼs structure. Therefore, maintaining the integrity of the genetic structure of sperm is essential for successful fertilization and the subsequent development of the embryo and fetus [3].
A lack of efficient DNA repair mechanisms can lead to increased DNA damage in germ cells, resulting in abnormal spermatogenesis and infertility. Proper functioning of DNA repair is crucial for preserving the integrity and quality of the germ cell genome. Therefore, the DNA repair system plays a critical role in sperm production. Dysfunction of genes involved in DNA damage repair within germ cells can reduce sperm quantity and contribute to abnormalities in sperm quality [4].
Therefore, this study was conducted to investigate the relationships between common single-nucleotide polymorphisms (SNPs) of DNA damage repair genes in the base excision repair (BER) pathway, specifically: XRCC1 (p.Arg399Gln), OGG1 (p.Ser326Cys), MUTYH (p.Gln324His), and APEX1 (p.Asp148Glu), and SDF results and other semen parameters.
Until now, there have been limited studies in our region that have investigated the association between specific genetic variants and SDF. Therefore, the aim of this study was to examine the relationship between selected gene polymorphisms and SDF in the population of the Gaza Strip. Identifying genetic variants associated with SDF may provide insight into the underlying mechanisms and support the development of targeted interventions to improve sperm quality.
This case–control study included 75 semen samples from males with abnormal results of the SDF test (cases) and 74 semen samples from males with normal SDF levels (controls). The study was ethically approved by the Ethical Research Committee at the Islamic University of Gaza (App. ID 28-2022); this study has also complied with the Declaration of Helsinki besides approved by the ethics committee, and all subjects gave their consent to participate in the study.
The reports of semen analysis, results of the SDF test, and general characteristics for all participants in this study were obtained from the records of Al-Basma IVF center in the Gaza Strip. Semen analysis included sperm count, liquefaction time, volume, morphology, motility, viscosity, and the percentage of abnormal sperm. The Spectrum Technologies (USA) HaloSperm kit was used to assess the level of DNA damage in semen samples. Three to four hundred sperm were examined, and an SDF of ≥ 30% was considered abnormal.
The genomic DNA was isolated from semen samples using the Wizard Genomic DNA Purification Kit (Promega, USA) following the manufacturerʼs instructions. The SNPs of the selected genes were genotyped using the allele-specific polymerase chain reaction (AS-PCR) technique. Specific PCR primers were designed using a web-based AS primer design application (http://bioinfo.biotec.or.th/WASP). Primer sequences are available from the corresponding author upon request. The PCR products were separated by running them on ethidium bromide-stained 3% agarose gels and visualized on a gel documentation system.
Data was collected, summarized, categorized, and analyzed using Statistical Package for Social Sciences (SPSS) software version 25. The frequencies of alleles and genotypes were compared between cases and controls by standard odds ratio (OR) at 95% confidence intervals (CIs) using an online calculator (https://www.medcalc.org/calc/odds_ratio.php). Hardy-Weinberg equilibrium (HWE) was tested using the online tool (https://wpcalc.com/en/equilibrium-hardy-weinberg/). All results were considered significant if the P-value was < 0.05. The multifactor dimensionality reduction (MDR) software (v.3.0.2) (http://www.epistasisblog.org/) was utilized to evaluate SNP-SNP interactions [5]. Interaction entropy graphs were created based on MDR results to identify synergistic and non-synergistic interactions among the variables.
As reported in our earlier work [6], the demographic characteristics and routine seminal parameters of the study population were as follows (Table 1): there were no statistically significant differences between the two groups in terms of mean age, BMI, smoking status, or IVF technique, as all P-values were greater than 0.05. The liquefaction time of the cases was significantly longer than that of the controls (P-value < 0.001), and the total motility of the sperm in the cases was significantly lower than that in the controls (P-value < 0.001). The sperm normal form in patients was also significantly different from that in controls (P-value = 0.049), with a lower proportion of patients having a normal form. However, there were no significant differences between cases and controls in terms of other seminal parameters, including viscosity, volume, and sperm count.
Comparison of the seminal parameters in the study population.
| Seminal parameters | Cases (n = 75) | Controls (n = 75) | t/χ2 | P-value | |
|---|---|---|---|---|---|
| Viscosity | |||||
| Normal | 69 (92.0%) | 70 (93.3%) | 0.098 | 0.817 | |
| Abnormal | 6 (8.0%) | 5 (6.7%) | |||
| Liquefaction time (min) | 23.60 ± 5.76 | 20.30 ± 1.51 | 4.682 | < 0.001‡* | |
| Volume (mL) | 2.76 ± 1.34 | 2.83 ± 0.95 | 0.367 | 0.714‡ | |
| Normal form | |||||
| More than 4% | 67 (89.3%) | 73 (97.3%) | 3.857 | 0.049* | |
| Less than 4% | 8 (10.7%) | 2 (2.7%) | |||
| Total motility | |||||
| More than 42% | 33 (44.0%) | 68 (90.7%) | 37.13 | < 0.001* | |
| Less than 42% | 42 (56.0%) | 7 (9.3%) | |||
| Sperm count | |||||
| More than 39 × 106/ejaculate | 19 (25.3%) | 26 (34.7%) | 1.556 | 0.279 | |
| Less than 39 × 106/ejaculate | 56 (74.7%) | 49 (65.3%) | |||
The reference values for seminal parameters were based on the WHO laboratory manual for the examination and processing of human semen [7]. ‡: Unpaired t-test results; *: statistically significant (P-value < 0.05). Bolded values indicate statistical significance. The control group initially consisted of 75 samples, but one sample was excluded prior to the remaining experiments.
Statistical analysis of genotypic frequencies for the investigated SNPs showed no significant differences between SDF patients and controls in relation to the OGG1 and APEX1 gene polymorphisms (P-values > 0.05). However, for the XRCC1 rs25487 polymorphism, a significant difference (P-value < 0.05) was observed between SDF patients and controls. The CC genotype was significantly more common (P-value = 0.004) in the control group, while the TC genotype was significantly less common (P-value = 0.024) in the control group. Additionally, a significant difference (P-value < 0.05) was found between SDF patients and controls for the MUTYH rs3219489 polymorphism, with the GC genotype being significantly more frequent (P-value = 0.025) in the control group (Table 2).
Genotype frequencies of the investigated polymorphisms.
| Genes/SNPs | Genotypes | Cases(n = 75) | Controls(n = 74) | Odds ratio | 95% CI | P-value |
|---|---|---|---|---|---|---|
| XRCC1rs25487T > C | TT | 6 (8%) | 2 (3%) | 3.130 | (0.610–16.040) | 0.171 |
| TC | 37 (49%) | 23 (31%) | 2.159 | (1.106–4.214) | 0.024* | |
| CC | 32 (43%) | 49 (66%) | 0.379 | (0.195–0.738) | 0.004* | |
| OGG1rs1052133C > G | CC | 33 (44%) | 34 (46%) | 0.924 | (0.485–1.763) | 0.811 |
| CG | 12 (16%) | 13 (18%) | 0.894 | (0.378–2.112) | 0.798 | |
| GG | 30 (40%) | 27 (36%) | 1.160 | (0.599–2.248) | 0.659 | |
| MUTYHrs3219489G > C | GG | 29 (39%) | 25 (34%) | 1.236 | (0.632–2.413) | 0.535 |
| GC | 23 (31%) | 36 (49%) | 0.467 | (0.239–0.912) | 0.025* | |
| CC | 23 (31%) | 13 (18%) | 2.075 | (0.957–4.501) | 0.064 | |
| APEX1rs1130409G > T | GG | 15 (20%) | 12 (16%) | 1.292 | (0.558–2.986) | 0.549 |
| GT | 23 (31%) | 34 (46%) | 0.520 | (0.266–1.017) | 0.056 | |
| TT | 37 (49%) | 28 (38%) | 1.599 | (0.833–3.071) | 0.158 |
*: Statistically significant (P-value < 0.05). Bolded values indicate statistical significance. SNPs: single-nucleotide polymorphisms; CI: confidence interval.
Statistical analysis of allelic frequencies of the tested SNPs revealed no significant difference in the gene polymorphisms of OGG1, MUTYH, and APEX1 between SDF patients and controls (P-value > 0.05). However, the allele frequency analysis for the XRCC1 rs25487 polymorphism between SDF patients and controls showed a significant difference between the two groups (P-value = 0.004) (Table 3). Statistical analysis of XRCC1 rs25487 T > C SNP revealed a significant difference between the two groups under all inheritance models, except for the recessive model, with all P-values < 0.05 (data not shown). The statistical analysis of OGG1 rs1052133 C > G and APEX1 rs1130409 G > T SNPs indicated no significant differences between the two groups according to all models (P-value > 0.05) (data not shown). The statistical analysis of the MUTYH rs3219489 G > C SNP revealed a significant difference between the two groups only under the overdominant model, with a P-value of 0.02 (data not shown). Analysis of the observed and calculated expected genotype frequencies in the control group showed that the distribution of genotypes was in HWE for all tested polymorphisms except for OGG1 rs1052133 C > G SNP (data not shown). The findings indicated no statistically significant relationships between any SNPs and conventional semen parameters (sperm count, motility, and normal sperm form), as all P-values were greater than 0.05 (data not shown).
Allele frequencies of the investigated gene polymorphisms.
| Genes/SNPs | Alleles | Cases (N = 150) | Controls (N = 148) | Odds ratio | 95% CI | P-value |
|---|---|---|---|---|---|---|
| XRCC1T > C | T | 49 (33%) | 27 (18%) | 2.174 | (1.268–3.727) | 0.004* |
| C | 101 (67%) | 121 (82%) | ||||
| OGG1C > G | C | 78 (52%) | 81 (55%) | 0.896 | (0.568–1.413) | 0.637 |
| G | 72 (48%) | 67 (45%) | ||||
| MUTYHG > C | G | 81 (54%) | 86 (58%) | 0.846 | (0.535–1.338) | 0.475 |
| C | 69 (46%) | 62 (42%) | ||||
| APEX1G > T | G | 53 (35%) | 58 (39%) | 0.848 | (0.529–1.357) | 0.491 |
| T | 97 (65%) | 90 (61%) |
N = number of investigated alleles in the study sample. *: Statistically significant (P-value < 0.05). Bolded values indicate statistical significance. SNPs: single-nucleotide polymorphisms; CI: confidence interval.
The highest risk combinations of the genotypes for the four SNPs, XRCC1 rs25487, OGG1 rs1052133, MUTYH rs3219489, and APEX1 rs1130409, are presented in Table 4.
The highest risk combinations of the genotypes for the investigated SNPs.
| SNPsMUTYH, XRCC1, APEX1, OGG1 | Frequency(case:control) | High/Low-risk | OR | 95% CI | P-value |
|---|---|---|---|---|---|
| CC, CC, GT, CG | 3:0 | High | 7.193 | (0.365–141.700) | 0.194 |
| CC, TC, GT, GG | 4:2 | High | 2.028 | (0.360–11.420) | 0.422 |
| GG, CC, TT, CC | 5:3 | High | 1.690 | (0.389–7.344) | 0.483 |
MUTYH (G > C), XRCC1 (T > C), APEX1 (G > T), OGG1 (C > G). SNPs: single-nucleotide polymorphisms; OR: odds ratio; CI: confidence interval.
The entropy values reflect the level of interaction among a set of SNPs, indicating how closely the analyzed SNPs are related to SDF (Figure 1). The Fruchterman-Reingold plot revealed the most significant effect, with higher entropy observed for XRCC1 (4.35%). The impact of polymorphisms can be ranked in the following ascending order of entropy: OGG1 rs1052133 (0.10%), APEX1 rs1130409 (1.80%), MUTYH rs3219489 (2.90%), and XRCC1 rs25487 (4.35%).
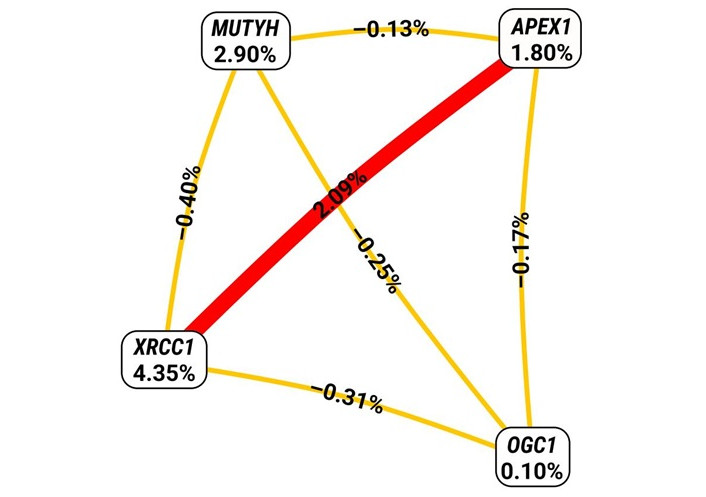
Interaction entropy diagram for gene-gene interactions and SDF risk. The red line represents the synergistic interaction between XRCC1 rs25487 and APEX1 rs1130409, which could affect an individualʼs susceptibility to DNA fragmentation in sperm. The gold-colored line represents the redundant effect among the remaining gene-gene interactions.
Accumulating evidence supports the crucial role of the BER pathway in the DNA repair system. Therefore, polymorphisms in BER pathway genes are potential risk factors for various diseases. In the present study, the main objective was to investigate whether specific genetic variations (SNPs) in XRCC1, OGG1, MUTYH, and APEX1 were linked to the quality of seminal fluid, particularly an increased risk of SDF in a selected male population from the Gaza Strip. The study focused on the analysis of the XRCC1 (p.Arg399Gln) rs25487, OGG1 (p.Ser326Cys) rs1052133, MUTYH (p.Gln324His) rs3219489, and APEX1 (p.Asp148Glu) rs1130409 SNPs in a group of 75 men with positive SDF (cases) and compared the results to those of a group of 74 men with negative SDF (controls).
Analysis of seminal parameters revealed a statistically significant difference in liquefaction time between the groups (P-value < 0.05). This finding suggests that abnormal SDF may be associated with delayed or impaired semen liquefaction, affecting the release and motility of sperm. This result is consistent with a study conducted in Iraq [8].
The link between abnormal SDF and prolonged liquefaction time may be attributed to oxidative stress [9], inflammation, or other underlying factors associated with DNA damage. These factors can affect the composition and functionality of seminal fluid, ultimately resulting in impaired liquefaction.
One of the studyʼs key findings was a significant decrease in total sperm motility among cases compared with controls (P-value < 0.001). Specifically, 56% of cases had sperm motility below 42%, compared with only 9.3% of controls. This significant difference highlights the impact of abnormal SDF on spermatozoaʼs overall motility. The observed decrease in total motility in cases compared with controls aligns with previous research that reported similar findings [10, 11].
The exact mechanisms connecting elevated SDF with decreased sperm motility are still not fully understood. However, it is possible that the DNA damage seen in sperm with abnormal fragmentation could disrupt the cellular processes necessary for efficient motility. Previous research has indicated that DNA damage in sperm could result in oxidative stress and mitochondrial dysfunction, both of which are associated with impaired sperm motility [12, 13].
In addition, the present study revealed a significant difference in the normal form of sperm between the cases and controls. This finding suggests that abnormal SDF may contribute to variations in sperm morphology. Previous research has established a link between abnormal sperm morphology and impaired sperm function, reducing fertility [14]. The association between abnormal SDF and altered sperm morphology can be attributed to several underlying mechanisms. DNA fragmentation in sperm may arise from diverse factors, including oxidative stress, exposure to environmental toxins, or genetic abnormalities. Additionally, sperm DNA damage has been associated with compromised chromatin compaction, which can influence the overall structure and morphology of the sperm [15].
Finally, there were no statistically significant differences between cases and controls regarding other seminal parameters, including viscosity, total semen volume, and sperm count. This result aligns with previous studies that reported no significant correlation between sperm count or semen viscosity and SDF [16, 17].
XRCC1, a crucial enzyme in the BER process, functions as a scaffolding protein that is essential for maintaining the stability of the BER pathway. Its main role is to recruit other necessary enzymes to the abasic site and coordinate their activities to enhance the efficiency of the pathway. The XRCC1 gene is highly expressed in the testes, particularly in pachytene spermatocytes and round spermatids. This consistent expression pattern is important for supporting spermatogenesis by aiding in DNA damage repair during meiosis and recombination in germ cells. As a result, mutations or variations in XRCC1 have the potential to disrupt the normal process of spermatogenesis, which is crucial for proper sperm production [18]. The rs25487 (p.Arg399Gln) polymorphism is a common genetic variation in the XRCC1 gene that has been extensively researched. This variation changes the amino acid from arginine, which is basic and positively charged, to glutamine, which is polar but uncharged at a conserved residue within the poly (ADP-ribose) polymerase binding domain of XRCC1 [19].
Genotypic analysis revealed that the TC genotype was significantly more prevalent in SDF-positive individuals (49%) than in controls (31%) (OR = 2.159; P-value = 0.024), suggesting an increased risk of SDF. Conversely, the CC genotype was significantly more frequent in the control group (66%) than in cases (43%) and was associated with a protective effect against SDF (OR = 0.379; P-value = 0.004). Although the TT genotype showed a higher prevalence in cases (8%) compared to controls (3%), this difference was not statistically significant (P-value = 0.171). These findings indicate a statistically significant association between the XRCC1 rs25487 polymorphism and susceptibility to SDF, particularly under codominant and dominant inheritance models. Our results are consistent with previous studies that reported associations between XRCC1 gene polymorphisms and idiopathic male infertility [18, 20, 21]. However, contrasting findings from other populations may be attributed to ethnic variability or differences in study design and sample size. Notably, the genotype distribution in the control group conformed to the HWE, suggesting the sample was genetically stable with respect to this SNP.
In spermatozoa, 8-oxoguanine DNA glycosylase 1 (OGG1) is an essential enzyme in the BER pathway. Both the sperm nucleus and mitochondria rely on this glycosylase, which actively removes 8-hydroxy-2ʼ-deoxyguanosine (8OHdG) and releases the adduct into the extracellular space [22]. Among many OGG1 variants, the most extensively studied one is rs1052133 C > G (p.Ser326Cys) [23]. This polymorphism has been linked to a decreased repair capacity [24].
In our study, there were no statistically significant differences in genotypic or allelic frequencies between SDF cases and controls (P-value > 0.05). However, both the CG and CC genotypes showed ORs below 1, suggesting a potential protective role. These findings align with those reported from Barcelona in 2018 [20]. A study from China revealed a significant association between GG and SDF [25]. The observed variations may be attributed to differences in sample sizes or ethnic diversity among the participants.
The distribution of OGG1 rs1052133 (C > G) genotypes in the control group deviated significantly from HWE, most likely due to the small sample size (n = 74).
The MUTYH gene encodes an adenine DNA glycosylase that removes mispaired adenines from the newly synthesized DNA strand when they are incorrectly paired with 8-oxoguanines.
The MUTYH gene variant rs3219489 G > C (p.Gln324His) results from an SNP causing an amino acid change from glutamine (Gln) to histidine (His) at position 324 of the MUTYH protein. This alteration in the nature of the two amino acids can impact the enzymeʼs function.
The CC genotype was more prevalent in SDF patients (31%) than in control men (18%), although the difference did not reach significance (P-value = 0.064). Similarly, the GG genotype did not significantly differ between the two groups, although it was less common in the control men (39% and 34%, respectively). However, the GC genotype significantly varied between the SDF patients and the controls (31% and 49%, respectively), with a P-value of 0.025. To date, no reports have investigated the potential impacts of the MUTYH Gln324His (G > C) SNP on seminal parameters or SDF. However, the results suggest that genetic SNPs can influence the efficiency of DNA repair processes, leading to the accumulation of DNA damage, potentially contributing to the development of various complex diseases and affecting semen quality and SDF.
In addition, a significantly increased risk of SDF with MUTYH, rs3219489 G > C, was observed under an overdominant genetic model (GG + CC versus GC) with a P-value = 0.02. Overdominant genetic inheritance implies that the heterozygous genotype confers a different phenotype than its homozygous counterpart does.
Furthermore, the genotype distribution in the control group adhered to HWE (P-value = 0.995), suggesting no major genotyping errors or population stratification.
APEX1 plays a crucial role in repairing apurinic/apyrimidinic sites that result from DNA cleavage by OGG1 and MUTYH while also aiding in the recruitment of other players in the BER process, such as DNA polymerase β and DNA ligase III [26]. Multiple genetic variations have been identified in this gene, including a G > T alteration in exon 5 that leads to the substitution of aspartic acid with glutamic acid (p.Asp148Glu; known as rs1130409) [23].
The distribution of APEX1 rs1130409 (G > T) genotypes in the control group was in HWE, as there was no significant deviation (P-value = 0.757) between the observed and expected genotypes. These findings suggest that the alleles for APEX1 rs1130409 (G > T) are randomly distributed in the study cohort.
The distribution of genotypes was as follows: GG genotype in 15 cases (20%) and 12 controls (16%); GT genotype in 23 cases (31%) and 34 controls (46%); and TT genotype in 37 cases (49%) and 28 controls (38%), with no significant differences (all P-values > 0.05). However, when the GG genotype was used as a reference, the OR for the GT genotype was 0.520 (P-value = 0.056), indicating a trend toward a decreased risk of SDF. These findings suggest that the APEX1 rs1130409 SNP may be associated with the risk of SDF, although the observed associations did not reach statistical significance. The trend toward a decreased risk of SDF in individuals with the GT genotype suggests that the heterozygous genotype may confer some protective effect against SDF.
To the best of our knowledge, there are no published studies on this polymorphism in relation to infertility or SDF. However, many studies have explored the associations between the rs1130409 SNP in APEX1 and various cancers [27–34].
The examined polymorphisms were not significantly associated with traditional semen parameters, such as motility, sperm count, or sperm morphology.
Importantly, the lack of statistical significance could be attributed to the relatively small sample size used in this study. A larger sample size would offer greater statistical power to identify significant associations. Additional factors, including environmental influences, lifestyle, and genetic variations, may play a role in the overall risk of SDF and should be taken into account in future studies.
The genotypic combinations with the highest risk for the four SNPs—MUTYH (G > C), XRCC1 (T > C), APEX1 (G > T), and OGG1 (C > G)—based on OR were (CC, CC, GT, CG) (OR = 7.193), followed by (CC, TC, GT, GG) (OR = 2.028), and finally (GG, CC, TT, CC) (OR = 1.690). However, these risk combinations did not show significant differences between the study groups.
The Fruchterman-Rheingold plot was utilized to assess the level of interaction among SNPs through entropy measurements [35]. As depicted in Figure 1, a synergistic interaction was observed between XRCC1 rs25487 and APEX1 rs1130409, potentially impacting an individualʼs vulnerability to DNA fragmentation in sperm. The figure also suggests a redundancy effect among the remaining SNPs.
This investigation involved Palestinian men residing in the Gaza Strip and experiencing increased SDF. The study focused on genetic variations in four SNPs: XRCC1 (p.Arg399Gln) rs25487, OGG1 (p.Ser326Cys) rs1052133, MUTYH (p.Gln324His) rs3219489, and APEX1 (p.Asp148Glu) rs1130409. The major outcomes of the study can be summarized as follows: elevated SDF is significantly correlated with low sperm motility, a low level of normal sperm form, and a high liquefaction time. Based on the frequencies of genotypes and alleles, the OGG1 C > G and APEX1 G > T polymorphisms are not linked to SDF in our study population. The MUTYH G > C polymorphism was associated with a lower risk for SDF, meaning individuals carrying the MUTYH (G/C) genotype may have a protective advantage against SDF. The XRCC1 rs25487 T > C polymorphism was associated with a greater risk for SDF in the study population with the (T/C) genotype. On the other hand, the genotype (C/C) is associated with a lower risk for SDF. The associations between XRCC1, rs25487, and SDF were significant under codominant, dominant, overdominant, and log-additive models, whereas the MUTYH rs3219489 polymorphism is significant under the overdominant model. There was no significant relationship between XRCC1 (T > C), OGG1 (C > G), MUTYH (G > C), or APEX1 (G > T) and the semen parameters (sperm count, motility, and sperm form) in our study sample. The highest SDF risk combination of the genotypes MUTYH (G > C), XRCC1 (T > C), APEX1 (G > T), and OGG1 (C > G) was CC, CC, GT, and CG, respectively.
The sample size in this study is relatively small, which limits the generalizability of the findings. Therefore, further validation using a larger and more diverse sample is necessary, particularly for the OGG1 C > G and APEX1 G > T gene polymorphisms.
AS-PCR: allele-specific polymerase chain reaction
BER: base excision repair
HWE: Hardy-Weinberg equilibrium
MDR: multifactor dimensionality reduction
OR: odds ratio
SDF: sperm DNA fragmentation
SNPs: single-nucleotide polymorphisms
This work was performed at the genetic diagnosis laboratory of the Islamic University of Gaza.
FAS: Supervision, Conceptualization, Writing—original draft. MMA and MJA: Resources, Supervision. HNA: Data curation, Formal analysis, Writing—review & editing. MAH: Investigation, Writing—original draft. All authors read and approved the submitted version.
The authors declare that there are no conflicts of interest.
The study was ethically approved by the Ethical Research Committee at the Islamic University of Gaza (App. ID 28-2022). This study has complied with the Declaration of Helsinki besides approved by the ethics committee.
Informed consent to participation in the study was obtained from all participants.
Not applicable.
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 894
Download: 17
Times Cited: 0
