Affiliation:
Department of Infectious Diseases and Microbiology, School of Public Health, University of Pittsburgh, Pittsburgh, PA 15261, USA
ORCID: https://orcid.org/0000-0003-0802-3111
Affiliation:
Department of Infectious Diseases and Microbiology, School of Public Health, University of Pittsburgh, Pittsburgh, PA 15261, USA
Email: velpandi@pitt.edu
ORCID: https://orcid.org/0000-0002-9043-0885
Explor Neuroprot Ther. 2024;4:119–147 DOI: https://doi.org/10.37349/ent.2024.00075
Received: February 18, 2023 Accepted: March 05, 2024 Published: April 16, 2024
Academic Editor: Vijay K. Sharma, National University of Singapore, National University Health System, Singapore
The article belongs to the special issue The Future of Biomarkers in CNS Diseases
Degeneration and dysfunction of neurons in the brain are hallmarks of neurodegenerative diseases. Over the past decades, significant efforts have been devoted to the development and validation of biomarkers for neurodegenerative diseases. The range and diversity of biomarkers for central nervous system (CNS) diseases has continued to expand, encompassing biofluid-based sources such as blood or cerebrospinal fluid (CSF), nucleic acids, tissues, and imaging. While imaging and tissue biopsy-based markers are continually being identified and their applications expanding, they do have limitations compared with RNA and protein biomarkers. This review comprehensively summarizes various biomarkers, including microRNA (miRNA), long noncoding RNA (lncRNA), circulating miRNA (cimiRNA), and proteins, in the context of CNS disorders. In addition, the review emphasizes the existing limitations and challenges associated with the use of biomarkers in both clinical practice and research on neurodegenerative diseases. In conclusion, this review provides an insightful overview of the identified biomarkers for neurodegenerative diseases, underscoring the crucial role of biomarker research in combating these debilitating conditions. The article also highlights future challenges related to the implementation of novel biomarkers in clinical practice and trials, thereby contributing to the ongoing efforts to advance the understanding and management of neurodegenerative diseases.
Neurodegenerative diseases result from a complex interplay of genetic, environmental, and aging factors. These conditions entail progressive dysfunction and gradual loss of specific neuronal subsets, leading to the accumulation of misfolded proteins, heightened oxidative stress, mitochondrial dysfunction, compromised bioenergetics, and neuroinflammatory processes [1]. The overall process of neurodegeneration is illustrated in Figure 1. Neurodegenerative disorders have a profound impact on an individual’s quality of life, marked by a gradual decline in cognitive and physical functions [2, 3]. The prevalence of such diseases, exemplified by Alzheimer’s disease (AD), is anticipated to rise with increasing life expectancy, posing a substantial clinical and economic burden for the US and other nations, surpassing the costs associated with cancer [4]. By 2037, Parkinson’s disease (PD) is projected to affect 1.6 million people in the USA [5], while AD is expected to surge by approximately 130 million by 2050 [6].
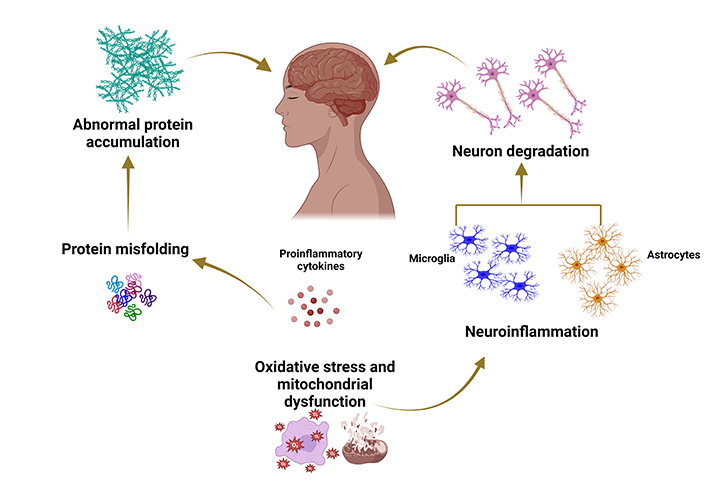
Typical mechanisms underlying the pathophysiology of neurodegenerative diseases. Created with BioRender.com
Therapeutic approaches for neurodegenerative diseases encompass a spectrum of strategies, ranging from physical therapy and medications to surgical interventions. Currently, however, there is a scarcity of drugs for many neurodegenerative conditions, and available treatments are often limited to managing symptoms and slowing disease progression. The precise mechanism for identifying neurodegenerative diseases remains elusive, presenting a significant challenge for clinicians [7–9]. Consequently, there is an unmet need to develop diagnostic kits capable of identifying biomarkers for early detection. The utilization of biomarkers holds substantial promise in advancing the diagnosis and treatment of central nervous system (CNS) diseases. Biomarkers offer the potential for early detection, improved diagnostic accuracy, and the development of personalized patient-specific treatment options [10]. In the context of the CNS, biomarkers are measurable indicators that convey information about normal or pathological processes within the brain and spinal cord. These biomarkers can be identified in diverse biological samples, including blood, cerebrospinal fluid (CSF), and through imaging studies [11, 12]. Biomarkers play a crucial role in enhancing diagnostic processes in clinical settings and are instrumental in the development and monitoring of effective disease-modifying treatments [13]. Their application extends to the diagnosis, monitoring, and assessment of treatment efficacy in a range of neurological diseases and disorders, including AD, multiple sclerosis (MS), and PD [2, 14–16]. Various body fluids, such as blood, CSF, urine, and saliva, contain detectable biomarkers that can be analyzed using diverse molecular approaches. This underscores the versatility and potential of biomarkers in advancing the understanding and management of neurological conditions (Figure 2).
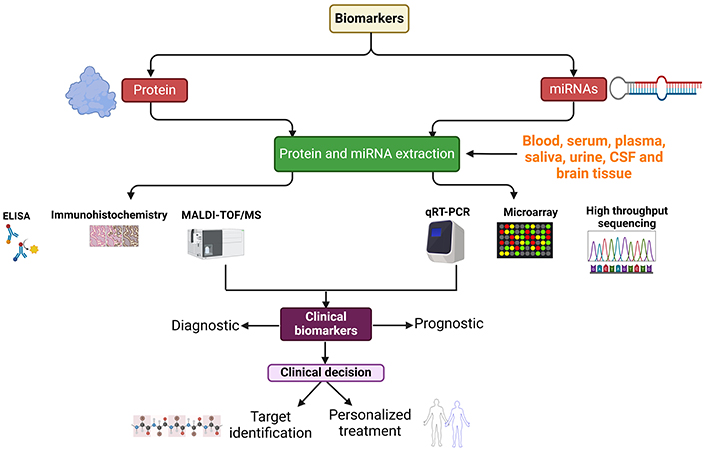
A schematic representation showing miRNA and protein biomarker collection from different sources, extraction, and quantitative approaches. miRNA: microRNA; ELISA: enzyme-linked immunosorbent assay; MALDI-TOF/MS: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; qRT-PCR: quantitative reverse transcription-polymerase chain reaction. Created with BioRender.com
In recent years, the exploration of biomarkers has emerged as a promising avenue for gaining a deeper understanding of neurodegenerative disorders. microRNAs (miRNAs) and protein biomarkers have garnered significant attention due to their intricate involvement in the molecular mechanisms underlying neurodegeneration. Proteins, acting as pivotal players in cellular functions, are implicated in various physiological and pathological processes within the CNS. Alterations in their expression, post-translational modifications, or aggregation can lead to synaptic dysfunction, neuronal cell death, and the progression of neurodegenerative disorders [17] making them valuable candidates for biomarker discovery. In addition to proteins, miRNAs, a class of small RNA molecules, play a crucial role as regulators of gene expression. Dysregulation of miRNAs has been implicated in neurodegenerative processes [18], offering a unique perspective on the intricate molecular landscape of these disorders. The exploration of both proteins and miRNAs as biomarkers provides a multifaceted approach to understanding and potentially diagnosing neurodegenerative conditions.
The CNS, comprised of the brain and spinal cord, constitutes a complex and sophisticated system responsible for regulating and coordinating body activities. CNS diseases encompass a broad spectrum of disorders wherein the functioning of the brain or spinal cord is compromised, leading to diminished motor, sensory, or cognitive capabilities [19]. These conditions may arise from various factors such as infection [20], injury [21], degeneration, or structural defects [22]. Common CNS diseases include stroke, MS, PD, and AD.
Biomarkers play a crucial role in diverse settings for identifying and managing individuals at high risk of developing various diseases, including AD, PD, Huntington’s disease (HD), dementia, and different types of cancers such as lung cancer, colorectal cancer, breast cancer, liver cancer, and esophageal cancer [23, 24]. Within the context of CNS disorders, a variety of biomarkers can be employed, encompassing genetic, molecular, functional, and image-based biomarkers [11, 25]. This multifaceted approach underscores the potential for biomarkers to contribute significantly to the understanding and management of CNS-related conditions.
Biomarkers serve as measurable biological molecules or factors that can indicate normal or disease states. Extensive research over the past decades has aimed at developing reliable biomarkers for neurodegenerative diseases. These biomarkers can be employed independently or in conjunction with other factors to enhance the reliability and precision of diagnosis, prognosis, and treatment planning in CNS diseases [26–28]. The exploration of biomarkers through miRNA expression holds immense clinical potential due to the non-invasive nature of studying gene expression using plasma samples, facilitated by advancements in microarray and high-throughput sequencing technology. miRNAs are being investigated as early biomarkers and diagnostic tools for various diseases, including cancer and Alzheimer’s, gaining strong interest in recent years [29]. They play a significant role in regulating processes such as cell proliferation, differentiation, maturation, apoptosis, and other developmental activities [30]. Numerous studies have demonstrated that miRNA dysregulation is associated with various diseases, suggesting their potential for diagnosis and treatment [29, 31, 32]. More recently, serum miRNAs and blood cell-derived miRNAs are being explored as non-invasive diagnostic biomarkers for PD, glioblastoma, and other diseases [33–35].
In addition to miRNAs, long noncoding RNAs (lncRNAs) have been implicated in the pathogenesis of neurodegenerative diseases, suggesting that their dysregulation could trigger neuronal death through unexplored RNA-based regulatory mechanisms [36, 37]. Studies have reported on the involvement of lncRNAs in various molecular processes that regulate gene expression in the CNS, ranging from neural stem cell differentiation, predominantly through chromatin remodeling, to the control of neuronal activities [38, 39].
Another class of noncoding RNAs, circulating miRNAs (cimiRNAs), holds promise as biomarkers for CNS disorders. These miRNAs are found in various body fluids, including CSF, blood, urine, breast milk, tears, and saliva [40, 41]. cimiRNAs originate in the nucleus, transfer to the cytoplasm, and are either actively released into body fluids or passively released without requiring any energy [42]. They exhibit exceptional stability, lacking loose ends that are accessible to canonical RNA decay pathways [43]. Abundant in the brain, cimiRNAs display cell- and tissue-specific expression and often remain unaltered, in contrast to proteins that undergo various post-translational modifications. This stability makes them readily detectable using laboratory techniques such as RT-PCR [42]. Moreover, cimiRNAs can endure harsh environmental conditions, including high temperature, pH levels, long-term storage, and repeated defrost cycles, showcasing greater resistance to degradation through endogenous RNA activity [44]. Their quantitative evaluation can be performed using various approaches, including next-generation sequencing (NGS), microarrays, and qRT-PCR. A notable study by Dube et al. [45] recently identified a significant correlation between circular RNA (circRNA) expression and the diagnosis, clinical dementia severity, and neuropathological severity in AD patient brain tissue samples. This highlights the potential utility of cimiRNAs as valuable biomarkers in understanding and diagnosing CNS disorders.
The past few decades have witnessed a rapid expansion in the discovery and application of protein biomarkers for neurological disorders. The complex network of protein interactions within the nervous system provides a nuanced framework for comprehending underlying pathophysiology, encompassing synaptic plasticity, cognition, and long-term potentiation [46]. Exploring protein biomarkers not only unveils molecular signatures associated with neurological disorders but also holds the potential to revolutionize clinical approaches by enabling early detection, accurate prognosis, and the development of targeted treatments [47]. The discovery of protein biomarkers for neurological disorders has seen promising developments, especially concerning the use of neurofilament proteins (NFPs) and neurogranin (Ng), amyloid-beta 42/40 (Aβ42/40), and tau. Examples of some miRNA and protein biomarkers are summarized in Tables 1 and 2. The comparatively slower progress of biomarkers for CNS disorders, compared to other areas of research, can be attributed to intricate processes, challenges in obtaining CNS tissue samples, and the complexities of the brain’s structure and functions [48, 49]. Additionally, numerous external factors can influence the design and outcomes of studies, including the interplay between genes and the environment, the diversity of CNS disorders, and variations in patient populations.
Examples of miRNAs biomarkers for various neurological disorders
| S.No | Disease | Sample source | miRNA | Method | Reference |
|---|---|---|---|---|---|
| 1. | AD | Plasma | miR-342-5p | qRT-PCR | [50] |
| 2. | AD | Brain tissue | miR-31 | qRT-PCR | [51] |
| 3. | AD | Brain tissue | miR-132 | qRT-PCR | [52] |
| 4. | AD | Brain tissue | miR-212-5p | qRT-PCR | [53] |
| 5. | AD | Cell line | miR-124 | qRT-PCR | [54] |
| 6. | AD | Brain tissue | miR-146a | qRT-PCR | [55] |
| 7. | PD | Tissues | miR-34c | qRT-PCR | [56] |
| 8. | PD | Brain tissue | miR-599 | qRT-PCR | [57] |
| 9. | PD | Tissue and cell line | miR-21 | qRT-PCR | [58] |
| 10. | PD | Brain tissue | miR-185 | qRT-PCR | [59] |
| 11. | PD | Cells | miR-34a | qRT-PCR | [60] |
| 12. | PD | Serum | miR-218-5p, miR-320a-5p | qRT-PCR | [61] |
| 13. | PD | Brain tissue | miR-543 | qRT-PCR | [62] |
| 14. | PD | Brain tissue | miR-10b-5p | Illumina sequencing | [63] |
| 15. | PD | Brain tissue | miR-7 | qRT-PCR | [64] |
| 16. | PD | Plasma | miR-23b-3p | Illumina sequencing | [65] |
| 17. | PD | Serum | miR-214 | qRT-PCR | [66] |
| 18. | PD | Plasma | miR-133b, miR-221-3p | qRT-PCR | [67] |
| 19. | PD | Serum | miR-223-3p, miR-7-1-5p | qRT-PCR | [35] |
| 20. | PD | Saliva | miR-29a-3p, miR-29c-3p | Microarray/qRT-PCR | [68] |
| 21. | AD | Serum | miR-93 | qRT-PCR, Solexa sequencing | [69] |
| 22. | AD | Cell culture | miR-298 | qRT-PCR | [70] |
| 23. | AD | PBMC | miR-150-5p | qRT-PCR | [71] |
| 24. | AD | Serum | miR-28-3p | qRT-PCR | [72] |
| 25. | AD | Serum | miR-128 | qRT-PCR | [73] |
| 26. | AD | Serum | miR-455-3p | qRT-PCR | [74] |
| 27. | AD | Brain tissue | miR-129-5p | qRT-PCR, Illumina sequencing | [75] |
| 28. | AD | Serum | miR-331-3p | qRT-PCR | [76] |
| 29. | AD | Plasma | miR-103, miR-107 | qRT-PCR | [77] |
PBMC: peripheral blood mononuclear cells
Examples of protein biomarkers for various neurological disorders
| S.No | Disease | Sample source | Protein | Method | Reference |
|---|---|---|---|---|---|
| 1. | AD | Plasma | NFL | ELISA | [78] |
| 2. | AD | CSF | NFL | ELISA | [79] |
| 3. | AD | CSF | NFL | ELISA | [80] |
| 4. | PD | Serum | NFL | Electrochemiluminescence immunoassay | [81] |
| 5. | PD | CSF | NFL | ELISA | [82] |
| 6. | PD | CSF | Ng | ELISA | [83] |
| 7. | HD | CSF | Ng | ELISA | [84] |
| 8. | AD | Blood, CSF | Ng, synaptotagmin 1 | ELISA | [85] |
| 9. | AD | CSF | Ng | ELISA | [86] |
| 10. | AD | CSF | Ng | ELISA | [87] |
NFL: neurofilament light
miRNAs are short non-coding RNA molecules, typically 22–24 nucleotides in length, playing a crucial role in diverse biological processes, including cellular proliferation, differentiation, cell cycling, and the maintenance of immune homeostasis [88, 89]. Abundantly expressed in neurons, miRNAs are essential for various neurobiological functions, such as synaptogenesis, neural patterning, neuronal differentiation, the formation and maintenance of cell identity, and neuronal plasticity [90]. miRNAs have been implicated in various human diseases, including neurological disorders like AD, PD, and MS. They have the potential to serve as biomarkers for different neurological disorders and are released as circulating molecules into body fluids such as CSF, blood, and urine, making them valuable for early detection of neurodegenerative disorders [91].
miRNAs can be collected from various sources, including CSF, blood, and other biofluids like serum, plasma, and urine, and utilized as diagnostic biomarkers for neurological disorders [74, 92–94]. Ideal biomarkers need to be easily accessible and extracted through liquid biopsies from different bodily fluids [95]. The circulating levels of miRNAs from these fluids can be characterized and used as non-invasive diagnostic biomarkers [96, 97]. Various specific and sensitive methods, including qRT-PCR, microarray, and NGS, are employed to detect miRNAs. For example, in a study on PD and multiple system atrophy (MSA), miRNA biomarkers were collected from CSF, and their expression levels were quantified using quantitative PCR [98]. Recently, dysregulation of miRNA and non-coding RNA expression has been reported in diverse neurological diseases, such as AD, PD, and HD [99, 100]. The role of miRNA biomarkers has been particularly emphasized in the early stages of AD [51, 101]. Novel plasma miRNAs (miR134-3p, miR-22-5p, miR-1185-2-3p, miR-1909-3p, and miR-107) have been identified as potential therapeutic targets for AD at the prodromal stage [102].
The investigation of specific miRNAs in the context of AD and PD has provided insights into their potential roles and therapeutic implications:
miR-98 in AD: This effect was mediated via the Notch signaling pathway through binding to hairy and enhancer of split-related with YRPW motif protein 2 (HEY2). It was found that miR-98 decreased the production of Aβ-protein and significantly increased oxidative stress while modifying mitochondrial functioning in scopolamine-treated AD mice. Detection of miR-98 and the protein levels of HEY2 and β-amyloid precursor protein (APP) was performed in brain tissues using RT-PCR and western blot analyses [103].
miR-153 in AD: miR-153 expression was reported to be downregulated in the brains of individuals with advanced-stage AD. This miRNA was found to obstruct the expression of Aβ-precursor protein in cultured human fetal brain cells [104].
miR-128 in AD: The expression of miR-128 was upregulated, and peroxisome proliferator-activated receptor gamma (PPAR-gamma) expression was downregulated in the plasma of AD patients. miR-128 was shown to suppress Aβ-mediated neurotoxic effects by binding to PPAR-gamma and blocking the intracellular NF-κB pathway [105]. miR-128 directly inhibits the expression of tau phosphorylation by targeting glycogen synthase kinase 3β (GSK3β), modulators APP binding protein 2 (APPBP2) and the mammalian target of rapamycin (mTOR). Upregulation of miR-128 in the hippocampus of five familial AD (5XFAD) mice ameliorates learning and memory impairments, tau phosphorylation, Aβ secretion, and enhances autophagic flux. While downregulation of miR-128 in the late stage of AD increases the concentration of Aβ in brain [106].
miR-7 in PD: miR-7, a brain-enriched miRNA, plays a significant role in brain development and neurological disorders, including PD [107, 108]. It directly downregulates α-synuclein expression in dopaminergic neurons, providing cellular protection [109]. Abnormal aggregation and expression of the α-synuclein protein in the brain are key features of PD [110–112]. miR-7 regulates α-synuclein expression by interacting with the 3’-untranslated region (UTR) region of the synuclein alpha non A4 component of amyloid precursor (SNCA) gene, blocking the conversion of the gene’s information into a protein. Reduced levels of miR-7 in the substantia nigra have been observed in PD patients, emphasizing its role in regulating α-synuclein expression and suggesting its potential as a therapeutic target for PD [113].
miR-124 is one of the most widely expressed miRNAs in the brain and is implicated in various crucial processes, including neurogenesis, synaptic architecture, apoptosis, inflammation, neurotransmission, and mitochondrial function [114, 115]. In the context of PD, chronic inflammation in the CNS driven by microglial cells plays a vital role in disease progression. Studies have suggested the following roles for miR-124 in neuroinflammation and neurodegenerative diseases. Overexpression of miR-124 in the brain has been associated with protection of SH-SY5Y cells from apoptosis induced by microglial activation triggered by lipopolysaccharide exposure. This finding suggests a potential key role for miR-124 in inhibiting neuroinflammation in the context of PD [116]. Downregulation of miR-124 expression has been linked to increased Aβ deposition, angiogenesis, and microvascular dysfunction in the hippocampus and cerebral cortex of AD mice. This effect is mediated by the control of complement C1q-like protein 3 [117]. Brain-specific miR-124 has been implicated in inhibiting neuroinflammation during the development of PD by targeting p38, p62, and autophagy. This suggests that miR-124 may serve as an effective therapeutic target for controlling the inflammatory response in PD patients [116]. Furthermore, a clinical study has demonstrated that small extracellular vesicles (sEVs) loaded with miR-124-3p protect dopaminergic neurons in the substantia nigra and striatal fibers, counteracting motor behavior symptoms in a PD model, and induce neuronal differentiation under physiological conditions in vitro. These findings suggest that miR-124-3p-enriched sEVs could offer a promising therapeutic strategy for halting PD neurodegeneration and improving motor function [118]. The multifaceted roles of miR-124 underscore its significance in various processes relevant to neurodegenerative diseases, particularly PD and AD.
HIV-associated neurocognitive disorders (HAND) impact a significant percentage of people living with HIV (PLWH), ranging from 15% to 60% of individuals with HIV infection. PLWH refers to individuals who have been diagnosed with HIV and are living with the virus. These individuals may be receiving treatment and care for their HIV infection. Though effective therapy for HAND is currently lacking, management strategies involve a combination of antiviral drugs, cognitive therapy, and other supportive therapies. Identifying biomarkers for cognitive health in PLWH is crucial for effective interventions to prevent and delay the onset of HAND. Substance abuse and comorbid conditions can complicate the detection of these biomarkers in PLWH [119, 120]. Several studies have explored the potential of miRNAs as biomarkers for HAND. Microarray-based analysis of plasma samples from two cohorts of HIV-positive patients, with and without HAND, identified three upregulated miRNAs (miR-3665, miR-4516, and miR-4707-5p) in patients with HAND. This suggests that plasma miRNA profiling holds potential for predicting and understanding HAND [121]. Investigation of miRNAs isolated from the CSF of HIV-positive patients indicated that 11 miRNAs were significantly upregulated in HIV encephalitis [122]. HIV-1-encoded viral proteins of HIV-1 transactivator of transcription (Tat), has attracted significant attention due to its toxicity to many CNS cells [123]. A study investigated the effects of HIV-1 Tat and cocaine co-exposure on two miRNAs (hsa-miR-2355 and hsa-miR-4726-5p), finding that their expression was significantly downregulated in human primary astrocytes exposed to both substances [124].
HIV-1 Tat-mediated upregulation of miR-34a targets the 3’-UTR of NOD-like receptor family CARD domain-containing 5 (NLRC5) in microglial cells. Dysregulated NLRC5 inflammasome leads to decreased activation of the NF-κB p65 signaling axis, resulting in increased expression of pro-inflammatory cytokines [125]. miR-34a and miR-138 negatively regulated the expression of astrocytic SIRT1 in astrocytes exposed to HIV-1 Tat, leading to astrogliosis, a characteristic feature of aging in the CNS [126]. Seventeen miRNAs were differentially expressed in the brain tissue of HAND patients. Some of these miRNAs were predicted to target peroxisome biogenesis factors, suggesting a new mechanism by which HIV-1 may cause neurocognitive dysfunction [127]. The advantages of using miRNAs as biomarkers include their early detection potential, stability in various bodily fluids, and the ability to enable larger studies and retrospective analyses [128].
lncRNAs are a class of non-coding RNA molecules longer than 200 nucleotides, located in the nucleus or cytoplasm [129]. They play various roles in cellular processes, including transcriptional regulation, chromatin modification, and post-transcriptional regulation. lncRNAs can mediate pathogenesis through mechanisms such as decoy, scaffold, histone modifiers, miRNA sequestration, and transcriptional interference [130, 131]. Their levels can be determined using techniques like qRT-PCR, NGS, and microarray, making them potential non-invasive prognostic and diagnostic markers [100, 130]. In the context of AD, several of the following lncRNAs have been implicated.
Aβ accumulation in the brain, associated with neurodegeneration in AD, is linked to altered expression levels of the lncRNA beta-site APP cleaving enzyme1-antisense (BACE1-AS). Higher expression of BACE1-AS is observed in both APP mouse models and Alzheimer’s patients, suggesting its potential as a diagnostic biomarker for AD and a therapeutic target [132]. Knockdown of early B cell factor 3 antisense (EBF3-AS) was shown to inhibit apoptosis induced by Aβ25–35 and okadaic acid in SH-SY5Y human cells. This suggests that EBF3-AS could be a potential therapeutic target for AD treatment [133]. Small nucleolar RNA host gene 1 (SNHG1) expression was found to be upregulated in an in vitro model of AD cells. SNHG1 may contribute to cell damage by controlling the miR-361-3p/zinc-finger protein 217 (ZNF217) pathway, providing a potential conceptual framework for AD treatment [134]. The expression of lncRNA 51A was found to be upregulated in AD-affected brain tissues compared to control tissues. 51A, originating from the initial intron of the Sortilin-related receptor 1 (SORL1) gene (a recognized risk factor for AD), facilitates alternative splicing of SORL1, enhancing the formation of Aβ [135]. These findings highlight the diverse roles of lncRNAs in the pathogenesis of AD and their potential as diagnostic markers and therapeutic targets.
lncRNAs have been identified as potential diagnostic markers and therapeutic targets for PD, with aberrant expression detected in brain tissues, peripheral blood, and CSF of PD patients [136–138]. lncRNAs are widely expressed in the brain and play various roles in biological functions such as cell transcription, histone modification, and DNA methylation [139]. In the context of PD, significant changes in the expression of lncRNAs have been observed in the substantia nigra of PD patients [140]. In the substantia nigra of PD patients, non-coding RNA AL049437 was found to be upregulated, while lncRNA AK021630 was downregulated compared to normal tissues. Knockdown of AL049437 increased cell viability, mitochondrial mass, mitochondrial transmembrane potential, and tyrosine hydroxylase secretion in a cell model of parkinsonism, while AK021630 had the opposite effect. This suggests that lncRNA AK021630 might inhibit the occurrence of PD, while lncRNA AL049437 may contribute to the risk of PD [140]. lncRNA GAS5 has been shown to stimulate microglial inflammation by activating the NLRP3 pathway by sponging miR-223-3p in a PD mouse model [141]. Elevated levels of lncRNA HAGLROS have been found in both SH-SY5Y cells and a mouse model of PD. HAGLROS has been linked to the inhibition of cell death and autophagy through the stimulation of the phosphoinositide-3 kinase/protein kinase-B/mTOR (PI3K/Akt/mTOR) pathway and regulation of the miR-100/ATG10 pathway [142]. Suppressing lncRNA NEAT1 prevents 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) from inducing autophagy in vivo through regulating the PTEN-induced kinase 1 (PINK1) protein. Additionally, lncRNA LINC-PINT through RNA interference results in higher cell death in cultures exposed to oxidative stress, suggesting a potential neuroprotective role in PD pathogenesis [143]. Up-regulated lncRNA-p21 indirectly increases α-synuclein expression by sponging miR-1277-5p, inhibiting viability, and promoting apoptosis in SH-SY5Y cells. This indicates that lncRNA-p21 might be a novel therapeutic target in the context of PD [144]. lncRNA GAS5 has been linked with inflammatory effects in LPS-treated microglial cells and rotenone-treated PD mice, activating NLRP3 expression by sponging miR-223-3p, thus suggesting a role in the progression of PD [141, 145]. The major advantages of lncRNAs include their association with disease pathogenicity, diverse mechanisms of action, and tissue-specific expression, making them valuable for identifying specific neurological disorders [100, 146]. The exploration of lncRNAs in PD may provide insights into disease mechanisms and potential therapeutic strategies.
Early diagnosis of CNS disorders using cimiRNAs holds significant promise for providing patients with more specialized and efficient therapy choices [147, 148]. One particular miRNA family, the miR-29 family, consisting of miR-29a, miR-29b, and miR-29c, plays a crucial role in neuronal maturation and dendritic spine morphology [149, 150]. The downregulation of miR-29 has implications in aging and various neurological disorders, including AD [151], HD [152], and MS [153]. The expression of the miR-29 family, particularly miR-29a and miR-29c, is significantly downregulated in the blood serum of patients with PD compared to healthy controls [154]. Upregulation of miR-29a contributes to axonal outgrowth and neurological recovery through targeting PTEN/PI3K/Akt pathway after intracerebral hemorrhage insults in rats and cortical neurons [155]. A novel cimiRNA, cimiRNA-132, has been identified to regulate crucial pathways related to neuronal plasticity, including long-term potentiation and DNA methylation. It was found to be upregulated in the serum of AD patients, suggesting its potential as a biomarker for diagnosis [156]. Increased expression of miR-221-3p was found to activate the Akt/mTOR pathway, indicating a protective effect in PD. This miRNA could be considered a target for PD treatment [157]. The major advantages of miRNA biomarkers include their potential for use in multimarker models for accurate diagnosis, guided treatment, and monitoring the course of a disease. miRNA biomarkers offer high specificity and sensitivity, low baseline variability, and their non-invasive nature makes them cost-effective for use in personalized patient profiles, allowing for more specific therapeutic interventions [95]. cimiRNAs are stable in biofluids and their expression levels may not be significantly affected by basic characteristics like gender, age, and smoking status, making them reliable for monitoring and early diagnosis of diseases. Additionally, miRNAs can identify a relatively early response to toxic exposure, even in benign situations [158, 159]. In summary, the use of miRNAs in various diseases, including CNS disorders, holds significant potential as biomarkers due to their remarkable tissue specificity, early appearance, and stability in biological fluids.
Ng is a neuron-specific and post-synaptic protein expressed in granule-like structures in pyramidal cells of the hippocampus and cortex. It plays a crucial role in synaptic plasticity, synaptic regeneration, and long-term potentiation mediated by the calcium- and calmodulin-signaling pathway [160]. Ng influences these processes by regulating the level of calmodulin and the pattern of calcium/calmodulin-dependent postsynaptic signaling at dendritic spines [161]. As a calcium-binding protein, Ng can bind to calcium ions during neuronal activity, triggering conformational changes that modulate its interactions with other proteins [87, 162]. Several key points regarding Ng and its role in neurodegenerative diseases are highlighted.
The expression of Ng is reduced in the brains of individuals with neurodegenerative diseases such as AD, PD, HAND, and schizophrenia [44, 163, 164]. Ng levels significantly decrease in the cortex and hippocampus of AD patients and transgenic AD mouse models. Cognitive decline and Ng deficiency in a mouse model (5XFAD mice) were improved by increasing Ng expression through stereotaxic hippocampal injections of a lentivirus. Upregulation of Ng levels in the hippocampus of 5XFAD mice led to an increased postsynaptic density protein-95 (PSD-95), indicating potential therapeutic implications [165, 166].
CSF and blood levels of Ng are closely related to the occurrence and progression of AD, making it a potential biomarker for the diagnosis of AD and mild cognitive impairment (MCI) [167]. Ng has been associated with synaptic dysfunction in AD, and its expression is reduced after controlled cortical impact in rats, suggesting its potential as a reliable biomarker for synaptic dysfunction in AD [80]. Lack of Ng in mice results in a remarkable decline in hippocampus-dependent spatial memory and deficits in hippocampal long-term potentiation [168]. Increasing Ng levels has been shown to alleviate Aβ-induced impairments in synaptic function, including reductions in long-term potentiation and synaptic transport of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid glutamate receptors (AMPARs) in neurons expressing the APP [169]. In summary, Ng plays a crucial role in synaptic function, and its dysregulation is associated with various neurodegenerative diseases. Monitoring Ng levels in CSF and blood shows promise as a diagnostic biomarker for AD, and therapeutic strategies aimed at increasing Ng levels may have potential in mitigating cognitive decline associated with neurodegenerative disorders.
The involvement of Ng in synaptic function and the impact of inflammatory processes on Ng expression underscore its potential as a biomarker and therapeutic target in the context of neurocognitive disorders. The role of Ng in neurodegenerative diseases and conditions such as HAND is an area of active research. Neurocognitive impairment in individuals with HAND is associated with neuronal injury, synaptic disruption, and inhibition of long-term potentiation [160, 170, 171]. A prospective study indicated that CSF Ng levels were similar in both HIV-infected individuals and uninfected controls. This suggests that either synaptic injury may not play a significant role in HIV neuropathogenesis, or CSF Ng may not be sensitive to the synaptic impairment found in HIV-associated neurocognitive deficits [172]. Guha et al. [170] studied the expression of Ng in frontal cortex tissue from HIV-1 positive patients with and without HAND. The study confirmed a considerable decrease in Ng expression in frontal cortex tissues of HIV-1 individuals with neurocognitive disorders. This decrease was attributed to the overexpression of pro-inflammatory cytokines IL-1β and IL-8 [170].
The research on Ng in various neurodegenerative disorders, including AD, PD, Creutzfeldt-Jakob disease (CJD), and HAND, highlights its potential as a biomarker for these conditions. However, there are complexities and debates surrounding its specificity and diagnostic relevance. The cognitive decline is indicating that connection between Ng loss in synapses and age-related cognitive decline [173]. In PD, CSF Ng levels were found to be decreased compared to control, suggesting synaptic dysfunction in the parkinsonian disorder. Concentrations of Ng in CSF were reported to be increased in PD in a disease stage-specific manner and were associated with cognitive decline and the severity of motor symptoms [174]. In PD, CSF Ng levels were found to be decreased compared to control, suggesting synaptic dysfunction in the parkinsonian disorder. In CJD, CSF Ng levels were found to be higher in individuals with clinical CJD compared to healthy controls. This contradicts the reduction in Ng levels observed in AD and highlights the complexity of Ng’s role in different neurodegenerative conditions [175, 176]. In conclusion, while Ng shows promise as a biomarker for certain neurodegenerative disorders, including AD and PD, challenges remain in determining its specificity and diagnostic significance across diverse neurological conditions.
NFPs play a crucial role in providing structural support within neuronal axons, contributing to the proper functioning of the nervous system. They are composed of four subunits: neurofilament light (NFL-L), neurofilament heavy (NFL-H), neurofilament middle (NFL-M), and α-internexin or peripherin. NFPs confer structural support for neurons and are essential for the proper functioning of the nervous system [177, 178]. Abnormal accumulations of NFPs in the brain are observed in neurodegenerative disorders such as AD, PD, MS, and HD, potentially leading to neuronal injury and contributing to the progressive loss of nerve function in these conditions [164, 178, 179]. NFPs have emerged as promising blood biomarkers in neuroscience, offering new perspectives on various neurological disorders and accelerating clinical trials. They are characterized by a diameter of 10 nm and a length of several micrometers, constituting the neuronal cytoskeleton along with microfilaments and microtubules [180, 181].
Studies, such as that by Niemelä et al. [182], suggest that levels of fragmented CSF NFPs have a stronger correlation with disease development in HD compared to CSF tau levels. NFPs may offer superior tracking of HD disease progression in clinical trials [182]. In the context of persistent neuronal damage in HIV-infected patients, plasma NFP levels have been found to be highly associated with CSF NFP levels. This suggests that plasma NFP could serve as a feasible biomarker of CNS injury in HIV infection [183]. Studies, including one by Anderson et al. [184], indicate that higher concentrations of plasma NFP in HIV-infected adults are linked to poorer neuropsychological function. Additionally, NFP levels in the CSF may indicate the presence of neurocognitive impairment in HIV-positive individuals receiving antiretroviral therapy (ART).
In AD, abnormal processing of APP leads to the accumulation of Aβ peptides in the brain, forming plaques that are a hallmark pathological feature of the disease [185]. Aβ40 and Aβ42 naturally exist in the brain under normal physiological conditions, and their interaction is widely thought to play a crucial role in the development of AD. The ratio of Aβ40 to Aβ42 may be crucial for understanding the dynamics of Aβ deposition and its impact on neuronal function. An imbalance in the Aβ40/42 ratio could contribute to the formation of more toxic amyloid aggregates, ultimately leading to neuronal dysfunction and cognitive impairment in AD [186, 187].
Combining the Aβ42/40 ratio with other CSF biomarker profiles has greater predictive value for underlying AD in patients with MCI than does combining Aβ42 alone [188]. The CSF Aβ42/40 ratios were quantified using Elecsys immunoassay platforms, which predicted the Aβ status in all stages of AD with similar accuracy in a validation cohort [189]. A newly developed LC-MS/MS assay effectively determined the plasma Aβ42/40 concentration ratio, which serves as a reliable indicator of brain amyloid status. In addition, incorporating additional risk factors for amyloid pathology, such as age and Apolipoprotein E4 (APOE4) copy number, into the model improved the accuracy of identifying amyloid positivity compared with APOE and age alone [190]. Age, the APOE4 gene and white matter lesions may be associated with lower plasma levels of Aβ42/40. Individuals who have abnormal levels of plasma Aβ42/40 but normal Aβ positron emission tomography (PET) scans may be in the early stages of Aβ pathology and exhibit significant accumulation of Aβ in typical cortical regions associated with AD [191]. The presence of human APOE isoforms modifies the accumulation of Aβ in the brains of mouse models during aging. Brain Aβ42 levels were significantly lower in mice that expressed human APOE isoforms than in age-matched control mice. The accumulation of Aβ42 started at 5 months of age in mice with APOE4, whereas mice with APOE3 experienced a significant increase in Aβ42 levels at 21 months of age. At 6–7 months of age, the level of Aβ42 in the cerebroventricular fluid was greater in APOE3 mice than in APOE4 mice, indicating that APOE3 is more effective at removing Aβ42 than is APOE4 at this stage of development [192]. Another clinical study demonstrated that CSF Aβ42 levels and the Aβ42/40 ratio are decreased in APPNL-F knock-in mice, indicating the occurrence of early pathological events before amyloid deposition becomes widespread in the brain [193]. Patients with amyloid PET-positivity and PET-negativity could be clearly distinguished from one another using the Aβ42/40 ratio, indicating that the CSF Aβ42/40 ratio may be more precise for differentiating amyloid-positivity from amyloid-negativity than Aβ42 alone [194]. Furthermore, the Aβ42/40 ratio is considered to be more resilient than Aβ42 alone to a number of preanalytical factors that could produce false-positive results [195].
Early detection of AD is vital for prompt intervention, including disease-modifying therapy. Due to the high cost and invasiveness of obtaining CSF or amyloid PET, there is a demand for a safe and convenient screening method. The plasma Aβ42/40 ratio was measured by an antibody-free mass spectrometric method to determine the early stages of AD, which could facilitate clinical trials and preventive measures for AD [196]. A combination of plasma Aβ42/40 and p-tau217 discriminated Aβ status with relatively high accuracy, with area under the curve (AUC) values ranging from 0.83 to 0.86 in cognitively unimpaired (CU) individuals and AUCs ranging from 0.86 to 0.88 in patients with MCI. The combination of plasma Aβ42/40 and p-tau217 biomarkers holds promise for the early detection of amyloid positivity in preclinical and prodromal AD [197].
Cross-sectional and longitudinal analyses revealed that the Aβ42/40 plasma ratio was inversely associated with cortical Aβ burden. When used as a screening tool, TP42/40 had a positive predictive value of 81% for high cortical Aβ burden, which is 110% greater than the population incidence of cortical Aβ positivity [198]. In a more recent investigation employing the EUROIMMUN® ELISA, the ability of the plasma Aβ42/40 ratio to distinguish individuals with Aβ pathology assessed by CSF Aβ levels exhibited a decreased performance, with an AUC of 0.79. However, this performance was enhanced (AUC = 0.86) by incorporating a combination of the plasma Aβ42/40 ratio, p-tau217, NFL, and glial fibrillary acidic protein (GFAP) [199]. Hu et al. [200] reported good accuracy of the Aβ42/40 ratio using other analytical platforms. Another study revealed that a plasma Aβ42/40 ratio threshold of 0.089 yielded an AUC of 0.79, with a sensitivity of 85%, specificity of 63%, and positive and negative predictive values of 81% and 70%, respectively. Plasma concentrations of the Aβ1–40/Aβ1–42 ratio, measured using the single-molecule array (Simoa) immunoassay, predicted the brain Aβ positron emission tomography status in a large-scale longitudinal monocentric cohort (n = 276) of older individuals with subjective memory complaints [201]. A recent study demonstrated that plasma Aβ42/40 levels assessed using the fully automated high-sensitivity chemiluminescence enzyme (HISCL) immunoassay in a memory clinic memory cohort exhibited outstanding discriminatory capability for visually determined amyloid PET positivity, achieving an AUC of 0.949 [202].
Individuals with brain amyloidosis and normal cognition exhibited higher levels of CSF Aβ42 than did those with MCI or AD, suggesting that higher levels of CSF Aβ42 were associated with preserved cognitive function in individuals with brain amyloidosis [203]. Microglia treated with Aβ42 fibrils exhibited upregulated gene expression compared to those treated with Aβ40 fibrils. Specifically, 251 and 2,133 genes were upregulated in astrocytes and microglia treated with Aβ42 fibrils, respectively, while 191 and 251 genes were upregulated in astrocytes and microglia treated with Aβ40 fibrils, respectively. Compared with those in response to Aβ40 fibril treatment, the Aβ42 fibrils in glial cells generally stimulate similar but stronger amounts of fibrils [204].
However, the varying performance of the different Aβ assays and platforms across the studies could be at least in part due to the differences in the cohort characteristics (sample size, diagnostic groups, and outcome measures) and preanalytical sample handling. All these biases were minimized by a head-to-head comparison of Aβ assays in the same cohort of individuals with early AD from the Swedish BioFINDER study [205]. A meta-analysis involving thirteen studies on salivary biomarkers for AD revealed that patients with AD exhibited significantly greater salivary Aβ1–42 levels than did controls. However, there were no notable differences in salivary t-tau, p-tau, or acetylcholinesterase (AChE) levels between AD patients and controls. These findings suggest that salivary Aβ1–42 could be a sensitive biomarker for AD, but larger cohorts are necessary to confirm its diagnostic sensitivity and specificity [206]. Despite their diagnostic potential, several challenges remain in the clinical implementation of Aβ42/40 biomarkers, including standardization of measurement techniques, variability in sample collection and processing, and the influence of confounding factors such as age, sex, and genetic background. Future research efforts should focus on refining Aβ42/40 assays, validating their utility across diverse populations, and exploring their prognostic value in predicting disease progression and response to therapy.
In AD and other tauopathies, hyperphosphorylated tau, the formation of paired helical filaments, and neurofibrillary tangles are believed to be caused by pathological modifications in the tau protein that lead to conformational changes in the tau structure [207], a hallmark pathological feature of the disease. Tau proteins primarily reside in the axonal region of neurons and play a key role in maintaining the stability and organizing the assembly of microtubule proteins [208].
Tau is also found in the soma, dendrites, and astrocytes [209]. An increasing amount of evidence indicates that oxidative stress contributes to tauopathies and plays a crucial role in the hyperphosphorylation, polymerization, and toxicity of tau proteins [210].
CSF p-tau231 has been widely reported as a biomarker for detecting AD at both the MCI and dementia stages. CSF p-tau231 and p-tau217 predicted the presence of Aβ and tau to a similar extent. However, before p-tau231 reached abnormal levels, the earliest changes in Aβ levels were detected in specific parts of the brain, such as the medial orbitofrontal precuneus and posterior cingulate cortex, even before Aβ was detected globally via PET scans. Hence, CSF p-tau231 may be a potential candidate for detecting the initial stages of Aβ pathology and could be used in therapeutic trials for this disease [211]. Plasma p-tau, which has shown specificity for AD compared to other neurodegenerative diseases, will be highly valuable for guiding clinical diagnosis and determining eligibility for recently approved therapies [212]. Plasma p-tau231 and p-tau217 levels effectively revealed the earliest changes in cerebral Aβ levels in a preclinical population of patients with AD. Plasma p-tau231 may be a more suitable biomarker for clinical trials in middle-aged individuals with soluble Aβ changes [213]. Recent research indicates that the longitudinal measurement of plasma p-tau217 could reflect the connection between amyloid pathology and tau deposits [214], rendering it a viable biomarker for tracking the progression of both amyloid and tau pathologies in the disease. Elevated levels of plasma p-tau181 were observed in individuals who were positive for Aβ and who were experiencing cognitive impairment, as well as in carriers of the APOE4 carrier, and were notably linked to older age, poorer cognitive function, and higher levels of CSF-phosphorylated tau181 [215]. Furthermore, the correlations between plasma p-tau181 levels and decreased brain metabolism, particularly in individuals with cognitive impairment and Aβ positivity, suggest the utility of plasma p-tau181 as a simple, cost-effective, minimally invasive, and readily available means to evaluate both current and future metabolic dysfunction linked with AD compared with PET and magnetic resonance imaging (MRI) methods [215]. In individuals with cognitive impairment, plasma p-tau181 concentrations exhibited negative correlations with both gray and white matter volume as early as 12 months baseline, and neurodegeneration progressively increased over a maximum of a 4-year period [216].
In those with familial AD, plasma p-tau levels increase more than ten years before the patient becomes symptomatic. Plasma p-tau181 increased 16 years before the onset of cognitive impairment, but plasma p-tau217 significantly increased approximately 20 years before the expected year of MCI onset [217, 218]. A recent finding revealed that elevated levels of plasma p-tau217 correlated with elevated tau PET signals in the entorhinal cortex among individuals initially exhibiting normal tau PET scans. Longitudinal increases in the p-tau217 marker have been linked to deteriorated cognition and brain atrophy [214]. The blood p-tau181 assay detected the early stage of AD with high accuracy, with stepwise increases throughout the disease continuum. The blood p-tau181 assay was found to be specific to AD, distinguishing it from other neurological disorders with outstanding accuracy compared with mid-region CSF p-tau181. Furthermore, during a one-year period, blood p-tau181 levels predict hippocampal atrophy and cognitive impairment, suggesting that it is an appropriate marker of the course of AD [219]. Similarly, plasma p-tau181 is linked to cognitive decline, hippocampal atrophy, temporal cortical thinning, and cortical hypometabolism in individuals with a positive amyloid PET scan [220]. A study by Suárez-Calvet et al. [221] revealed that CSF levels of p-tau181, p-tau217, and p-tau231 in the ALFA+ cohort increased notably early in the preclinical stage of AD when only minimal Aβ pathology was detected, and these three tau proteins could accurately distinguish between Aβ-positive and Aβ-negative cognitively normal individuals. In addition, Schindler et al. [222] recently showed the predictive accuracy of plasma p-tau231 and p-tau181 for identifying abnormal Aβ-PET and CSF Aβ42/40 results in participants who represented the two racial groups studied. These studies examined non-Hispanic White and African-American pairs of older adults with the same demographic characteristics (age, sex, cognition, and APOE ε4 genotype). In brief, the p-tau holds promise as a biomarker to monitor drug activity of disease modifying treatments in AD.
In the realm of CNS biomarker research, several challenges are anticipated in the future. A primary obstacle is the development of biomarkers with heightened sensitivity and specificity. Many existing biomarkers exhibit limitations in these aspects, posing difficulties in accurately diagnosing and monitoring certain CNS disorders. The intricate and multifaceted nature of CNS disorders adds to the complexity, making it challenging to pinpoint and validate biomarkers that genuinely encapsulate the underlying pathophysiology of these conditions. Moreover, addressing the heterogeneity of CNS disorders proves to be another intricate challenge, as it is difficult to identify biomarkers that can be consistently applied across various subtypes of the disorder. The diverse manifestations within CNS disorders necessitate a nuanced understanding for effective biomarker development and application. To overcome these challenges, longitudinal studies and the integration of big data will be indispensable in validating biomarkers and assessing their utility in the clinical setting. These approaches will enable researchers to gather comprehensive insights into the dynamic nature of CNS disorders and enhance the reliability and applicability of identified biomarkers [180, 223].
While numerous biomarkers have emerged from preclinical studies, their validation in larger and more diverse patient populations is a critical next step. Transitioning from preclinical validation to clinically useful tests presents a significant challenge, demanding the development of assays that are not only sensitive and specific but also user-friendly [31, 224]. Reproducibility and standardization emerge as major stumbling blocks in the field, with many RNA signatures entering clinical trials but failing to progress to practical clinical applications. The development and validation of miRNA biomarkers can be financially burdensome, necessitating a careful consideration of costs versus benefits to justify their implementation in terms of improved diagnosis and treatment of diseases [225]. In addition to financial considerations, the expansive data generated in miRNA biomarker research poses challenges related to data sharing and collaboration. Balancing the imperative for collaborative efforts with concerns surrounding patient privacy and data security is essential for maximizing the potential of data-driven biomarker discovery. Overcoming these challenges is pivotal for translating promising preclinical findings into clinically relevant applications and advancing the field of miRNA biomarker research [226].
The field of image-based biomarkers is rapidly advancing, driven by recent technological breakthroughs. Various imaging techniques are currently being employed to gather valuable insights into biological processes and disease states. These include different imaging techniques are currently being utilized such as functional MRI (fMRI), quantitative MRI (qMRI), PET, and single photon emission computed tomography (SPECT) [8, 227, 228]. The utilization of these advanced imaging techniques offers a non-invasive and highly detailed view of biological structures and functions. These image-based biomarkers have the potential to enhance early detection, diagnosis, and monitoring of various diseases, providing valuable information for personalized medicine and treatment planning. Continued technological advancements in imaging modalities are expected to further broaden the scope and impact of image-based biomarkers in medical research and clinical practice [229, 230]. Addressing these challenges requires careful attention to standardizing imaging protocols, implementing correction techniques, and employing advanced imaging technologies that minimize the impact of patient-related factors. Quality control measures and standardized imaging procedures can also help enhance the reliability and reproducibility of imaging biomarkers. As technology advances, efforts to develop imaging techniques that minimize patient discomfort, reduce radiation exposure, and account for potential sources of variability [227, 231, 232]. Additionally, ongoing research and improvements in imaging protocols aim to mitigate the impact of these factors, ensuring that imaging biomarkers remain a valuable tool in medical diagnosis and research.
Tissue biopsies in the CNS are typically acquired through neurosurgical removal or stereotactic tissue biopsy procedures. While these methods are valuable for diagnosing various CNS conditions, they come with inherent risks and limitations due to their invasive nature [233]. Brain biopsy, for example, involves collecting small pieces of brain tissue for microscopic examination to diagnose abnormalities such as tumors, inflammation, AD, infections, and other brain disorders. However, this approach poses several challenges [234]. Neurosurgical removal or stereotactic tissue biopsy is inherently invasive, carrying risks for the patient. This invasiveness can result in pain, discomfort, and a small but notable risk of complications such as bleeding or infection. This method is only feasible when the targeted tissue is accessible. In the case of certain diseases or specific regions of the body, especially within the CNS, obtaining a biopsy may be challenging or impractical. The amount of tissue that can be obtained through biopsy may be restricted, impacting the ability to conduct multiple tests or replicate results. This limitation is particularly relevant in cases where larger samples are required for comprehensive analysis. The process of obtaining and isolating tissue for analysis can be costly and time-consuming. This makes it difficult to perform multiple tests efficiently and may hinder the rapid replication of results.
Given these challenges, the development and utilization of non-invasive or minimally invasive diagnostic methods, such as imaging and liquid biopsy techniques, are gaining prominence. These approaches aim to provide valuable diagnostic information while minimizing the risks and limitations associated with traditional tissue biopsies, especially in the sensitive and critical context of the CNS [235, 236].
Stereotactic brain biopsy is indeed a minimally invasive procedure designed to extract tissue samples from specific areas within the brain suspected of harboring infections or tumors. This procedure involves the utilization of a stereotactic device, which employs a coordinate-based navigation system integrated with the patient’s brain images, typically obtained through MRI or computed tomography (CT) [237, 238]. This technique has become a valuable diagnostic tool, providing important information for treatment planning while minimizing the impact on the patient.
The ethical considerations surrounding tissue sample collection, particularly in the context of neurodegenerative disease research, are indeed critical and multifaceted [239]. Proper storage and handling of biopsy samples are vital for maintaining the integrity of biomarkers. Challenges arise in preventing contamination or degradation of samples, especially during long-term storage. Robust protocols for sample preservation, including temperature control and adequate labeling, are crucial to ensure the reliability of biomarker analyses [240, 241]. At present, the development of economically viable biomarkers for widespread use in the early diagnosis of neurodegenerative diseases is a persistent challenge [13, 25, 242]. While many CNS biomarkers show promise in preclinical studies, transitioning them to clinical practice remains a significant hurdle. Biomarkers for disease prognosis, treatment, and outcome assessment often undergo extensive preclinical evaluation, but their translation into practical clinical applications requires rigorous validation and approval processes [243].
A mounting body of evidence supports the application of biomarkers in diagnosing and treating CNS disorders. Nevertheless, additional research is imperative to comprehensively grasp the clinical efficacy of these markers and to pinpoint novel biomarkers. It is pivotal to underscore that the biomarker discovery process should unfold concurrently with clinical validation. Specifically, the validation of biomarkers ought to transpire in both retrospective and prospective manners, utilizing independent sample cohorts, ideally procured by multiple hospitals. Exploring and elucidating the regulatory mechanisms of miRNAs, lncRNAs, and cimiRNAs in neurodegenerative disorders is essential to enhance the applicability and accuracy of these molecules as biomarkers for clinical diagnoses. To augment the sensitivity and specificity of protein detection in biological samples, advancements in testing methods, such as mass spectrometry and immunoassays, are imperative. Additionally, there remains much to unravel about the dynamics of protein levels in the body, encompassing genetic and environmental factors, and how these levels evolve over time in both healthy individuals and those with neurological disorders. In conclusion, the integration of biomarkers into CNS research holds the potential to revolutionize the field by enhancing diagnostic precision, predicting disease progression and treatment response, ultimately culminating in the development of more effective therapies.
5XFAD: five familial Alzheimer’s disease
AD: Alzheimer’s disease
Akt: protein kinase-B
APOE4: Apolipoprotein E4
APP: amyloid precursor protein
AUC: area under the curve
Aβ: amyloid-beta
cimiRNAs: circulating microRNAs
CJD: Creutzfeldt-Jakob disease
CNS: central nervous system
CSF: cerebrospinal fluid
ELISA: enzyme-linked immunosorbent assay
HAND: HIV-associated neurocognitive disorders
HD: Huntington’s disease
lncRNAs: long noncoding RNAs
MCI: mild cognitive impairment
miRNA: microRNA
MRI: magnetic resonance imaging
MS: multiple sclerosis
mTOR: mammalian target of rapamycin
NFPs: neurofilament proteins
Ng: neurogranin
NGS: next-generation sequencing
PD: Parkinson’s disease
PET: positron emission tomography
PLWH: people living with HIV
qRT-PCR: quantitative reverse transcription-polymerase chain reaction
Tat: transactivator of transcription
VA: Conceptualization, Writing—original draft, Writing—review & editing. SS: Writing—original draft, Writing—review & editing. Both authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Fatemeh Afrashteh, Rayan Rajabi
Amanda C. Y. Chan ... Joy Vijayan
Grace YY Chia ... Benjamin YQ Tan
