Affiliation:
INRAE, Centre Île-de-France Jouy-en-Josas Antony, 78350 Jouy-en-Josas, France
Email: naima.cortes-perez@inrae.fr
ORCID: https://orcid.org/0000-0003-4189-7304
Explor Immunol. 2023;3:28–39 DOI: https://doi.org/10.37349/ei.2023.00087
Received: June 29, 2022 Accepted: December 01, 2022 Published: February 26, 2023
Academic Editor: Nitin Saksena, Victoria University, Australia
The article belongs to the special issue Immunology, Immunopathology and Genomics of SARS-COV-2
Coronavirus disease caused by the recently emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) represents a major public health that has submerged the world into a crisis unprecedented in the modern era. A better understanding of the innate immune response could help to fight this pandemic and be better prepared for potential future outbreaks. Interestingly, innate immune cells can develop a non-specific memory termed trained immunity. This review details recent evidence concerning the interaction of SARS-CoV-2 with innate immune cells, in particular those in which the trained immunity activity has been demonstrated.
Socioeconomic and ecological changes over the centuries, such as urbanization and globalization, anticipate the emergence of new pandemics [1]. In recent years, newly emerging or re-emerging infections had outbroken worldwide such as severe acute respiratory syndrome (SARS), Zika, and Ebola virus [2], among others. However, the recent SARS coronavirus 2 (SARS-CoV-2) pandemic has generated a major emergency reminding us that the pandemic remains unpredictable. Thus, dealing quickly and effectively with such situations is a major international challenge. In this context, Tomalka et al. [3] highlight how a better understanding of innate immunity may be useful for the development of more generalized vaccines able of tackling the current pandemic and being prepared for future outbreaks. Innate immunity is considered the first line of defense and it regroups both physical and chemical components and involves hematopoietic and non-hematopoietic cells [4]. The innate immune response is triggered by innate immune cells upon sensing [via pattern recognition receptors (PRRs)] of conserved molecular signals from the pathogen-associated molecular patterns (PAMPs), damage-associated molecular patterns (DAMPs) or lifestyle-associated molecular patterns (LAMPs) [5, 6]. Hematopoietic cells related to the innate immune response are predominantly of myeloid origins such as monocytes/macrophages, dendritic cells (DCs), neutrophils, eosinophils, basophils, mast cells, or of lymphoid origins such as natural killer (NK), innate lymphoid cells (ILCs), and others such as γδ T cells [7–9]. However, it should be noted that there is increasing evidence about the role of erythrocytes in innate immunity as explained by Anderson et al. [10].
The inherent characteristics of the innate immune response, such as the rapid action, the absence of specificity, and the lack of T cell receptor on innate cells, seemed incompatible with the notion of immunology memory or, at least, with that known in the adaptive immune response. As a result, until several years ago, immune memory was assumed to be an exclusive feature of the adaptive immune response; however, recent findings, in particular, those performed by Netea et al. [8, 11–13] have revealed that the innate immune cells develop a non-specific memory called trained immunity. Netea et al. [8] described trained immunity as “long-term functional reprogramming of innate immune cells, which is evoked by exogenous or endogenous insults and which leads to an altered response towards a second challenge after the return to a “non-activated state”. According to Fanucchi et al. [14], this memory event is maintained by epigenetic and metabolic reprogramming of innate immune cells. This review focuses on the interaction of SARS-CoV-2 with innate immune cells (especially those with recognized evidence of trained immunity) and discusses some aspects recently described in the literature about the virus entry, replication and sensing mechanisms.
The SARS-CoV-2 virus belongs to the family Coronaviridae of the order Nidovirales. The order Nidovirales reassembles positive-sense single-stranded genome-enveloped RNA viruses and contains a total of 109 viral species distributed among fourteen viral families [15–19]. A simplified version of the SARS-CoV-2 taxonomic classification has been shown in Table 1.
Summarized classification of SARS-CoV-2 virus
| Taxonomic rank | Classification |
|---|---|
| Order | Nidovirales |
| Suborder | Cornidovirineae |
| Family | Coronaviridae |
| Subfamily | Orthocoronavirinae |
| Genera | Betacoronavirus |
| Subgenera | Sarbecovirus |
| Specie | SARS-CoV-2 |
In general, SARS-CoV-2 causes an infection in the respiratory tract (the main entry site for the virus), which may be asymptomatic or present symptoms such as fever and cough. However, in some patients, the problems can be more serious and lead to death. According to the World Health Organization (WHO), 6 million deaths have been reported around the world [20, 21]. The factors governing the severity of the disease are not fully known, however, certain factors such as the patient’s age and association with other health problems (obesity or diabetes) may promote the severity of the disease [22].
The life cycle of SARS-CoV-2 infection has not been fully elucidated; however, several characteristics of the infectious process are currently known [23]. Three important proteins are included in the SARS-CoV-2 structure the membrane (M) protein involved in virus assembly, envelope (E) protein that forms ion channels in the cell membrane, and the spike (S) protein involved in cell entry [24].
In the first step of cell infection, the S protein (a glycoprotein) plays a central role, as the key that allows virus entry into the cell by binding to the receptor (Figure 1). Although the mechanism of action of the S protein may seem similar to that of other viruses such as human immunodeficiency virus (HIV) or influenza virus and other beta-coronavirus, the S protein of SARS-CoV-2 has some specific traits, elegantly described by Jackson et al. [25]. One important feature is that the SARS-CoV-2 S protein is sheltered by sugar molecules that protect it and give it more flexibility, as described by Scudellari [23]. S protein undergoes some modifications and cleaves into two subunits, S1 and S2 [25, 26]. This pre-activation of the S protein occurs inside the virus-producer cells performed by the protein convertase furin [27, 28]. To enter the cell, the S1 subunit of the SARS-CoV-2 S protein is recognized by a cellular receptor called angiotensin-converting enzyme 2 (ACE2) (Figure 1), while the S2 subunit participates in the fusion with the cell membrane [29]. However, in addition to the ACE2 receptor, other elements such as transmembrane protease serine 2 (TMPRSS2, a protease present on the cell surface) and cathepsin L (an internal protease) are also involved in SARS-CoV-2 entry, notably on the cleavage of the fusion S2 subunit at the site S2’, rendering these proteolytic enzymes interesting targets for antiviral drug development [30, 31].
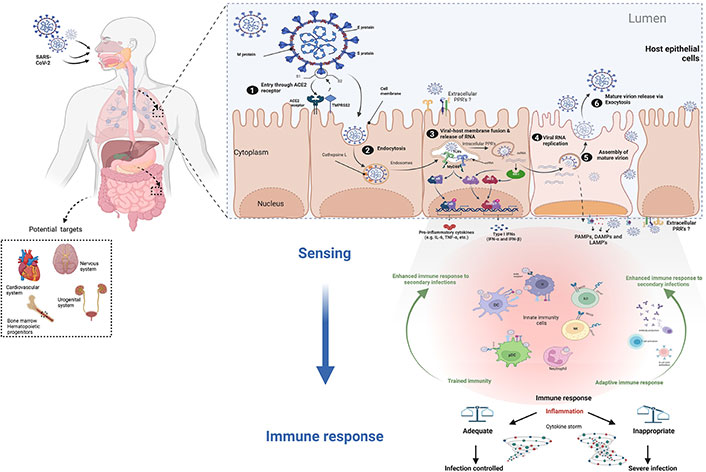
The infection cycle of SARS-CoV-2 in cells expressing receptor. ACE2 is not only largely expressed in the lung and intestinal epithelium, but also in other organs and systems where the virus can cause damage (dotted square). SARS-CoV-2 proteins involved in virus entry into cells are the M protein, E protein, and S protein. SARS-CoV-2 infection respiratory epithelium: 1) the S protein binds to the receptor and splits into two subunits (S1 and S2). The S1 subunit is recognized by the cellular receptor ACE2. 2) TMPRSS2 and cathepsin L are other receptors involved in SARS-CoV-2 endocytic entry. 3) viral RNA is released into the cell following membrane fusion. 4) SARS-CoV-2 utilizes the host cell machinery to replicate and produce viral proteins. 5) RNA and viral proteins assemble to form new virions. 6) the mature viruses are then released by exocytosis. The first line of defense against infection is innate immune cells that detect PAMPs, DAMPs, and/or LAMPs through intracellular and/or surface-exposed PRRs. Activation of PRRs will trigger cellular signals to shape the appropriate immune response (e.g., induction of cytokines, primarily IFN.) As a result, innate immune cells will undergo certain changes (trained immunity) and initiate the assembly of an adaptive immune response. An adequate and balanced immune response leads to disease resolution. CD147: cluster of differentiation 147; IRF 3: interferon regulatory factor 3; IFN: interferon; ISRE3/7: IFN-stimulated response element 3/7; MyD88: myeloid differentiation factor 88; NF-kB: nuclear factor-kappa B; NKG2D: NK group 2, member D; pDCs: plasmacytoid DCs; TLRs: toll-like receptors; IL-6: interleukin-6; RIG-I: retinoic acid-inducible gene I; TNF-α: tumor necrosis factor-α. The Figure was created with BioRender software (https://biorender.com/) using pre-generated templates
Cells that play a role in the innate immune response can come from hematopoietic or non-hematopoietic sources. Hematopoietic includes cells from myeloid and lymphoid origins. This review will focus on innate immune cells in which the process of trained immunity has been well described. The mechanisms of trained immunity are not fully understood; however, it has been noted that epigenetic alterations (such as DNA methylation, chromatin organization changes, and transcription of long non-coding RNAs) associated with metabolic reprogramming are important markers of trained immunity [8, 14]. In this context, Acevedo et al. [32] published a comprehensive review describing the process of trained immunity in several cell subsets, such as neutrophils, monocytes, macrophages, DCs, NK cells, ILCs, and hematopoietic stem cells.
However, it is not clear how myeloid and lymphoid innate immune cells can interact with SARS-CoV-2 or which ones are able to initiate an inflammatory response. Therefore, some important questions need to be addressed, for example: can ACE2 and TMPRSS2 play a role in innate immune sensing? are all innate immune cells subsets susceptible to interacting with the virus? which ones develop a productive infection and which ones are refractory? which can sense the virus and react appropriately? on the other hand, the proprotein convertase furin has been linked to the regulation of the immune response [33], so what role might it play in virus-infected innate immune cells?
As recently highlighted by Diamond and Kanneganti [22] in 2022, the innate immune response represents the first line of defense against SARS-CoV-2. Indeed, You et al. [34] provide evidence of the development of trained immunity by observing chromatin remodeling in immune cells from convalescent SARS-CoV-2 patients [35]. The innate immune response is triggered by sensing specific signals (i.e., PAMPs, DAMPs, and LAMPs) by cellular PRRs (Figure 1). Thus, activation of the PRRs will trigger a series of signals that will help to mount the most appropriate immune response. Among the PRRs exhibiting an interaction with SARS-CoV-2, we can especially mention those linked to intracellular detection of RNA viruses, including TLRs, RIG-I-like receptors, and nucleotide-binding oligomerization domain (NOD)-like receptors. The signaling cascade following activation is routed toward the induction of inflammatory cytokines, especially the production of IFN [22, 36–38]. However, as demonstrated by Zheng et al. [39], in addition to endosomal and cytosolic detection of SARS-CoV-2, other surface-exposed PRRs may also be involved (Figure 2), such as TLR2 that can sense the E protein [40]. In this context, recently Szabo et al. [41] hypothesized that TLR2 may act as a virus sensor at the central nervous system level and thus play a key role in central nervous system-related pathologies. On the other hand, an in-silico study performed by Choudhury and Mukherjee [42] suggests that TLR4, TLR1, and TLR6 may be involved in the S protein recognition. Indeed, all this has been finely detailed in the review by Diamond and Kanneganti [22]. Moreover, other studies reinforce the important role of TLRs activation in SARS-CoV-2 infection and even consider them as potential therapeutic targets [43–46]. Zhao et al. [47] demonstrated in 2021 that IL-1β induction by SARS-CoV-2 is TLR4-dependent, and showed evidence of physical interaction between TLR4 and the S protein. On the other hand, the idea that the virus may bind to other molecules and thus indirectly activate TLRs cannot be ruled out. Interestingly, work performed by Petruk et al. [48] in 2020 suggests a possible connection between the S protein and circulating lipopolysaccharides (LPS), which could be linked to the cytokine overstimulation observed in some SARS-CoV-2 infections.
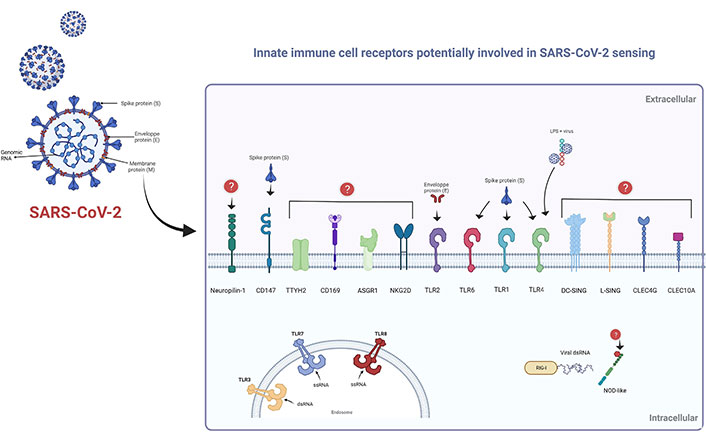
Cellular receptors of innate immunity are potentially involved in SARS-CoV-2 sensing. Schematic representation of cell receptors involved in SARS-CoV-2 detection. Surface-exposed receptors: TLR1, TLR6, and TLR4 can interact with the S protein; whereas TLR2 can bind to the E protein. Circulating LPS can bind to S protein, so the virus can indirectly bind to TLR4. S protein can also physically bind to CD147; however, the putative ligands of certain receptors remain unknown. Endosomal TLR3, TLR7, and TLR8 are involved in viral RNA recognition. Moreover, cytosolic RNA can be recognized by RIG-I-type receptors; however, the mechanism of NOD-like detection has not been fully elucidated. TTYH2: tweety family member 2; ASGR1: asialoglycoprotein receptor 1; DC-SING: DC-specific intercellular adhesion molecule-3-grabbing non-integrin; L-SING: liver/lymph node-specific intercellular adhesion molecule-3-grabbing integrin; CLEC4G: C-type lectin domain family 4, member G; ssRNA: single-stranded RNA; dsRNA: double-stranded RNA. This Figure was created with BioRender software (https://biorender.com/) using pre-generated templates
Further, in 2020, Wang et al. [49] were interested in studying the putative role of CD147 as a mediator of SARS-CoV-2 infection; they determined that the S protein interacts directly with CD147, their results also suggest that CD147 may act as an alternative receptor in cells where ACE2 expression is deficient, for example in T-lymphocytes. This is particularly interesting considering that in 2017 Geng et al. [50] found that CD147 deficiency in immature lymphocytes leads to their reprogramming into innate-like lymphocytes. Therefore, it would be interesting to evaluate the possibility of an innate-like reprogramming of T cells after SARS-CoV-2 infection. In this context, the epigenomic data of immune cells obtained by You et al. [34] may reveal useful information.
ACE2 is an important element that may help to understand virus tropism and susceptibility to infection, as suggested by Zheng [31]. ACE2 is highly expressed in the lung and intestinal epithelium but is also expressed (at different RNA and/or protein levels), in other organs and systems (e.g., cardiovascular, urogenital, and nervous) turning them into targets of infection and where the virus can potentially cause significant damage [51, 52]. Concerning the expression of ACE2 and/or TMPRSS2 in immune cells, Bao et al. [53] performed a large-scale integration analysis of the gene expression of these two receptors in 2020. From the analyzed databases and cohorts, they find no evidence of ACE2 or TMPRSS2 expression in immune cells. However, as stated by the authors, ACE2 is known to be induced by type I IFNs and this was not estimated in their analysis [53, 54]. Thus, if we consider this hypothesis under physiological conditions, immune cells are not susceptible to interacting with SARS-CoV-2 via a “classical” route of entry. However, interestingly CD147 (www.proteinatlas.org), another putative receptor [49, 55] is also expressed in immune cells [56].
On the other hand, as pointed out by Elahi [57] in an interesting review, some hematopoietic and erythroid progenitors can express ACE2, TMPRSS2, and also CD147. Hence, we cannot rule out the possibility of an impact of the virus at the level of hematopoietic stem cells directly in the bone marrow, which could be interpreted in epigenetic changes affecting cell differentiation and vigilance, especially under immunological stress conditions [58]. Further studies highlight the importance of other cellular receptors such as CD169 [59]; helper receptors such as C-type lectin receptors (DC-SIGN, L-SIGN, CD169) that act under low ACE2 expression [60]. Furthermore, Lu et al. [61] reported in 2021 a high-throughput screening approach to identify in myeloid cells, whether other cellular proteins are susceptible to bind SARS-CoV-2. As a result, they found five C-type lectins (DC-SIGN, L-SIGN, CLEC4G, ASGR1, C-type lectin domain containing 10A), and a chloride channel protein, TTYH2, which has been associated with cancer progression, but so far has not been directly related to an immunological process. Nevertheless, further research is needed to know more about this putative mechanism. The surface receptors potentially involved in SARS-CoV-2 entry into innate immune cells are shown in Figure 2.
Regarding virus replication in immune cells, not much information is available, the susceptibility of immune cells to SARS-CoV-2 and their putative ability to develop a productive infection has not yet been elucidated. In this context, Onodi et al. [62] studied the interaction of SARS-CoV-2 with human plasmacytoid pDCs. Interestingly, they show that pDCs are not productively infected by SARS-CoV-2 but can efficiently respond to viral infection. These observations were confirmed by the work of van der Sluis et al. [63] in 2022, who use an in vitro model to show evidence of efficient sensing of SARS-CoV-2 by pDCs without viral replication. They also highlight the role of neuropilin-1 in viral entry (as an alternative to ACE2), as well as of the TLR2/6 heterodimer as TLR7 for viral sensing and cytokine production. In a recent study, Percivalle et al. [64] explored the infectious process of some SARS-CoV-2 variants in monocytes and monocyte-derived macrophages. Their observations suggest that the virus can use other mechanisms (ACE2-independents) to enter the cell, however, more research is needed to elucidate these mechanisms [64]. They also show that SARS-CoV-2 provokes an abortive infection in monocytes and monocyte-derived macrophages demonstrating the absence of replication and productive infection. However, interestingly, SARS-CoV-2 maintains its infectious capacity in infected monocytes and monocyte-derived macrophages, so the authors suggest that these cells act as a trojan horse allowing virus dissemination [64].
In the same vein, in 2020, Dalskov et al. [65] were interested in better understanding the interaction of alveolar macrophages (AMs) with SARS-CoV-2. They observed that, unlike other viruses, SARS-CoV-2 does not induce IFN production by AMs. They also show that AMs do not express ACE2 on their surface; moreover, when they analyzed the efficacy of viral infection, they observed that the amount of the nucleocapsid protein decreases with time, while they found no viral proteins associating with viral replication, suggesting an absence of productive infection in these cells [65]. Altogether, these observations suggest that AMs can interact with the virus without triggering a productive infection, but are not able to sense it properly. Therefore, further research is needed to understand the underlying mechanisms [65].
Information from research with SARS-CoV-1, performed by Tseng et al. [66] in 2005 reveals that macrophages and DCs are not permissive to SARS-CoV-1 replication. In addition, studies also show that the interaction does not induce cell death and impairs, among other factors, their phagocytic capacity [66]. Recently, Boumaza et al. [67] performed in 2021 similar work using monocytes and monocyte-derived macrophages from SARS-CoV-2 patients. They show that these cells are non-productively infected with SARS-CoV-2. Interestingly, the study performed by Jalloh et al. [59] in 2022, shows that SARS-CoV-2 uses the CD169 receptor to enter macrophages in an ACE2-independent manner, resulting in a viral expression without virion production, which the authors call “restrictive infection”, also suggesting that this type of SARS-CoV-2 RNA expression may be important in maintaining the sensing and cytokine production capacity by macrophages, a condition that may contribute to a state of pathogenic hyperinflammation.
On the other hand, it has been previously shown by Shin et al. [68] that inhibition of SARS-CoV-2 papain-like protease disrupts viral replication. In addition, another study demonstrates that ILC2s are indispensable for the induction of T helper 2 (Th2)-related inflammation in response to papain (a protease-allergen) [69]. Thus, based on the above-mentioned observations, Gomez-Cadena et al. [70] studied the immune response of ILC2 to SARS-CoV-2; intriguingly, their results evidence the presence of NKG2D (a C-type lectin-like usually expressed by cytotoxic NK cells) in ILC2. Their results also suggest a protective role of this NKG2D+ ILC2s subset in SARS-CoV-2 infection. However, SARS-CoV-2 replication was not analyzed in these cells in that study, thus, it would be interesting to study the pattern of viral replication in these cells and to determine whether the infection is productive, restrictive, or abortive in ILCs.
As for the innate immune response, peripheral and tissue-resident NK cells (a subset of lymphocytes of innate immunity) play an important role in the control of viral infections, especially because of their cytotoxic potential [71, 72]. Furthermore, it has been shown that NK cells from healthy donors can directly reduce and control SARS-CoV-2 replication in human lung and kidney epithelial cell lines, whereas NK cells from SARS-CoV-2 patients are much less effective in controlling viral load [73]. The authors also show an unexpected role of transforming growth factor β (TGF-β) which, when presented at an inappropriate time, could interfere with the action of NK cells and thus favor the severity of the infection [73]. Besides that, several other findings show evidence of the role of NK cells in the immunopathology of SARS-CoV-2 [74–77]. Indeed, some viruses can directly infect them to interfere with NK cell function, as recently reported [78]. Some of them can even develop productively, such as HIV or varicella-zoster virus at least in in vitro models, however, as pointed out by the authors, more research is needed to understand whether infected NK cells can contribute to the disease severity in vivo [78]. Unfortunately, the infectivity of SARS-CoV-2 towards NK cells has not been addressed yet (or at least not yet published) but it could be an interesting research topic.
Interestingly, in human, an important role of neutrophils has been observed in SARS-CoV-2 pathology, notably relating the disease severity with the production of neutrophil extracellular traps (NETs) [79, 80]. This question was addressed by Veras et al. [81] in 2020. They found that human neutrophils release NETs in response to interaction with SARS-CoV-2, more importantly, they found that this reaction requires viral replication within the neutrophil, at least partially, since their data did not allow them to ensure that a complete replication cycle occurs in the neutrophil. Their results also indicated that the virus can use the ACE2/TMPRSS2 pathway to enter the cell. Thus, the authors bring to light the impact of the direct interaction of the neutrophil with the SARS-CoV-2 (entry and replication) on disease development [81]. However, as pointed out by the authors, the mechanisms of sensing and activation of NETs are not completely deciphered [81]; one candidate is probably TLR7 which detects single-stranded RNA [81, 82], however, we cannot rule out the participation of other receptors.
Despite numerous advances in this area, the mechanisms associated with the immune-sensing of SARS-CoV-2 remain to be fully elucidated and emphasize the need for further research on the interaction between SARS-CoV-2 and immune cells to better understand the immunopathology of the disease. Indeed, in addition to the mechanism of the cytosolic and endosomal RNA sensing, other pathways such as extracellular sensing and/or the potential role of virus-entry-receptors in the immune process need to be further studied. Undoubtedly, some important questions remain to be answered, and the proprieties that make an immune cell susceptible to developing a productive response, as well as the downstream elements responsible for the selection of the correct immune response, need to be further studied.
ACE2: angiotensin-converting enzyme 2
AMs: alveolar macrophages
CD147: cluster of differentiation 147
DAMPs: damage-associated molecular patterns
DCs: dendritic cells
DC-SING: dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
E: envelope
IFN: interferon
ILCs: innate lymphoid cells
LAMPs: lifestyle-associated molecular patterns
LPS: lipopolysaccharides
L-SING: liver/lymph node-specific intercellular adhesion molecule-3-grabbing integrin
M: membrane
NETs: neutrophil extracellular traps
NK: natural killer
NKG2D: natural killer group 2, member D
PAMPs: pathogen-associated molecular patterns
pDCs: plasmacytoid dendritic cells
PRRs: pattern recognition receptors
RIG-I: retinoic acid-inducible gene I
S: spike
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2
TLRs: toll-like receptors
TMPRSS2: transmembrane protease serine 2
The author thanks Luis Bermudez Humaran for his help in the preparation of the figures.
The author contributed solely to the work.
The author declares that he has no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 3716
Download: 22
Times Cited: 0
Nitin Saksena ... Thyago H. Cardoso
Sneha Das ... Rupesh K. Srivastava
Junming Chen ... Jianshe Yang
Hana Ratnawati ... Steven Felim
