Affiliation:
1Antibody & Product Development Lab, Wenzhou-Kean University, Wenzhou 325000, Zhejiang Province, China
Affiliation:
1Antibody & Product Development Lab, Wenzhou-Kean University, Wenzhou 325000, Zhejiang Province, China
2Zhejiang Bioinformatics International Science and Technology Cooperation Center, Wenzhou-Kean University, Wenzhou 325000, Zhejiang Province, China
3Wenzhou Municipal Key Lab of Applied Biomedical and Biopharmaceutical Informatics, Wenzhou-Kean University, Wenzhou 325000, Zhejiang Province, China
4James Cook University, Singapore 387380, Singapore
Email: samgan@apdskeg.com
ORCID: https://orcid.org/0000-0001-9936-5090
Explor Immunol. 2022;2:783–793 DOI: https://doi.org/10.37349/ei.2022.00083
Received: August 11, 2022 Accepted: December 01, 2022 Published: December 29, 2022
Academic Editor: Buyong Ma, Shanghai Jiaotong University, China
Aim: As the primary response antibody with increasing use as a therapeutic immunoglobulin (Ig) format, IgM is also the largest antibody structure among the five major human isotypes. Spontaneously formed pentamers and hexamers of IgM have avidity effects that could compensate for weaker interactions in monomeric Igs. However, this advantage is counterbalanced by potential steric clashes when binding to multiple large antigens. Recent findings have challenged the expected canonical independence of Fc receptor (FcR) binding at the heavy chain constant (C)-region where the heavy chain C-region isotypes affected antigen binding at the variable (V)-regions, and the variable heavy (VH) families of the V-region affected FcR engagement at the antibody C-regions. With such effects found on other Ig isotypes, IgM candidates need to be investigated with regards to such effects, especially when considering its natural oligomerisation at the C-region that can amplify or modulate such allosteric effects.
Methods: Through a panel of 14 recombinant complementarity determining regions (CDRs)-grafted trastuzumab and pertuzumab VH1-7 IgMs subjected to bio-layer interferometry measurements, the interactions with the antigen human epidermal growth factor receptor 2 (Her2), Fc-mu receptor (FcμR), and superantigen Protein L (PpL) were investigated.
Results: Significant effects from the V-regions to mitigate FcμR binding and the IgM C-region bidirectional effect modulating Her2 antigen engagements at the V-regions were found. Additional modulatory effects from superantigen PpL binding on the V-region of the kappa chain (Vκ) mitigating antigen binding were also found, revealing possible novel mechanisms of antibody superantigens that can be moderated by the antibody VH frameworks.
Conclusions: These findings show that the oligomerisation of IgMs plays a significant role in FcμR, antigen, and superantigen binding that made IgM distinct from the other antibody isotypes and how these features should be considered during further development and protein engineering of IgM therapeutics.
Immunoglobulin M (IgM) is the primary response antibody isotype produced in the adaptive immune response [1] and is structurally the largest multimeric antibody among the five major human isotypes (IgM, IgA, IgD, IgG, and IgE). IgM oligomerises at its constant mu (Cμ) 4 domain of the heavy chain constant (C)-region tailpiece to form pentamers in the presence of joining (J)-chains and hexamers in the absence of J-chains [2, 3]. Through oligomerisation, IgM has avidity effects that can compensate for weaker antigen binding in monomeric antibody isotypes. In a study comparing pertuzumab and trastuzumab IgG1 and IgM binding to human epidermal growth factor receptor 2 (Her2) antigen, trastuzumab IgG1 showed stronger interactions with Her2 antigen than pertuzumab IgG1 [4, 5], but pertuzumab IgM showed stronger interaction with the Her2 antigen than trastuzumab IgM [6] due to the absence of significant steric clashes with binding multiple Her2 antigens [7]. Such effects may therefore limit the total occupancy of many multimeric IgM.
Apart from antigen binding, IgM further activates the downstream immune responses via cell surface receptors. There are a few receptors with an affinity for IgM. Two receptors, polymeric Ig receptor (pIgR) and receptor for IgA or IgM Fc-alpha/mu receptor (Fca/μR), have multiple affinities for other antibody isotypes, while only one receptor, Fc-mu receptor (FcμR), has a sole affinity for IgM. FcμR [also known as TOSO or Fas apoptotic inhibitory molecule 3 (FAIM3)] is expressed on lymphoid cells such as B cells, T cells, and natural killer (NK) cells [8–10], and binds to the constant heavy (CH) 4 domain of IgM [11] to activate the immune effect via its Ig tail tyrosine (ITT) phosphorylation motif [12]. Recent studies showed that FcμR regulates cell survival when cross-linked by the IgM-antigen complex with other B-cell receptors [13].
With recent evidence demonstrating that antibody regions can exert allosteric effects on a distal region, where the variable (V)-regions, particularly the framework regions (FWRs) can mitigate the Fc receptor (FcR) binding sites (and thus FcR binding) at the heavy chain C-regions (CH) for IgA [14, 15], IgE [16], and IgG1 [17], a holistic [18] investigation for multimeric IgMs is timely. Considering the effects of non-antigen interacting regions such as the signal peptide [19] and variable heavy (VH)-variable light (VL) pairing [20], playing a role in antibody production and how the different regions, such as within the V- and C-regions, can come together to form amino acid stretches and pockets that can bind non-conventional antigens [21, 22], such interactions may underlie the mechanism of bacterial superantigen function on antibodies. Since microbial antibody superantigens bind to antibody regions [23, 24] to modulate antigen and receptor engagement [24], where Protein L (PpL) binds the variable region of the kappa (Vκ) light chain to elicit eventual immune escape, an in-depth investigation of such distal effects would also allow for future interventions to microbial superantigens permit immune escape.
Most current antibody therapeutics are IgGs, but the interest in the other isotype formats, such as IgM, has steadily increased in recent years. The contributory role of IgM in viral immunity was highlighted in the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic [25–28], and the promise of IgM as a therapeutic antibody was shown through the listed company IGM Biosciences currently with therapeutic IgM candidates undergoing phases 1 and 2 clinical trials (ClinicalTrials.gov Identifier: NCT04553692 and CT04082936) among others.
Given the promise of IgM antibodies and how such distal effects can eventually influence the development of IgM-based therapeutics, there is a need for whole antibody-level investigations. Thus, to investigate these effects, a panel of 14 recombinant VH family variants of trastuzumab and pertuzumab IgMs [where the complementarity determining regions (CDRs) of the two clinical antibodies were grafted onto the VH1-7 family FWRs] [17, 19] were expressed, with their interactions to Her2 antigen and FcμR with PpL binding measured using bio-layer interferometry (BLI) for a systematic analysis of the V-region effects.
The recombinant IgM variants with trastuzumab [Protein Data Bank (PDB) 1N8Z] and pertuzumab (PDB 1S78) sequences were generated by subcloning the V-regions from the respective IgG1 VH1-7 variants from previous work [17, 19] to the IgM CH [5] and signal peptides [19, 29, 30]. The light chains were created by grafting the respective pertuzumab and trastuzumab Vκ1 sequence with the constant kappa (Cκ). Briefly, the plasmids (one for the heavy chain and one for the light chain) were transformed into DH5-alpha (DH5α) competent Escherichia coli [31] separately for plasmid production and purified using the column-based plasmid extraction [32].
The heavy and light chain plasmids (without J-chain) were then co-transfected into HEK293 cells using Polyethylenimine (PEI) MAX® [Catalogue (Cat): 24765-1, PolyScience]. The supernatant was harvested 14 days post-transfection and purified using the AKTA equipped with a PpL affinity column (Cat: 17547815, Cytiva) as previously optimised [20, 33]. The purified recombinant IgMs were then subjected to HiLoad Superdex 200 pg preparative size exclusion chromatography (SEC) columns (Cat: 28989335, Cytiva) to collect the predominate hexamer fractions as previously described [5], where only the multimeric fraction was collected for analysis.
Equilibrium dissociation constant (KD) measurements of Fc-tagged human FcμR to recombinant pertuzumab and trastuzumab IgM variants were performed using the Octet Red96® system. Fc-tagged FcμR was immobilised on anti-human IgG Fc (AHC; Cat: 18-5060, Sartorius) biosensors with a 1.0 nM loading threshold setting. The loaded biosensors were then subjected to five concentrations (12.5, 25, 50, 100, and 200 nM) of multimeric IgM. For background noise subtraction, measurements were also performed without multimeric IgMs.
KD measurements of recombinant pertuzumab and trastuzumab IgM variants to Her2 antigen were performed using the Octet Red96® system using two capture methods. Recombinant pertuzumab and trastuzumab IgM variants were immobilised on PpL (Cat: 18-5085, Sartorius) biosensors with a 1.0 nM loading threshold setting. Similarly, recombinant pertuzumab and trastuzumab IgM variants were immobilised on streptavidin (SA; Cat: 18-5085, Sartorius) biosensors offline loaded with anti-IgM conjugate (Cat: 7102892500, Thermofisher Scientific) with 0.5 nM loading threshold setting (the lowest acceptable threshold due to the large size of the molecules and its cost). The loaded biosensors were then subjected to five concentrations (12.5, 25, 50, 100, and 200 nM) of Her2 antigen. For background noise subtraction, measurements were also performed without Her2.
The program and steps used were described as follows. The Octet Acquisition version 10.0 program was used to set the following steps for KD measurements: pre-conditioning [0.2 mol/L glycine, pH 1.52 and 10 × kinetic buffer (KB; Cat: 18-1105, Sartorius), 30 s]; initial baseline (10 × KB, 60 s); loading (ligands with specific threshold); baseline (10 × KB, 120 s); association (analyte, 300 s); dissociation (10 × KB, 600 s); regeneration (0.2 mol/L glycine, pH 1.52 and 10 × KB, 30 s). The KD values were calculated and populated based on the association rate constant (ka) and dissociation rate constant (kd) values using the data analysis v10.0 program using the 1:1 fitting model as was performed in all our previous work on the other isotypes, including that of IgM [5]. KD, ka, kd, and R2 analysis were populated by the program. All measurements were performed in triplicates [4, 5, 14–17, 19, 21, 22, 24].
To measure the distal allosteric effects of the VH FWRs family to the FcμR binding site of IgM CH, seven VH family variants of both trastuzumab and pertuzumab were recombinantly stitched to the IgM C-region (Figure S1). Given that no J-chain was co-expressed, hexamers were expected to be the predominant oligomers, and this was also supported by the relatively single monomeric peak from the SEC (Figures S2 and S3) and electron microscopy (EM) data (Figure S4).
All the pertuzumab VH1-7 variants (Figure 1) bound to FcμR except for Pertuzumab (P) VH2, which had a very poor response R2 value of 0.216. For the responsive PVH IgMs, the KD ranged from 29.73 nM to 267.6 nM, with the strongest binder as PVH1, followed by PVH7, 4, 3, 6, and PVH5 as the weakest binder.
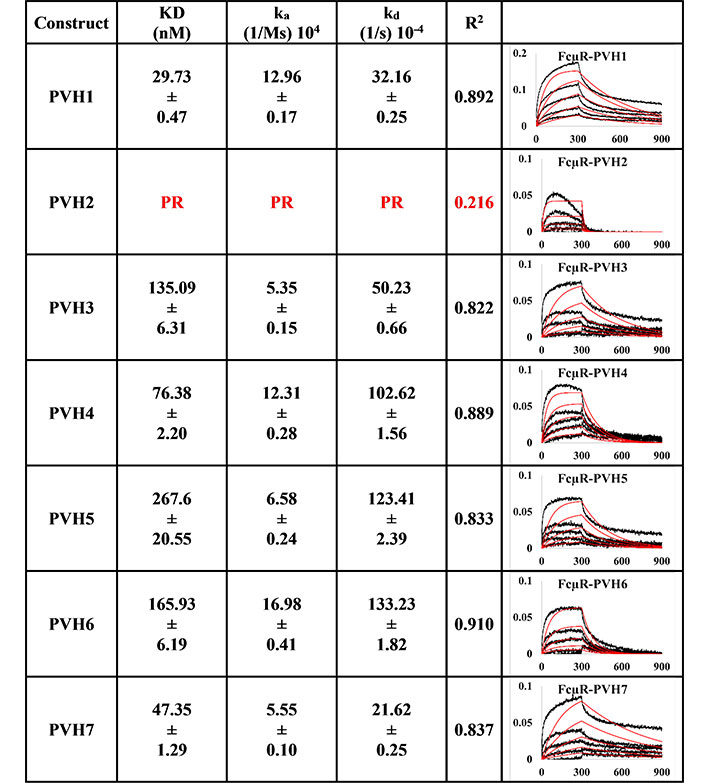
BLI measurement of pertuzumab IgM variants (PVH1-7) interacting with FcμR bound to AHC biosensor. KD, ka, kd, R2, and fitting curves are shown. PR indicates poor responses where the measurements have a poor R2 value are shown in red. The x-axis and y-axis of the graph represent time (s) and response (nM), respectively. All values are rounded off to 2 decimal places. Triplicate experiments were performed, and standard errors were reported. PR: poor responses
The trend for trastuzumab HVH1-7 variants (Figure 2) was different from the pertuzumab variants despite having very minor differences of only a few amino acids at the CDRs. For example, Trastuzumab HVH1 IgM did not bind to FcμR, having a poor response R2 value of 0.585. The rest of the HVH IgM KDs ranged from 23.29 nM to 210.47 nM (relatively similar to the PVH IgMs). The best to the weakest binder for the HVHs were in the order of HVH7, 3, 5, 6, 4, and 2.
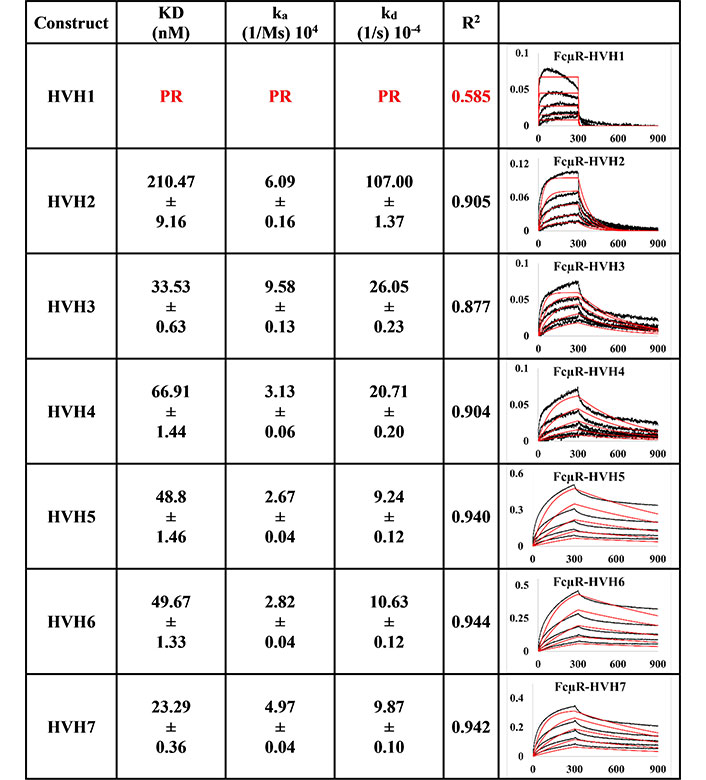
BLI measurement of trastuzumab IgM variants (HVH1-7) interacting with FcμR bound to AHC biosensor. KD, ka, kd, R2, and fitting curves are shown according to the 1:1 model. PR indicates poor responses where the measurements have a poor R2 value are shown in red. The x-axis and y-axis of the graph represent time (s) and response (nM), respectively. All values are rounded off to 2 decimal places. Triplicate experiments were performed, and standard errors were reported
As a control for proper protein folding of IgM, two immobilisation methods: i) PpL binding to Vκ1 and ii) anti-IgM binding to the Fc region of IgM were used to evaluate the binding at the V-regions to the known antigen Her2 for KD measurement.
The measurements of the anti-IgM captured method (Figures 3 and 4) were more consistent than those of PpL-captured pertuzumab IgMs to Her2 (Figures 5 and 6), showing possible modulating effects from the PpL engagement.
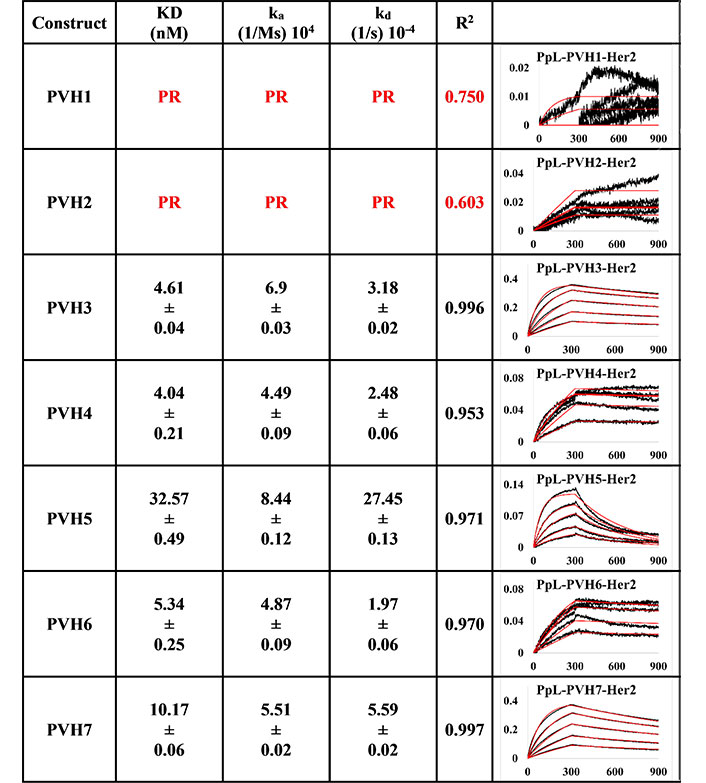
BLI measurement of pertuzumab IgM variants (PVH1-7) interacting with Her2 using Anti-IgM bound on SA biosensor to capture the Fc region of IgM. KD, ka, kd, and R2 and fitting curves are shown according to the 1:1 fitting model. The x-axis and y-axis of the graph represent the time (s) and response (nM), respectively. All values were rounded off to 2 decimal places. Triplicate experiments were performed, and standard errors are shown
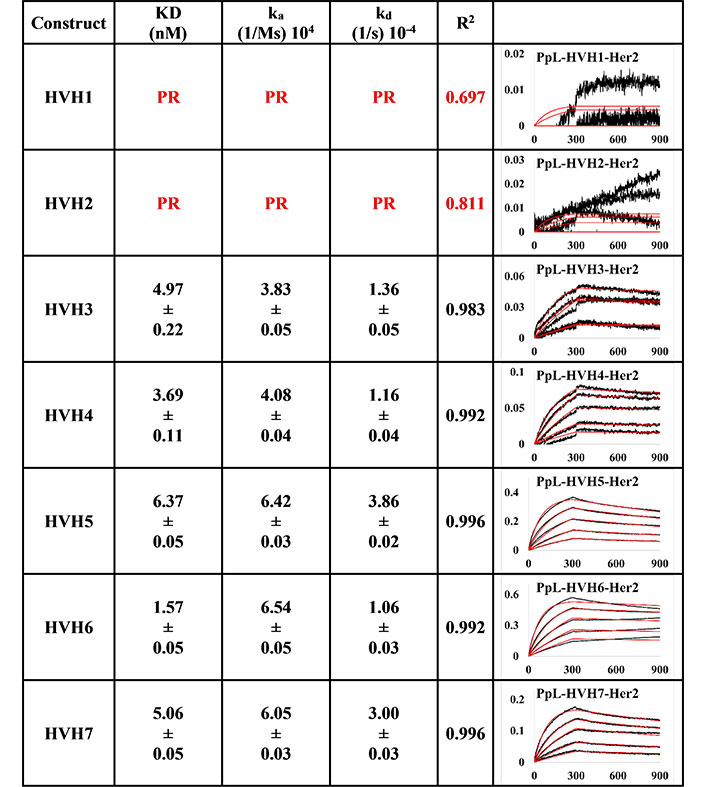
BLI measurement of trastuzumab IgM variants (HVH1-7) interacting with Her2 using Anti-IgM bound on SA biosensor to capture the Fc region of IgM. KD, ka, kd, and R2 and fitting curves are shown according to the 1:1 fitting model. The x-axis and y-axis of the graph represent the time (s) and response (nM), respectively. All values were rounded off to 2 decimal places. Triplicate experiments were performed, and standard errors are shown
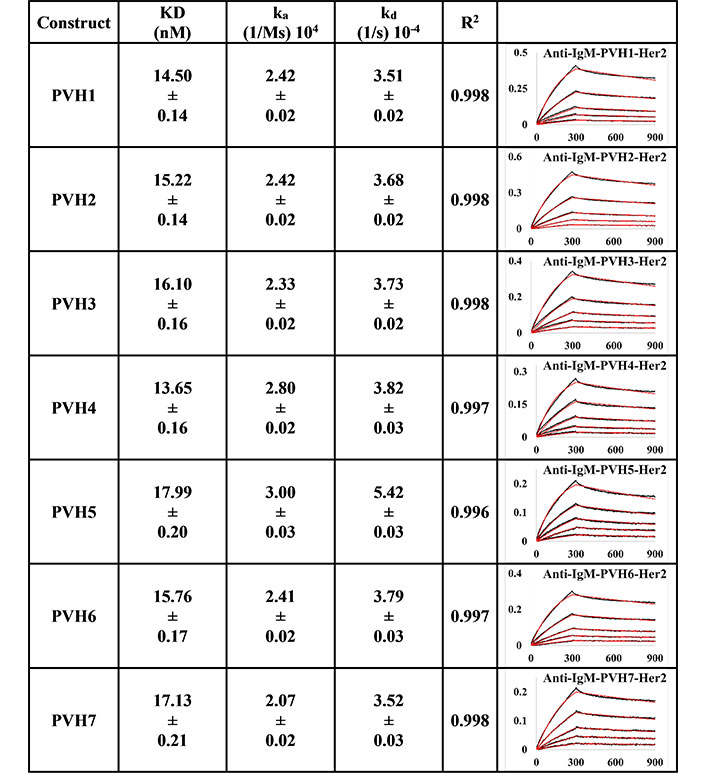
BLI measurement of pertuzumab IgM variants (PVH1-7) interacting with Her2 using PpL biosensor to capture the light chain of IgM. KD, ka, kd, and R2 and fitting curves are shown according to the 1:1 fitting model. PR indicates poor responses where measurements with a poor R2 value are shown in red. The x-axis and y-axis of the graph represent time (s) and response (nM), respectively. All values are rounded off to 2 decimal places. Triplicate experiments were performed, and standard errors were reported
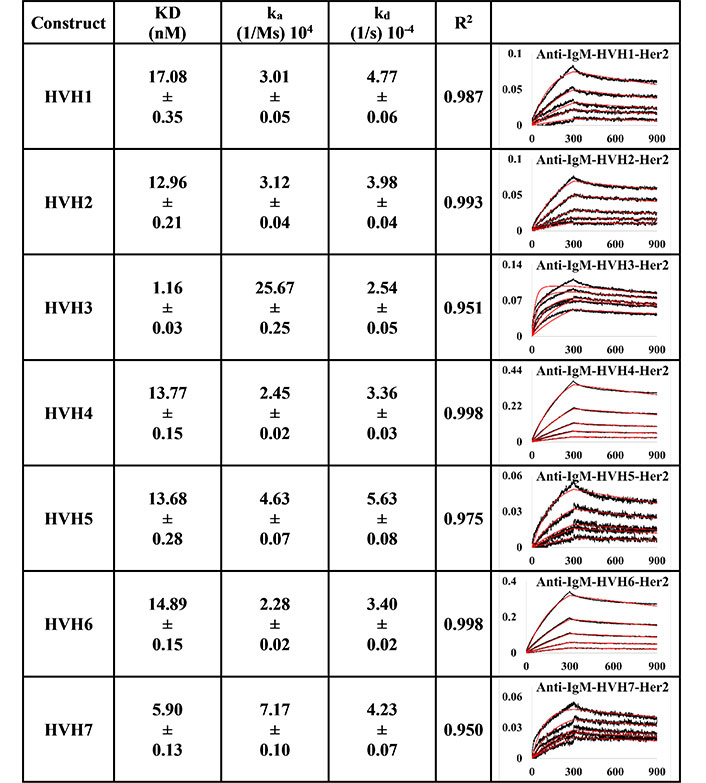
BLI measurement of trastuzumab IgM variants (HVH1-7) interacting with Her2 using PpL biosensor to capture the light chain of IgM KD, ka, kd, and R2, and fitting curves are shown according to the 1:1 model. PR indicates poor responses where measurements with a poor R2 value are shown in red. The x-axis and y-axis of the graph represent time (s) and response (nM), respectively. All values are rounded off to 2 decimal places. Triplicate experiments were performed, and standard errors were reported
Anti-IgM immobilisation method showed that all pertuzumab (Figure 3) and trastuzumab (Figure 4) VH IgM variants bound to Her2. The pertuzumab variants showed a consistent KD value range between 13.65 nM and 17.99 nM, while the trastuzumab variants had KD values that ranged from 1.16 nM to 17.08 nM. While trastuzumab had better binders, both antibody models yielded a similar range of KDs. Ranking the best binders to the weaker ones, pertuzumab variants are in the order of PVH4, 1, 2, 6, 3, 7, and 5, while trastuzumab variants are ranked in the order of HVH3, 7, 2, 5, 4, 6, and 1. It is noteworthy that the specific VH families from pertuzumab or trastuzumab had similar KD values despite the different ranking order. Two notable exceptions are seen for VH families: VH3 and 7, where trastuzumab VH3 and 7 are ten- and three-folds better binders than pertuzumab VH3 and 7, respectively.
Using the PpL immobilisation method shown in Figures 5 and 6, VH1 and 2 FWRs, regardless of whether with pertuzumab or trastuzumab CDRs, failed to bind to Her2 antigen with R2 values from 0.603 to 0.811. Generally, pertuzumab VH IgMs exhibited a more extensive range of KD values from 4.04 nM to 32.57 nM, compared to trastuzumab VH IgMs with KD values ranging from 1.57 nM to 6.37 nM. The best to the weakest binding IgMs for pertuzumab variants are in the order of VH4, 3, 6, 7, and 5, while for trastuzumab, the order is VH6, 4, 3, 7, and 5. Even though the interactions show that both pertuzumab and trastuzumab VH7 and 5 have the weakest interactions, the binding strengths of trastuzumab variants are still two to five-fold lower (better) than the pertuzumab variants.
To investigate the effect of VH families on the FcμR engagement and that of antibody superantigen PpL in IgMs, 7 VH families were grafted to the CDRs of two well-studied cancer therapeutic antibodies, pertuzumab and trastuzumab. Previously, the VH FWRs were found to affect the CH-FcR interaction on other human antibody isotypes [14, 16, 17]. Here our panel of whole IgMs was made by recombinantly stitching the previous VH variants from the IgG1 pertuzumab and trastuzumab variants onto the human IgM CH [5] (Figure S1) that formed multimers consistent with other published work on IgM [3, 34, 35] and with our SEC analyses of selected samples (Figures S2 and S3) and preliminary EM-based characterization (Figure S4).
Since binding to the respective Ig FcRs determines the downstream immune cells’ downstream activation, impaired receptor FcμR engagement by certain VH families, such as PVH2 and HVH1, can lead to lower efficacy in IgM therapeutics. Found on lymphocytes such as natural killer cells, B cells, and T cells [8–10], FcμR binds IgM at the CH [36]. In agreement with recent investigations that showed the V-regions (both VH and VL in combination) to influence FcR binding sites on the CH region for IgA [14, 15], IgE [16], IgG1 [17], similar effects were found here for IgM. From Figures 1 and 2, the best binders: PVH1 and HVH7 with 29.73 nM and 23.29 nM, respectively, had bindings to FcμR close to the reported literature of ~10 nM [10, 37] affinity, albeit in a different system. Changing the VH families modulated the binding strength up to 9-folds.
Despite the lack of clear patterns for the effects of the VH-FWR families on FcμR-CH engagement on both pertuzumab and trastuzumab, combinatorial contributions from both CDRs and VH contributed to the effects of FcμR engagement since the pertuzumab CDR-grafted variants showed more pronounced effects than the trastuzumab CDR-grafted counterparts. Since the light chain within each antibody remained constant, it was assumed that they had minimum impact on the fluctuating KDs within pertuzumab when compared to trastuzumab IgM models.
While the measurements with Her2, FcμR, and PpL were part of our intended measurements in studying the effects of the VH families, the binding with these proteins also demonstrated that our IgMs were correctly folded at their respective binding regions to be recognized biophysically. In establishing possible avidity effects, both PpL (Vκ capture) and anti-IgM (IgM CH capture) immobilisation methods, where the successful loading of the IgMs by the two capture methods (Figure S5–S8) demonstrated correctly folding IgM binding sites that together with the SEC and preliminary EM results, gave us confidence that our IgMs were functional and folded correctly.
The two immobilisation capture methods also allowed investigations into the possible proximal modulation effect of superantigen PpL binding the Vκ FWRs on antigen engagement of Her2. Although trastuzumab and pertuzumab bind Her2 as their antigen, they had different epitopes that was previously shown to affect accessibility and cause steric clashes [6, 7] for IgM. It is thus reasonable to expect the binding of superantigens [14, 38], particularly at the V-regions, to modulate antigen recognition due to the close packing of the V-regions for IgM to also result in steric clashes. It should be noted that the multimeric form of IgM locks the V-regions in spatial proximity, reducing the degrees of freedom of movement that would expectedly be further restricted by PpL binding.
Since antibody isotypes such as IgG1 were often immobilized at the Fc region using suitable capture proteins, e.g., Protein G in biophysical experiments to test the antigen binding affinity, we did not utilize natural antibody superantigens to capture the IgM Fc but used the two different capturing methods to measure the antigen binding affinity. From the PpL capture method (Figures 5 and 6), both pertuzumab and trastuzumab VH1 and VH2 gave poor responses to the Her2 antigen. Within the anti-IgM capture method (Figures 3 and 4), all the VH family variants bound to the Her2 antigen, including VH1 and 2. Such results showed that PpL binding to the VH1 and VH2 V-regions can abolish the binding to Her2. While this is due to the combinatorial effect of peculiar folding of specific VH-Vκ pairings, the findings provided an insight into the PpL immunomodulating the effects and the importance of the microbial superantigens from normal flora (discussed in [38] and [23]) which is expected to extend to other isotypes given the binding at the Vκ-FWR1 [21].
The log-fold difference between pertuzumab VH5 IgM and trastuzumab VH5 IgM within the PpL captured immobilisation results was in the same trend as the previously observed differences pertuzumab and trastuzumab IgG1s [4]. However, VH3 instead of VH5 IgM was seen with log-fold differences within the anti-IgM immobilisation results. Thus, holistically, PpL-captured IgM gave slightly better (lower) KD values than the anti-IgM capture method.
With the growing importance of IgM as a therapeutic antibody and its use in diagnostics (such as in haemagglutination kits), uncovering the mechanisms of these factors and how they can be mitigated by the presence of antibody superantigens and choice of FWR in CDR-grafting in humanisation [39] would be crucial for the efficacy of IgM for therapeutic and diagnostic uses.
Our panel showed the VH7 FWR to be a potential suitable FWR for CDR grafting for retaining binding strength for both the FcμR and antigen binding abilities. The amplification of the distal regional effects between the V- and C-regions due to the oligomerisation of IgM points to the need for future studies to investigate IgM candidates individually yet holistically to take into account the combinatorial factors involved. Such investigations would also need to include the effects of superantigens on antigen binding as found for IgEs and IgAs [5, 16]. Given that IgM is the primary response antibody, our findings here support the hypothesis that class-switching may be a mechanism for affinity maturation that ought to be considered during the humanisation step of antibody engineering [39].
In conclusion, we found exceptions within our recombinant IgMs that did not engage FcμR efficiently, likely caused by amplification of V-region distal effects on the C-region due to the oligomerisation. At the same time, the IgM CH was also found to influence antigen binding at the V-regions. With V-region superantigens such as PpL dampening antigen binding that the VH families could moderate, the interacting regions of the antibodies indeed demonstrate the complexity of IgM as an antibody format for future therapeutics showing the need for a personalised approach to antibody therapeutic development. In addition, our findings here support the use of CH engineering in antibody engineering, and that class switching may be a mechanism for affinity maturation in humans.
BLI: bio-layer interferometry
Cat: Catalogue
CDRs: complementarity determining regions
CH: constant heavy
C-region: constant region
EM: electron microscopy
FcμR: Fc-mu receptor
FcR: Fc receptor
FWRs: framework regions
Her2: human epidermal growth factor receptor 2
Ig: immunoglobulin
J-chains: joining chains
ka: association rate constant
KB: kinetic buffer
kd: dissociation rate constant
KD: equilibrium dissociation constant
PpL: Protein L
SEC: size exclusion chromatography
VH: variable heavy
V-region: variable region
Vκ: variable region of the kappa light chain
The supplementary material for this article is available at: https://www.explorationpub.com/uploads/Article/file/100383_sup_1.pdf.
We would like to acknowledge the A*STAR Microscopy platform for the preliminary EM and cryo-EM analysis of IgM structures.
WLL: Conceptualisation, Methodology, Investigation, Writing—Original Draft, Writing—Review & Editing. SKEG: Conceptualisation, Methodology, Investigation, Writing—Original Draft, Writing—Review & Editing, Funding Acquisition, Supervision. Both authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
The datasets GENERATED/ANALYZED for this study are available upon reasonable to the corresponding author (samgan@apdskeg.com).
This work was supported by the Joint Council Office, Agency for Science, Technology, and Research, Singapore [JCO1334i00050] and by the Wenzhou Science and Technology Bureau, Key Lab Program, Wenzhou Municipal Key Laboratory for Applied Biomedical and Biopharmaceutical Informatics, Wenke Jiji [2021] No.4 To Wenzhou-Kean University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2022.
Copyright: © The Author(s) 2022. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 3543
Download: 48
Times Cited: 0
