Affiliation:
1Department of Internal Medicine, Virginia Commonwealth University, Richmond, VA 23298, USA
ORCID: https://orcid.org/0000-0002-8441-0132
Affiliation:
2Department of Internal Medicine, George Washington University, Washington, DC 20052, USA
ORCID: https://orcid.org/0000-0002-6981-3427
Affiliation:
3Department of Internal Medicine, Eastern Virginia Medical School, Norfolk, VA 23507, USA
ORCID: https://orcid.org/0000-0001-9314-5361
Affiliation:
3Department of Internal Medicine, Eastern Virginia Medical School, Norfolk, VA 23507, USA
ORCID: https://orcid.org/0009-0003-7994-1371
Affiliation:
3Department of Internal Medicine, Eastern Virginia Medical School, Norfolk, VA 23507, USA
ORCID: https://orcid.org/0000-0002-0597-0247
Affiliation:
3Department of Internal Medicine, Eastern Virginia Medical School, Norfolk, VA 23507, USA
ORCID: https://orcid.org/0000-0003-3772-2616
Affiliation:
4Department of Gastroenterology, Carolinas Medical Center, Charlotte, NC 28203, USA
ORCID: https://orcid.org/0000-0002-8501-7922
Affiliation:
5Department of Gastroenterology, Eastern Virginia Medical School, Norfolk, VA 23507, USA
Email: dajevms@aol.com
ORCID: https://orcid.org/0000-0002-8737-0711
Explor Med. 2023;4:157–165 DOI: https://doi.org/10.37349/emed.2023.00130
Received: December 01, 2022 Accepted: February 02, 2023 Published: April 25, 2023
Academic Editor: Changqing Jing, Shandong Provincial Hospital Affiliated to Shandong University; Shandong Provincial Hospital Affiliated to Shandong First Medical University, China
The article belongs to the special issue The Role of Gut Microbiota and its Metabolites in Gastrointestinal Diseases
The gastrointestinal (GI) microbiome remains an emerging topic of study and the characterization and impact on human health and disease continue to be an area of great interest. Similarly, the coronavirus disease 2019 (COVID-19) pandemic has significantly impacted the healthcare system with active disease, lasting effects, and complications with the full impact yet to be determined. The most current evidence of the interaction between COVID-19 and the GI microbiome is reviewed, with a focus on key mediators and the microbiome changes associated with acute disease and post-acute COVID-19 syndrome (PACS).
The gastrointestinal (GI) microbiome plays a key role in the bidirectional signaling between the enteric nervous system (ENS) and central nervous system (CNS), suggesting that gut function is controlled through a microbiota-gut-brain (MGB) axis [1]. Gut bacteria produce metabolites, such as tryptophan, serotonin, and short-chain fatty acids (SCFAs) that can influence gut-brain signaling through this axis. Accordingly, disruptions in the GI microbiota composition have been shown to impact neural development, behavior, and GI motility [1, 2]. Emerging studies have supported that severe acute respiratory syndrome coronavirus 2 [SARS-COV-2, also known as coronavirus disease 2019 (COVID-19)] is associated with a disruption in the normal gut microbiome and may have wide-ranging effects on human physiology and clinically significant disease.
The distinct interaction between the GI microbiome and SARS-COV-2 starts with viral entry to the enterocytes. The presence of angiotensin-converting enzyme receptor (ACE-2R) along with co-expression of the transmembrane serine protease 2 (TMPRSS2) in the same cell are key elements for SARS-CoV-2 cell entry. To enter host cells, SARS-CoV-2 uses the ACE-2R [3]. The ACE-2R is present throughout the GI tract, with the highest concentration in the small intestine. As SARS-CoV-2 invades the intestinal mucosal cells, ACE-2R is activated and the resulting increase in angiotensin II leads to increased inflammation and damage to the intestinal mucosa. This increased damage, mediated via direct cytopathic effect of the virus, increased permeability secondary to invasion of enterocytes, and inflammatory response in the lamina propria, is the main suggested mechanism of the impact of SARS-Cov-2 on bowel physiology [4]. Conceptually, initiation and activation of these ACE-2 receptors on the surfaces of the enterocytes leads to GI inflammation and causes changes in microbiome as a result of viral invasion and amplification [5]. Furthermore, ileal and colon cells contain much larger concentrations of the receptor than lung cells which may offer an explanation to why some patients quickly recover from respiratory disease, but have prolonged manifestations of GI symptoms long after respiratory symptoms abate [6].
One study looking at COVID-19 patients during the first 24 months of the pandemic showed the GI microbiome composition of COVID-19 patients was found to differ significantly from non-COVID-19 patients’ regardless of whether they had received anti-viral or other proposed COVID-19 treatments [7]. Several gut bacteria with immunomodulatory potential, such as Faecalibacterium prausnitzii, Eubacterium rectale, and Bifidobacteria spp., were underrepresented in the acute phase of COVID-19 infection and remained underrepresented in samples taken 30 days after resolution of disease symptoms. Accordingly, these associations may be key in understanding how the microbiome can contribute to the severity of COVID-19 infection and the persistence of symptoms after disease resolution. This highlights the importance of understanding how GI microbes either predispose or contribute to inflammation and COVID-19 [7]. This can be termed as a bidirectional association where an insult (such as COVID-19) can alter signaling at the gut epithelium and generates a robust inflammatory response, which can further alter biochemical immune mechanisms and further predispose the entity to further disease.
The definition of dysbiosis involves shifts in commensal microbiota diversity compared to healthy patients and is a common finding in COVID-19 infected patients [7–10]. More specifically, GI dysbiosis in COVID-19 has been shown to have an enrichment of pathogenic bacteria and depletion of beneficial commensals and is closely related to COVID-19 disease severity. Analyzing serial stool samples collected from patients up to 30 days post-diagnosis showed GI microbial composition remained significantly altered when compared with healthy controls. Notably, the GI microbiome composition of 27 patients remained significantly distinct, regardless of whether they had received antibiotics. COVID-19 patients had an increase in Bifidobacterium dentium and Lactobacillus ruminis microbiota. They also showed a decrease in Eubacterium rectale, Ruminococcus bromii, Faecalibacterium prausnitzii, and Bifidobacterium longum [7]. Faecalibacterium prausnitzii and Eubacterium rectale, which are two known beneficial commensals, were depleted in fecal samples of COVID-19 cases [6]. F prausnitzii is a major producer of SCFAs (crucial for maintaining intestinal homeostasis) in the gut and has anti-inflammatory potential due to induction of interleukin (IL)-10 production [11]. It was found to be low in abundance in feces of COVID-19 patients and had an inverse correlation with disease severity. In contrast, Clostridium hathewayi and C. ramosum, both known to be associated with inflammation and bacteremia, were positively correlated with COVID-19 severity. Fecal samples of patients categorized as high severity and infectivity had a high abundance of opportunistic pathogenic bacteria (e.g., Collinsella species, Morganella morganii) and a lower abundance of SCFA-producing bacteria (e.g., Parabacteroides merdae, Lachnospiraceae bacterium) compared with those with lower severity and infectivity [12].
Defining COVID-19 severity is important to distinguish as it has been correlated with GI disease. Mild disease is defined as “no radiographic indications of pneumonia”, moderate is defined as “evidence of pneumonia with fever and upper respiratory infection (URI) symptoms”, and severe is defined as “respiratory rate (RR) was greater than 30 breaths per min” or “SpO2 was less than 93% on room air or if requiring mechanical ventilation or intensive care unit (ICU) level of care” (Table 1) [13].
Clinical characteristics of COVID-19 severity and dysbiosis effect [12]
| COVID-19 severity type | Criteria | Dysbiosis effect |
|---|---|---|
| Mild | Clinical symptoms minimal, no imaging consistent for pneumonia | Had viral clearance as evidenced by their fecal samples and these patients showed symptomatic recovery |
| Moderate | Fever, upper respiratory symptoms, and imaging evidence of pneumonia | |
| Severe | (1) Respiratory distress (i.e., RR > 30) (2) SpO2 less than 93% at rest (3) PaO2/FiO2 < 300 mmHg (1 kPa ≈ 7.50 mmHg) | Depletion of beneficial commensals, affecting the breakdown of SCFAs and L-isoleucine biosynthesis, which in turn impaired viral clearance and symptomatic recovery resulting in persistent metabolic dysfunction |
| Critically ill | (1) Respiratory failure requiring intubation and ventilation (2) Shock (3) Organ failure, requiring ICU monitoring and treatment |
Overall, those with mild or moderate disease had viral clearance as evidenced by their fecal samples and symptomatic recovery [7]. Those with severe or critical illness from COVID-19 had dysbiosis affecting the breakdown of SCFAs and L-isoleucine biosynthesis, which in turn impaired viral clearance and symptomatic recovery resulting in persistent metabolic dysfunction. Furthermore, the biosynthesis significantly correlated with disease severity and increased plasma concentrations of C-X-C chemokine ligand (CXCL)-10 (CXCL-10), N-terminal pro-B-type natriuretic protein (NT-pBNP), and C-reactive protein (CRP) [6]. Within a healthy gut, there are many microbes that indirectly help the host from being attacked by pathogens. For example, the microbiome has mechanisms for defense such as promoting T-cell development [14], production of IL-18 [15], and creating defensins [16] with the help from the production of SCFAs by the gut bacteria. The most common SCFAs produced within the microbiome include propionate (Bacteroidetes), butyrate (Firmicutes), and acetate (Table 2) [17, 18]. Butyrate has been shown to help regulate and differentiate T-regulatory cells. Butyrate producers were reduced significantly whereas opportunistic pathogens such as Enterococcus and Enterobacteriaceae were increased in COVID-19 infected patients [19]. Reduction in IL-22, which is necessary for gut and lung epithelial barrier integrity, has been associated with reduced butyrate-forming intestinal bacteria [20, 21]. Segmented filamentous bacteria have been shown to stimulate Immunoglobulin A (IgA) and T-helper type 17 differentiation [20]. Increased IgA has been effective to slow and/or stop COVID-19 mucosal infection of epithelial cells [21]. Furthermore, metabolites from Ruminococcus gnavus, Prevotella nigrescens, Bacteroides caccae, and Micrococcus luteus have an inhibitory effect on SARS-CoV2 activity [21, 22].
GI microbiota producers of SCFAs [17]
| Common SCFAs produced by microbiome | Common biome producers |
|---|---|
| Propionate | Bacteroidetes, Negativicutes class of Firmicutes, Lachnospiraceae, Roseburia inulinivorans, Blautia species |
| Butyrate | Ruminococcaceae species, Lachnospiraceae species, Firmicutes species, including Erysipelotrichaceae and Clostridiaceae |
The three microbiome-derived metabolites that prevented SARS-CoV-2 infection in vitro were N6-Δ2-isopentenyl-adenosine (IPA), 5-hydroxytryptamine receptor (5-HTR) agonist tryptamine, and pyrazine-2,5-bis(3-indolylmethyl)-pyrazine (BIP) [23]. Synthetic drugs for the treatment of COVID-19 show structural similarities to these molecules (Figure 1) [24]. It has been proposed that continued exploration of these and similarly phylogenetically diverse human-associated bacteria is likely to show other small molecules that inhibit SARS-CoV-2 and may be utilized to help combat severe COVID-19 infection. For example, there has been shown to be a demonstratable effect on the tryptophan metabolism pathway [25]. L-tryptophan biosynthesis during acute COVID-9 was decreased in patients with more severe disease when compared with those that had no GI symptoms. Increased plasma 5-hydroxytryptamine (5-HT) concentration was found to be correlated to GI-associated COVID-19 cases versus those without GI symptoms [25]. Future studies could investigate the modification of 5-HT signaling pathways to reduce protracted post-COVID symptoms.
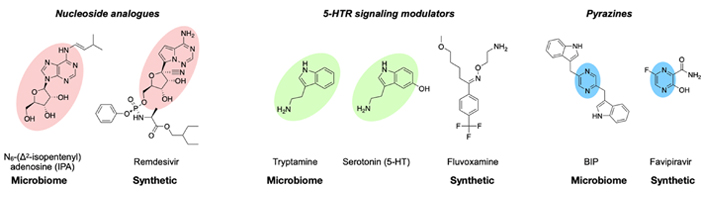
Microbiome-derived antiviral metabolites structurally resembling COVID-19 treatments [23]
Note. Reprinted from “Metabolites with SARS-CoV-2 inhibitory activity identified from human microbiome commensals” by Piscotta FJ, Hoffmann H, Choi YJ, Small GI, Ashbrook AW, Koirala B, et al. mSphere. 2021;6:e0071121 (https://journals.asm.org/doi/10.1128/mSphere.00711-21). CC BY.
Although lacking an identifiable biome alteration or a specific metabolite, the use of proton pump inhibitors (PPIs) has been shown to be associated with an increased risk of composite outcomes of critical care requirements, mechanical ventilation, and mortality in hospitalized COVID-19 patients [26, 27]. Hypothetically, the suppression of gastric acid may favor an increased viral load and enterocyte invasion by COVID-19 viral particles, thereby allowing a more robust cytokine storm [26]. Thus, PPI-related dysbiosis may play a key role in the clinical course and complications of COVID-19.
A post-acute COVID-19 syndrome (PACS) is defined by lasting symptoms and complications of the infection greater than 4 weeks from symptomatic onset of COVID-19 [28]. The mechanistic explanation has been suggested to be multifactorial, involving alterations of the immune and inflammatory mechanisms, cellular damage, or sequelae of critical illness which may contribute to symptom persistence long after the onset of disease. A cohort study of 1,733 patients found that 6 months after symptom onset, the most common symptoms reported by patients were sleep difficulties, muscle weakness, anxiety, depression, GI manifestations, and fatigue [29]. The microbiota composition of these recovered patients with COVID-19 remained significantly distinct compared with their healthy controls. Interestingly, the post-infection patients were found to have lower microbial richness, and was associated with worse pulmonary functions, higher level of CRP, and illness severity during the acute phase, suggesting close correlations between inflammatory response and GI dysbiosis in COVID-19 [30].
A prospective cohort study from China studied 106 patients and found that at six months, 76% of patients had PACS, and the GI microbiota composition at admission was associated with the occurrence of PACS [30]. Patients without PACS showed recovery of GI microbiome profile at six months comparable to that of non-COVID-19 controls. Microbial changes associated with PACS patients, had higher levels of Bacteroides vulgatus, Ruminoccus gnavus, and lower levels of Faecalibacterium prausnitzii. These are vitally important as they produce SCFAs, which have been shown to promote an anti-inflammatory state and optimal intestinal barrier function and integrity. The presence of SCFA, specifically butyrate-producing bacteria was associated with decreased development of PACS at 6 months [31]. Additionally, levels of Faecalibacterium sp. and Bifidobacterium sp. have been found to be reduced in patients with irritable bowel syndrome (IBS) compared with controls [4]. Furthermore, those with long-lasting pulmonary symptoms had higher levels of these opportunistic microbes. Neuropsychiatric symptoms and fatigue were also correlated with nosocomial GI pathogens, including Clostridium innocuum and Actinomyces naeslundii. These findings suggest that an individual’s GI microbiome profile has a profound effect on long-COVID and that these patterns may reflect different symptoms in patients with PACS.
Recently, albeit a case report, a patient with long-lasting severe GI symptoms was treated with a high-fiber formula for 2 months [32]. Endpoints of the report indicate that reported symptoms of loss of appetite, palpitations, and anxiety were significantly improved and associated with concomitant changes of the GI microbiota. Utilizing the hydrogen breath test in their patient cohort, they further found reduced microbial fermentation activity which correlated to small intestinal bacterial overgrowth. Moving forward, studies should focus on mitigation strategies such as nutritional modulation of the gut microbiota for the alleviation of GI symptoms in PACS. Microbiota-based prophylaxis and therapies for acute COVID-19 PACS, should evaluate the utility of fecal microbiota transplantation, pre- and pro-biotics, engineered symbiotic bacteria, and microbiota-derived metabolites. Additionally, GI microbiota-based profiles may be a tool of interest used for early risk stratification for occurrence of PACS and provide a therapeutic route towards targeting the GI microbiota.
A large variation in clinical presentation has been reported from patients dealing with PACS, with recent studies suggesting a significant prevalence of GI symptoms, including chronic abdominal pain and altered bowel habits. These closely align and overlap with IBS. In North America, it is currently estimated roughly 10–15% of the population suffers from IBS. The diagnosis as established by the Rome IV criteria involves recurrent abdominal pain as well as at least two of the following: abdominal pain related to defecation, associated with a change in stool frequency, and associated with a change in stool form or appearance [33]. These criteria must be met at least 1 day per week for 3 months for formal diagnosis. Many etiologies have been suggested, with most literature agreeing that the mechanisms involve impairment of intestinal secretion and motility, impaired brain-gut interaction, and psychosocial distress. Subtypes are based on altered bowel habits and include IBS with predominant constipation (IBS-C), IBS with predominant diarrhea (IBS-D), and IBS with mixed bowel habits (IBS-M). Recent studies have shown a new subtype of IBS, developing after acute gastroenteritis, known as PI-IBS. PI-IBS follows bacterial, viral, or protozoal infection, and patients typically present with IBS-D or IBS-M. It is estimated the prevalence of this subtype of IBS is roughly 11.5% [34]. This subtype is of increasing significance with the rise of PACS. A large subset of PACS patients develops GI symptoms with a prevalence of 30–40% [31]. In a recent study of 749 survivors of COVID-19, 29% had at least 1 new chronic GI symptom 6 months after infection, with the most common symptoms being heartburn, abdominal pain, constipation, and diarrhea. Of these patients with abdominal pain, 39% met Rome IV criteria for IBS [33].
The specific pathophysiology of developing PACS and specifically GI PACS is not clear, but it is thought to be multifactorial, involving inflammatory response from SARS-CoV-2 cell invasion, increased intestinal permeability, and intestinal dysbiosis [35].
Additionally, ACE-2 may also play a role in intestinal homeostasis. It can regulate blood flow perfusion by increasing mesenteric vascular resistance and enhancing mucosal nitric oxide production and mucosal bicarbonate secretion, all of which play a large role in motility. Though its effect on motility has yet to be confirmed, current evidence studying ACE-2 suggests a role in the pathophysiology of IBS via inflammation of the enteric nerve plexus [4]. One study demonstrated severe colitis in mice after fecal transplant of microbiota from mice with genetically inactivated of ACE-2 into germ-free wild-type animals [3]. Infection with SARS-CoV-2 also resulted in increased inflammatory markers persisting in patients with PACS [35]. A prospective, case-controlled study demonstrated that patients with PACS had increased serum levels of interferon (IFN)β, IFNλ1, IFNγ, CXCL-9, CXCL-10, IL-8, and soluble T cell immunoglobulin and mucin domain-containing protein 3 (TIM-3) at 4 months post-acute COVID-19 infection. Furthermore, patients who developed PACS had increased levels of IFNβ and IFNλ1 at 8 months post-infection, suggesting sustained inflammatory response after the initial infection [36].
Currently, there are no known treatments for PACS, GI PACS, or post-COVID-19 infection IBS (PC-IBS). One study examined 2 months of high-fiber formula in patients with post-acute COVID-19 with long-lasting severe GI symptoms. They found an improvement in GI symptoms, specifically nausea and appetite loss, thought to be secondary to increased SCFA production [32]. Additionally, there have been a few promising treatments for PI-IBS, such as glutamate and 5-aminosalicylic acid [37, 38]. The role of dietary glutamate for 8 weeks demonstrated significant improvement in IBS reported severity, changes in daily bowel movement frequency, stool form, and intestinal permeability. Another multicenter trial randomized placebo-controlled trial of mesalamine and demonstrated improvement in symptoms in patients with PI-IBS-D, while it had been previously ineffective in patients with IBS-D [39]. Currently, it appears that identification of patients at risk for developing post-COVID-19 IBS, may allow early intervention and potential prevention of development (or reduction) of related symptoms. Although, risk factors for GI PACS have been detailed, (including GI symptoms during active infection, pre- and post-COVID-19 psychiatric diagnoses, need for hospitalization, anosmia/dysgeusia), specific risk factors for PC-IBS have not been identified. The aforementioned risk factors increase chances for PI-IBS as well, though PI-IBS specific risk factors such as female gender are not associated with GI PACS [33]. Future studies are needed to evaluate the efficacy of PI-IBS treatments in the treatment and management of PC-IBS.
COVID-19 has been shown to have wide-ranging health effects on a variety of normal physiologic cascades that are important for health immunomodulation. There may be a bi-directional association as COVID-19 can alter signaling at the gut epithelium and generates a robust inflammatory response. This can set off a cascade of resulting changes in the microbiome and further alter biochemical immune mechanisms which allows for the risk of further disease. The associated GI dysbiosis has shown to correlate with abnormal immune response in patients with severe COVID-19 disease and may further exacerbate inflammatory and immune effects to impede the resolution. More information is needed to better understand the relationship/impact of acute PACS on the GI microbiome. Enhanced understanding will hopefully better direct the most appropriate mitigation strategies for treatment and prevention.
5-HT: 5-hydroxytryptamine
ACE-2R: angiotensin-converting enzyme receptor
COVID-19: coronavirus disease 2019
CXCL: C-X-C chemokine ligand
GI: gastrointestinal
IBS: irritable bowel syndrome
IBS-D: irritable bowel syndrome with predominant diarrhea
IFN: interferon
IL: interleukin
PACS: post-acute coronavirus disease 2019 syndrome
PC-IBS: post-coronavirus disease 2019 infection irritable bowel syndrome
PI-IBS: post-infection irritable bowel syndrome
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2
SCFAs: short-chain fatty acids
KVH and DAJ: Methodology, Writing—original draft, Writing—review & editing. AP, MS, AV, ARVS, SMD, and BSY: Writing—original draft, Writing—review & editing.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 2903
Download: 30
Times Cited: 0
Giuseppe Merra ... Marco Marchetti
Maciej Chichlowski ... Neeraj Pandey
Yuka Ikeda ... Satoru Matsuda
Lindsey B. Cundra ... David A. Johnson
Byung Soo Yoo ... David A. Johnson
Shymaa M. Al-Jabri ... Awatif A. Al-Judaibi
