Affiliation:
1Department of Pharmacology, Jiangsu Provincial Key Laboratory of Critical Care Medicine, School of Medicine, Southeast University, Nanjing 210009, Jiangsu, China
†These authors share the first authorship.
Affiliation:
1Department of Pharmacology, Jiangsu Provincial Key Laboratory of Critical Care Medicine, School of Medicine, Southeast University, Nanjing 210009, Jiangsu, China
†These authors share the first authorship.
Affiliation:
1Department of Pharmacology, Jiangsu Provincial Key Laboratory of Critical Care Medicine, School of Medicine, Southeast University, Nanjing 210009, Jiangsu, China
ORCID: https://orcid.org/0000-0001-8170-498X
Affiliation:
1Department of Pharmacology, Jiangsu Provincial Key Laboratory of Critical Care Medicine, School of Medicine, Southeast University, Nanjing 210009, Jiangsu, China
2Co-innovation Center of Neuroregeneration, Nantong University, Nantong 226019, Jiangsu, China
3Institute of Life Sciences, Key Laboratory of Developmental Genes and Human Disease, Southeast University, Nanjing 210009, Jiangsu, China
Email: yaohh@seu.edu.cn
ORCID: https://orcid.org/0000-0002-2128-0529
Explor Med. 2023;4:471–486 DOI: https://doi.org/10.37349/emed.2023.00157
Received: December 10, 2022 Accepted: March 08, 2023 Published: August 31, 2023
Academic Editor: Jun Ren, Fudan University Zhongshan Hospital, China
Stroke, a central nervous system (CNS) injury, is responsible for the second leading cause of death in the world, bringing a great burden on the world. Stroke is normally divided into ischemic and hemorrhagic stroke, among which ischemic stroke takes up 87% proportion. Accumulating evidence has denoted a rather pivotal role for autophagy in the pathogenesis of ischemic stroke, which is activated in neuronal cells, glial cells, and endothelial cells. Besides, circular RNAs (circRNAs), a novel type of epigenetic regulation, are highly expressed in the CNS and are involved in the process of CNS diseases, which is regarded as an important molecular mechanism in ischemic stroke. Meanwhile, circRNA and autophagy have a significant correlation. The intracellular signaling pathways regulating autophagy can either restrain or activate autophagy. However, under the circumstances of ischemic stroke, the precise communication between circRNA and stroke is largely unknown. This review aims to provide a summary of the relationship between circRNA, autophagy, and ischemic stroke, as well as the current research advancements in understanding how circRNA regulates autophagy in the context of stroke.
Globally, stroke is responsible for roughly 6.5 million deaths and more than 10% of all mortalities each year, which is a leading cause of disability and death and can be pathologically classified into ischemic stroke and hemorrhagic stroke [1, 2]. Ischemic stroke is caused by the infarction of cerebral vessels due to thromboembolism and thrombus formation and takes up 87% proportion of all stroke cases [3]. The occlusion of the cerebral artery leads to a deficiency of oxygen, glucose, and lipids, causing the necrosis of brain parenchyma with cascade responses, such as neuroinflammation, excitotoxicity, oxidative stress, and autophagy [4]. The word “autophagy” first appeared in the 1960s, which originated from the Greek roots “auto” (self) and “phagy” (eating). Just take it literally, cells can eat their cytoplasmic components and organelles by autophagy, referred to as a common cellular catabolic process nowadays [5].
Autophagy is a strictly controlled multi-stage and highly dynamic process. Phagophore corresponds to the autophagic compartment, which a called pre-autophagosomal [6]. Then the double-membrane autophagosome is generated by the isolation of the phagophore membrane [7]. The outer autophagosomal membrane fuses with the simple-membrane lysosome, turning into an autolysosome to degrade useless substances and release macromolecules to the cytoplasm [8]. Similar to other cellular pathways, it can be modulated positively or negatively by variable regulatory constituents [9]. Based on the mode of cargo delivery to the lysosomal lumen and physiological functions, three different kinds of autophagy have been described, namely microautophagy, chaperone-mediated autophagy (CMA), and macroautophagy [5]. Macroautophagy, a non-selective process, forms a double-membrane compartment sequestering cytoplasmic material or organelles and maturing into autophagosomes [10]. The mixture of autophagosome and lysosome yield autolysosome, in which cargo is degraded. The resulting production is released to the cytoplasm and reutilized [11]. By contrast, microautophagy and CMA do not form autophagosomes like macroautophagy, in which redundant cytoplasmic components directly contact with lysosomes and are consumed. What the difference between the two processes is that the entry of substrates into lysosomes relies on specialized cytoplasmic proteins to bind with the receptor protein of the lysosomal membrane without which redundant contents accumulate in the cytoplasm and cause cell dysfunction. Thus, the protein is referred to as the chaperone of cell autophagy, and the process of CMA is regarded as the only process to allow selective degradation. However, microautophagy is also a non-selective autophagy process in which components directly invaginate into lysosomes through membranes. It can maintain and regulate membrane homeostasis and lysosome size via degradation of long-live and membrane proteins [12]. Selective and non-selective autophagy are evolutionarily necessary and occur in parallel to jointly regulate body homeostasis [13]. In terms of studying autophagy, the process of macroautophagy is relatively the main and best way, so this review will refer to “macroautophagy” as autophagy in the following text [5].
Current evidence suggests that autophagy is a double-edged sword. As a basic part of cellular catabolism, autophagy provides and preserves energy balance by trying to promote survival through degrading protein aggregates, damaging mitochondria, and recycling amino acids, fatty acids, and glucose when the body encounters a variety of stressful conditions such as starvation, proteotoxicity, and organelle damage [14]. However, excessively induced autophagy may mediate cell death or apoptosis. On the one hand, physiological levels of basal autophagy are essential for the promotion of cytoprotection and survival. On the other hand, aberrant-induced autophagy can lead to various neurodegenerative diseases and cancers [11].
As a strictly multi-step process, almost all the possible regulatory mechanisms can control the completion of autophagy at transcriptional, post-transcriptional, and epigenetic levels [15]. According to recent studies, epigenetic disorder and subsequently inappropriate autophagy represent critical factors in the pathogenesis of related human diseases, including DNA methylation-related enzymes, histone-modifying enzymes, and non-coding RNAs [16].
Circular RNAs (circRNAs), a novel type of non-coding RNAs with covalently closed-loop structures, have drawn abundant interest over the past decade. Owing to the lack of 5’–3’ polarity or a polyadenylated tail, circRNAs are more stable than their linear counterparts and cannot be digested by ribonucleases (RNases), which are a suitable molecular target against various diseases [17]. In recent years, circRNAs have been identified in various species with the development of high throughput sequencing for non-coding RNA transcripts [18]. Whereafter, many researchers have explored the function of circRNAs in recent years. A handful of circRNAs have been confirmed to be involved in post-transcriptional regulation as microRNA (miRNA) sponges or competing endogenous RNAs (ceRNAs) [19, 20]. What’s more, some circRNAs have been shown to “sponge” other factors, like RNA binding protein, influencing the assembly of protein substructure [20]. Although circRNA is widely considered a type of non-coding RNA, increasing evidence has been indicated that at least some of them have translational functions [21]. Based on the unique tertiary structure, circRNAs can capture or sequester RNAs or proteins and release them in subcellular locations to mediate autophagy regulation [22].
Numerous lines of research suggest that autophagy plays a critical role in maintaining the homeostasis of the intracellular environment and dysregulated autophagy is involved in the pathogenesis changes of various diseases [13]. For example, autophagy can remove the accumulation of abnormal protein condensates or aggregates, which is hallmarks of neurodegenerative disease. Hence, deficits of autophagy may lead to neurodegenerative disease, and it has been testified that mutation of core autophagy-related proteins results in some neurodegenerative diseases, such as Alzheimer’s disease, Huntington’s disease, and Parkinson’s disease [23]. In neurodevelopment diseases, due to that autophagy plays a significant role in cellular processes in the developing brain, defects in core autophagy pathways components cause a number of disorders like sharing developmental delay [24]. Furthermore, autophagy-related signaling pathways have been indicated to be activated and autophagy activity regulation can influence the outcomes of ischemic stroke [25]. Nevertheless, whether autophagy is a friend or a foe of ischemic stroke is still under controversy [26]. During ischemic stroke, almost all types of cells are involved, which include neuron, glial, and brain microvascular cells, introducing a concept of neurovascular. What’s more, recent evidence has shown that autophagy is activated in those types of cells upon ischemic stroke [14]. Here, the protective and detrimental effects of autophagy in various cells will be discussed in the next sections.
Presently, the exact role of neuronal autophagy is still unclear when neuronal autophagy is induced by various intrinsic and extrinsic insults including ischemia/reperfusion (I/R), nutrient deficiencies, neurotoxins excitotoxic stimuli, and inflammation [13, 27, 28]. On the one hand, under these stressful conditions, mild to moderate induction of autophagy can maintain neuronal homeostasis, clear the aggregated protein and damaged mitochondria, preserve the energy balance, and alleviate the endoplasmic reticulum stress, referred to as adaptive autophagy [29]. On the other hand, excessive increases in autophagic activity might lead to enhanced degradation of essential cellular components and unrestrained induction of autophagy may be a causative factor for cell death, although the duration and extent of autophagy for cell death induction remain to be explored in depth [30, 31]. This unchecked autophagy is termed maladaptive autophagy.
Maladaptive autophagy in neurons is often triggered upon ischemic stroke and causes neuronal death [32, 33]. Many researches demonstrate that inhibiting neuronal autophagy progress significantly reduces the brain infarct volume in rat middle cerebral artery occlusion (MCAO) models. In addition, the blockade of neuronal autophagic cell death makes for the neuroprotective action of many compounds, including amlodipine, atorvastatin, lithium, N-acetyl-serotonin, and so on [34].
On the contrary, there are also many studies showing the association between autophagy and neuroprotection. Inhibition of autophagy by 3-methyladenine (3-MA), a specific inhibitor of autophagic/lysosomal protein degradation, reversed the neuroprotection of tunicamycin and thapsigargin, and enhance the I/R-induced release of cytochrome c and the downstream activation of apoptosis in neurons in oxygen glucose deprivation (OGD) and MCAO model [35].
Furthermore, many researches have supported that autophagy flux plays an important role in determining the neuroprotection or destruction aspect of autophagy. Autophagic responses and autophagy flux can be impaired in beclin 1 heterozygous mice compared to wild mice, which significantly lowered neuronal survivability and degraded neurological recovery score. Then, using rapamycin to enhance autophagy and autophagy flux on both the wild type and beclin 1 heterozygous mice. However, beclin 1 heterozygous mice did not produce any change in autophagy flux and neuronal survivability compared to untreated mice. Hence, disruption of autophagy and autophagy flux pathways is detrimental to the brain, and increasing autophagy is neuroprotective when autophagy flux is undisturbed [36, 37].
Compared to numerous studies around macroautophagy and stroke, the effect and mechanism of microautophagy and CMA in stroke are unknown, which is limited by immature technology and unidentified receptors. However, a growing body of evidence shows that CMA and microautophagy are crucial for neuronal proteostasis and the development of neurodegenerative diseases [38, 39].
Glial cells, including astrocyte, microglial, and oligodendrocyte, are the most abundant and widely distributed cells in the central nervous system (CNS), and activated glial cell autophagy has been well-proved to play critical roles in the regulation of neurological functions under stroke circumstances [40, 41].
Accumulating reports have proven that among all the cell types that can be targeted, astrocytes are undoubtedly the most promising [42], which may regulate synaptic transmission [43]. And, synaptic transmission is closely associated with neuron plasticity, playing an important part in the prognosis of patients with ischemic stroke. Meanwhile, astrocyte autophagy is common and can be divided into basal autophagy and induced autophagy under physiological and stressful conditions [44]. In the early times of ischemic stroke, activated autophagy is detrimental to astrocytes. Ischemic astrocytes in brain tissue can observe the accumulation of autophagy-like vacuoles containing electron-dense material. OGD could induce astrocyte autophagy and remarkably elevate the expression of early autophagy markers beclin 1 and light chain (LC) 3-II/LC3-I ratio. Inhibition of autophagy with 3-MA and bafilomycin AI (Baf-AI) mildly but significantly attenuated the death of damaged astrocytes under ischemic insult [45]. Said another report [46], the circular RNA HECTD1 (circHECTD1) significantly increased in ischemic brain tissue, and the knockdown of circHECTD1 restrained astrocyte autophagy and ameliorated neuronal damage. From another aspect, autophagy reportedly has a protective effect on astrocytes after ischemia and hypoxia. In a recent study, Kasprowska et al. [47] devoted to investigating whether autophagy activation is beneficial or harmful to astrocytes. After short-term OGD exposure (1, 4 h), compared with control cells, 3-MA-treated cells showed a higher level of cleaved caspase-3, showing the promotion of cell apoptosis. Nevertheless, exposed to OGD such 8 h or 24 h, astrocytes autophagy treated by 3-MA were significantly downregulated, thereby causing time-dependent inhibition of cleaved caspase-8 and tumor necrosis factor-α (TNF-α) related extrinsic apoptotic pathway and intrinsic apoptotic pathway (cleaved caspase-9) [47].
In the context of injury ischemia, studies on microglia and oligodendrocytes are limited. Microglial, the first immune defense of the CNS, is equivalent to a peripheral macrophage, involved in mediating neuroinflammation after ischemic stroke [48]. Yang et al. [49] took focal cerebral ischemia model by permanent MCAO in mice to mainly observe microglial autophagy and inflammation response in vivo. When using 3-MA (autophagy inhibitor), in addition to reduced microglial autophagy and inflammation, they also found significant reductions in infarct size, edema formation, and neurological deficits. Besides, Xia et al. [50] found an interesting phenomenon that the intensity of microglial autophagy was dependent on the time of ischemia. Activated microglia show a wide range of different functional states, mainly including pro-inflammatory (M1) and anti-inflammatory (M2) states. Moreover, inhibition of autophagic flux activated the NF-κB pathway, which is the main regulator of M1 microglial phenotype, and decreased the activity of cyclic adenosine monophosphate (AMP) response element-binding protein (CREB) participating in the polarization of M2 macrophage to modulate microglial phenotype between M1 and M2 phases.
The interaction of microglial and astrocytes jointly participates in the formation of glial scar which is the barrier for axonal regrowth and functional recovery in the late times of ischemic stroke [51], in which astrocytes are activated [52]. Wu et al. [53] reported that fused in sarcoma induced excessive autophagy and activated astrocytes which was a new target for improving the outcome after stroke. It is speculated that activated astrocytes autophagy exists which increase after stroke, nonetheless, researches on pathological change in stroke are more prevailing. If the hypothesis is true, autophagy facilitates axonal regrowth and attenuates glial scar. Transforming growth factor-beta inhibited autophagy pathway to induce the secretion of chondroitin sulfate proteoglycans of astrocytes which is responsible for glial scar [54].
Oligodendrocytes are the only cells capable of forming myelin in the CNS, which are abundant in both gray and white matter of the brain and spinal cord, especially white matter. In many clinical hypoxic-ischemic insults, white matter is the priority to be damaged. Similarly, making up the white matter myelin sheath-oligodendrocytes are also vulnerable to hypoxic-ischemic insults, so oligodendrocytes have more possibility to be explored [55]. Currently, the link between oligodendrocyte autophagy and neurodegenerative disease or spinal cord injury-related demyelination has been clarified. However, the role of oligodendrocyte autophagy in cerebral ischemia is under investigation, which is more sensitive and vulnerable to ischemic insults [56]. And hopefully, more knowledge of different aspects of oligodendrocyte autophagy will be shown in the future.
Since then, accumulating evidence has proven that autophagy regulation plays a dramatically vital role in neurodegenerative diseases and ischemic stroke. Meanwhile, endothelial cells, the major component of the blood-brain barrier (BBB), modulate themselves to maintain homeostasis by regulating the vascular tone and permeability [57]. And, ischemic stroke in the early stage is often accompanied by the disruption of BBB, and the inflammatory factors enter the brain parenchyma through BBB, which are doomed to cause irreversible damage to neurons. Above all, endothelial autophagy has the clinical potential to be a therapeutic target of ischemic stroke.
Anatomically, BBB is composed of self-fusion of brain microvascular endothelial cells (BMECs) with tight junctions (TJs) and the interaction of other components, including astrocytes, pericytes, and perivascular microglial [58]. In the pathological process of ischemic stroke, the interruption of blood flow in the brain leads to the inevitable formation of BMECs in the brain microenvironment [59]. Finally, TJ protein levels are downregulated, and BBB loses its protective function. Claudin 5 is specially involved in TJ proteins and participates in forming the backbone of TJ chain [60]. Under damaged circumstances, endothelial cells autophagy is activated to alleviate the hypoxia-induced BBB injury via regulating the redistribution and degradation of claudin 5 [6]. Induced by I/R or OGD/re-oxygenation (OGD/R), long noncoding RNAs (lncRNAs) metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is one of the highly upregulated endothelial lncRNA and acts as a protective role in BMECs against cerebral ischemic insults. Similar to circRNA, lncRNA MALAT1 served as an endogenous sponge with miR-26b and downregulated the expression of miR-26b, which inhibited BMEC autophagy and survival [61].
Here, this review will give more emphasis on specific autophagy, given the significance for neuropathies, cancer, and heart disease. In ischemia, mitochondria loss the ability to produce adenosine triphosphate for energy due to the lack of nutrients and oxygen, leading to adenosine triphosphate-dependent Ca2+ channel injury and Ca2+ load of cells and mitochondria [62]. Under the circumstances, mitochondrial autophagy, termed mitophagy, is emerging to clear out dysfunction or excessive mitochondria producing reactive oxygen species and inducing cell death to control the quality and quantity of mitochondria [63]. Mitophagy, a selective catabolism mechanism [64], modulates dynamic the process of mitochondria involved with fission and fusion and completes mitochondrial turnover in stroke which has the potential to achieve pre-diagnosis and prognosis of stroke and realizes clinical value [15]. Besides, owing to the disturbance of Ca2+ balance, the endoplasmic reticulum stress is triggered which refers to the homeostasis disorder and dysfunction of the endoplasmic reticulum and has tight relation with autophagy [56]. Autophagy inhibitor 3-MA used in vivo and in vitro, models tended to aggravate the endoplasmic reticulum stress and increased the level of caspase-12 and caspase-3 that induced apoptosis [65]. And the mechanism of some CNS protectants is inhibiting the endoplasmic reticulum stress level and enhancing autophagy to perform neuroprotective function [66]. Peroxisomes are selectively eliminated through autophagy in yeast mediated by PpAtg30, a Pichia pastoris protein. It involves the formation of pexophagy intermediates and mediates the selection of peroxisomes, which are delivered to the autophagy machinery for pexophagy [67]. Mature ribosomes are also selectively degraded via autophagy with the help of ubiquitin protease [68]. Compared with yeast, this specialized autophagy in mammalian systems is under-studied.
In different stages of stroke including hyperacute, acute, subacute, and chronic phase, activated autophagy plays different roles during the development of stroke. It has demonstrated that induction of adenosine 3’,5’-monophosphate (cyclic AMP)-activated protein kinase-dependent autophagy is involved in the ischemic preconditioning-inducing neuroprotection [69]. What’s more, in the acute stage, activation of autophagy with ataxia-telangiectasia mutated (ATM)/cell cycle checkpoint kinase 2 (CHK2)/beclin 1 axis can relieve oxidative stress to protect neurons against injury [70]. Hence, several studies suggest that activation of autophagy at the early or before stage of ischemic stroke is neuroprotective [14]. During the acute and subacute stages, activated autophagy tends to play a detrimental role, enlarging the ischemia area and promoting cell apoptosis. At 24 h after ischemia, Nogo-A/Nogo-66 receptor 1, an autophagy inhibitor, significantly reduced ipsilateral thalamus neuronal loss and gliosis [71]. In the chronic stage, autophagy is activated excessively, which is possible to bring secondary thalamic damage to influence functional recovery. The role of autophagy in different disease stages of stroke has summarized in Figure 1.
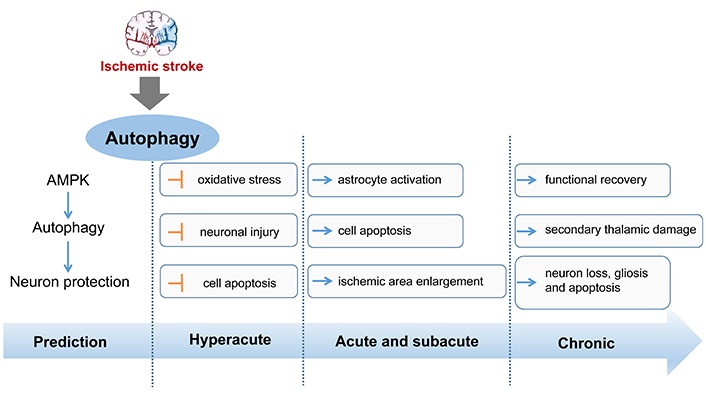
The role of autophagy in different stages of stroke. AMPK: AMP-activated protein kinase. Blue arrow represents the expression of up-regulation and the yellow symbol represents the expression of down-regulation
Recently, increasing evidence has shown that there is a significant correlation between circRNAs and autophagy. Autophagy is fundamental to cellular homeostasis under both normal and stressed conditions. When the extracellular and intracellular environments are stable and cellular homeostasis is maintained, autophagy signals of the cell are inclined to be inhibited.
The autophagy-associated pathways are complex and diverse, where circRNAs can activate or inhibit autophagy by affecting the key molecules in the process [72]. Based on their 3-dimensional covalent structure, circRNA effectively binds to signaling molecules, including autophagy-related (ATG) proteins and miRNAs to regulate autophagy [73]. Potentially, circRNAs are cleaved by autophagic degradation and also regulate autophagy in turn. For example, the circular RNA CDR1-AS (CDR1-AS) plays a role as a sponge for miR-7. CDR1-AS contains more than 70 selectively conserved miR-7 target sites. However, according to inhibit epidermal growth factor receptor (EFGR) expression, miR-7 can suppress cell viability and induce autophagy. Therefore, as a natural miR-7 sponge, CDR1-AS may regulate autophagy indirectly by perturbing the miR-7 function [74]. Several studies have suggested that circRNAs can regulate autophagy in different cell types, including cancer cells, neuronal cells, and liver cells [75–77]. These circRNAs can act as ceRNAs to sponge miRNAs, which in turn regulate the expression of autophagy-related genes. Moreover, some circRNAs can interact directly with autophagy-related proteins to modulate the autophagy process [78, 79]. Therefore, it is plausible that circRNAs may also regulate autophagy in the context of ischemic stroke. A summary of the functions of different autophagy-regulated circRNAs in various diseases has been represented in Table 1.
A summary of the functions of autophagy-related circRNAs
| circRNAs | Expression | Regulation targets and pathways | Influence on autophagy | Diseases | Refs |
|---|---|---|---|---|---|
| circCDYL | Up | circCDYL/miR-1275/ATG7/ULK1 axis | Enhancement | Breast cancer | [76] |
| circCUL2 | Down | circCUL2/miR-142-3p/ROCK2 axis | Enhancement | Gastric cancer | [80] |
| circHIPK3 | Down | circHIPK3/miR-124-3p/STAT3/PRKAA axis | Enhancement | Lung cancer | [81] |
| circHECTD1 | Up | circHECTD1/miR-142/TIPARP axis | Enhancement | Cerebral ischemic stroke | [46] |
| circSHOC2 | Up | circSHOC2/miR-7670-3p/SIRT1 axis | Suppression | Cerebral ischemic stroke | [75] |
| circ_002581 | Up | circ_002581/miR-122/CPEB1 axis | Suppression | Nonalcoholic steatohepatitis | [82] |
circCDYL: circular RNA CDYL; ULK1: UNC-51-like kinase 1; circCUL2: circular RNA CUL2; ROCK2: rho-associated coiled-coil-containing protein kinase-2; circHIPK3: circular RNA HIPK3; STAT3: signal transducer and activator of transcription 3; PRKAA: protein kinase AMP-activated catalytic subunit alpha 2; TIPARP: TCDD inducible poly[ADP-ribose] polymerase; circSHOC2: circular RNA SHOC2; SIRT1: sirtuin 1; CPEB1: cytoplasmic polyadenylation element-binding 1
Many studies have demonstrated that circRNAs microarrays or sequencing results relate to ischemic stroke with different techniques such as quantitative real-time polymerase chain reaction (qRT-PCR), gene ontology, and Kyoto Encyclopedia of Genes and Genomes analyses [83]. In the human sample, ischemic stroke affects the expression level of circRNAs in serum and plasma. For example, the ratio of serum circR-284: miR-221 was significantly elevated in acutely symptomatic patients of ischemic stroke within 5 days (P = 0.0002), which exhibited favorable characteristics as a biomarker indicative of carotid plaque rupture and stroke [84]. Ostolaza et al. [85] showed a set of differentially expressed circRNAs in peripheral blood from different stroke etiologic subtypes, which are useful as candidate biomarkers for stroke etiology. Furthermore, circular RNA FUNDC1 (circFUNDC1) was increased in ischemic stroke patients and its levels were correlated with baseline National Institutes of Health Stroke Scale (NIHSS) scores within 24 h and the 7th day. What’s more, survival Kaplan-Meier curves (P = 0.042) between ischemic stroke patients are significantly different with low (below cut-off) or high circFUNDC1 levels (above cut-off) after 18 months of follow-up, indicating that circFUNDC1 can serve as a diagnostic and prognostic biomarker [86].
Accumulating evidence shows that varieties of circRNA molecules can not only serve as biomarkers predicting stroke in advance but also play an important role in the mechanism and therapy of ischemic brain injury. Yang et al. [87] found that circular RNA SCMH1 (circSCMH1) levels were significantly decreased in patients’ plasma with acute ischemic stroke, inclined to become a new target for stroke treatment. To explore the specific function of circSCMH1 in the pathological process of stroke, circSCMH1 was precisely delivered to the brain via extracellular vesicle, realizing circSCMH1 overexpression and functional recovery in photothrombotic (PT) mice. The underlying cellular mechanism of circSCMH1 effects deserved investigation. Through extensive experiments, the role of circSCMH1 in promoting neuronal plasticity of the brain both in vivo and in vitro, and reducing glial activation and peripheral immune cell infiltration, therefore protecting normal brain function. Next, the molecular mechanism involved in neuronal plasticity was explored and methyl CpG binding protein-2 (MeCP2) was indicated as one of the targets of circSCMH1. Additionally, the function of circSCMH1 on brain repair in the nonhuman primate stroke model was evidenced. In conclusion, rabies viral glycoprotein peptide (RVG)-circSCMH1-extracellular vesicles (EVs) have been shown that, when used therapeutically, it can improve stroke recovery outcomes both in mice and monkeys after stroke, which indicates that the circSCMH1 can meet the requirements of restoring motor function and improving life quality in the stroke patient population [87]. Besides, circular RNA UCK2 (circUCK2) functions as a sponge to miR-125b-5p to inhibit its activity and leads to the increase of growth differentiation factor 11 (GDF11), ameliorating neuronal injury. The circUCK2/miR-125b-5p/GDF11 axis is an important signaling pathway during ischemic stroke, indicating that circUCK2 is a potential therapeutic target for ischemic stroke [88]. Based on numerous supporting evidences originating from animal studies, such as rodents and nonhuman primates, relevant human confirmation to corroborate the experimental findings is promising.
With the current exploration of the mechanism of circRNA and ischemic stroke, it is promising and deserving to further research other circRNAs expressing differently under ischemic stroke and investigate this mechanism. Future research in this area may focus on identifying specific circRNAs that are dysregulated in stroke and developing targeted interventions to restore normal circRNA expression and improve outcomes for stroke patients. Ultimately, targeting potential circRNAs and developing agents that regulate the expression of these and secondary control its downstream molecular pathways might be a new era in therapy for ischemic stroke.
A summary of the functions of different circRNAs in ischemic stroke has been represented in Table 2.
A summary of the functions of circRNAs in ischemic stroke
| circRNAs | Regulation | Regulation targets and pathways | Effects | Refs |
|---|---|---|---|---|
| circSCMH1 | Down | circSCMH1/MeCP2 axis | Enhance neuronal plasticity, inhibit glial activation and peripheral immune cell infiltration | [87] |
| circOGDH | Up | circOGDH/miR-5112/COL4A4 axis | Enhance COL4A4 expression to elevate neuron damage | [89] |
| circHECTD1 | Up | circHECTD1/miR-142/TIPARP axis | Promote astrocyte activation | [46] |
| circUCK2 | Down | circUCK2/miR-125b-5p/GDF11 axis | Improve the cell survival rate and ameliorate neuronal injury | [88] |
| circSHOC2 | Up | circSHOC2/miR-7670-3p/SIRT1 axis | Inhibit neuronal apoptosis via promoting autophagy and inhibit neuronal death | [75] |
| circCDC14A | Up | - | Promote astrocytes activation | [90] |
| circTLK1 | Up | circTLK1/miR-335-3p/TIPARP axis | Aggravate neuronal injury and neurological deficit. | [91] |
| circDLGAP4 | Down | circDLGAP4/miR-143/HECTD1 axis | Inhibite EndoMT and attenuate BBB damage | [92] |
circOGDH: circular RNA OGDH; COL4A4: Gallus collagen, type IV, alpha IV; circHECTD1: circular RNA HECTD1; circCDC14A: circular RNA CDC14A; circTLK1: circular RNA TLK1; circDLGAP4: circular RNA DLGAP4; EndoMT: endothelial-to-mesenchymal transition
Nowadays, the study on the function of autophagy and circRNA regulation in stroke is abundant and prevalent, typically through investigating them separately. However, due to the double-edged sword of induced autophagy upon stroke, it’s worth accurately regulating the duration and degree to which autophagy is activated in different cells of the neurovascular unit at each stage of ischemic stroke. Fortunately, circRNA shows its potential to modulate the progression of ischemic stroke through autophagy.
Based on reports, some of the circRNAs promote cell autophagy. Han et al. [46] observe that circHECTD1 expressed highly and increased dramatically in the transient MCAO group, coincident with the circRNA microarray and heat map. Served as an endogenous sponge, upregulated circHECTD1 can interact with miR-142, whose knockdown inhibited astrocyte activation induced by OGD-R in vivo and in vitro. And through the forecast, the TIPARP has a conserved miR-142 binding site within its 3’-untranslated region (UTR) in most species, which is the downstream target of the circHECTD1/miR-142 axis and promotes astrocyte activation proved by experiments. Meanwhile, the circHECTD1/miR-142 axis promoted astrocyte autophagy via downstream TIPARP, leading to activated astrocyte enhancement. In conclusion, circHECTD1 acted as a sponge of miR-142, and upregulated TIPARP expression, activating astrocytes via enhancing autophagy [46]. Besides, another report said, upregulated circular RNA FoxO3 (circFoxO3) activates BMECs autophagy. Mechanistic target of rapamycin complex 1 (mTORC1), a complex formed with mTOR, Raptor, and mammalian lethal with SEC13 protein 8 (mLST8) is involved in the negative regulation of autophagy. circFoxO3 indirectly regulates mTORC1 in two different ways. One way is that circFoxO3 competitively sequestered mTOR to inhibit mTORC1 activity, promoting autophagy activation. Under normal physiological conditions, E2F transcription factor 1 (E2F1), a transcriptional regulator of mTORC1, is obligated to enter the nucleus and bind to DNA for activation of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) transcription, facilitating the translocation of mTORC1 to lysosomes with the assistance of PFKFB3. However, the interaction between circFoxO3 and E2F1 blocked the pathway of E2F1 entering the nucleus, preventing its transcriptional regulation. Through the above two ways, mTORC1 activity is suppressed, promoting autophagy activation [79].
Moreover, the insight of Chen et al. [75] is focused on the role of the ischemic-preconditioned astrocyte-derived exosomes (IPAS-Exos) against ischemic stroke. They found that IPAS-Exos played a protective role in neuroprotection both in vitro and in vivo, with a decrease in the ratio of LC3-II/LC3-I and beclin 1 (two autophagy-related proteins) and an increase of p62. All the data clarified IPAS-Exos inhibited ischemia-induced neuronal death by regulating autophagy. How did IPAS-Exos regulate autophagy to protect neuronal cells? Through circRNA microarray, they found that circSHOC2 was highly expressed in IPAS-Exos and worked as a neuron protector. Mechanically, circSHOC2 acted as a molecular sponge for miR-7670-3p in neurons, inhibiting miR-7670-3p activity, thereby increasing the expression of SIRT1. In recent years, the downstream target molecule SIRT1 has been found to induce autophagy by regulating the tuberous sclerosis complex subunit 2 (TSC2)/mTOR/S6 kinase 1 (S6K1) signaling pathway, indicating the anti-autophagic role of circSHOC2 [75].
In the mice and cells with the I/R model, the expression levels of circ_016719 and mitogen-activated protein kinase 6 (Map2k6) were significantly increased and miR-29c was decreased. What’s more, the knockdown of circ_016719 expression reversed the increased expression of autophagy proteins, showing that circ_016719 regulates autophagy in ischemia by circ_016719/miR-29c/Map2k6 signaling pathway [93]. Additionally, Xu et al. [94] showed that circular RNA Akap7 (circAkap7) played a protective role in transient MCAO in mice and knockdown of ATG12 reversed the therapeutic effects of circAkap7, showing that circAkap7 attenuates I/R-induced cellular injury by promoting ATG12-mediated autophagy.
circRNAs not only influence disease development by promoting autophagy but also regulate by inhibiting autophagy [93]. In MCAO rats, circ_0025984 acts as a molecular sponge and absorbs miR-134-3p in astrocytes, and decreases the expression of ten-eleven translocation 1 (TET1). After translocating into the nucleus, TET1 can bind to the promoter of 150-kDa oxygen-regulated protein (ORP150) and converts 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC), leading to DNA demethylation and increased expression of ORP150, which eventually activates autophagy mediated by ATG7. Thereby, circ_0025984 protects astrocytes from ischemia-induced autophagy by targeting the miR-143-3p/TET1 pathway and might inhibit cerebral injury induced by ischemic stroke [95].
A summary of the functions of different autophagy-related circRNAs in ischemic stroke has been represented in Figure 2.
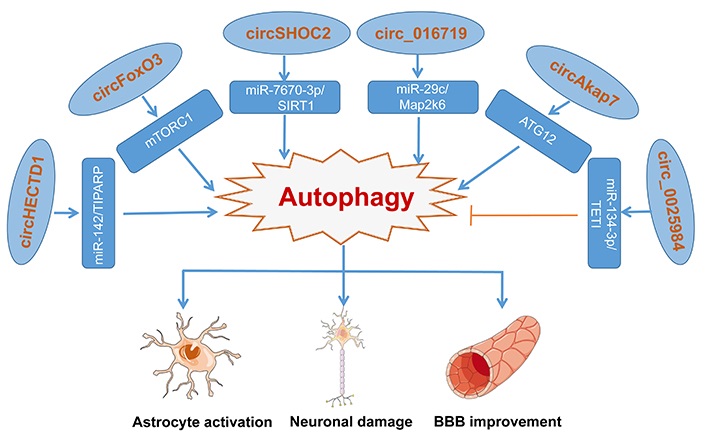
Various circRNA molecules act as an inhibitor or activator to modulate autophagy via different regulation pathways, making differences to astrocytes, glial, and neurons. Blue arrow represents the activating function and the yellow symbol represents the inhibiting function
All in all, autophagy plays a critical role in the pathogenesis of various diseases, and the involvement of circRNA in autophagy regulation has been gaining attention in recent years. Although the underlying mechanism of circRNA-mediated autophagy regulation in the prognosis of ischemic stroke remains uncertain, the potential of autophagy-related circRNAs as novel diagnostic biomarkers and therapeutic targets is promising. As research techniques advance, the identification of specific circRNAs and drugs targeting autophagy and circRNA could pave the way for more effective treatments of ischemic stroke.
3-MA: 3-methyladenine
AMP: adenosine monophosphate
ATG: autophagy-related
BBB: blood-brain barrier
BMECs: brain microvascular endothelial cells
CDR1-AS: circular RNA CDR1-AS
circAkap7: circular RNA Akap7
circFoxO3: circular RNA FoxO3
circFUNDC1: circular RNA FUNDC1
circHECTD1: circular RNA HECTD1
circRNAs: circular RNAs
circSCMH1: circular RNA SCMH1
circSHOC2: circular RNA SHOC2
circUCK2: circular RNA UCK2
CMA: chaperone-mediated autophagy
CNS: central nervous system
E2F1: E2F transcription factor 1
GDF11: growth differentiation factor 11
I/R: ischemia/reperfusion
IPAS-Exos: ischemic-preconditioned astrocyte-derived exosomes
LC: light chain
lncRNAs: long noncoding RNAs
Map2k6: mitogen-activated protein kinase 6
MCAO: middle cerebral artery occlusion
miRNA: microRNA
mTOR: mechanistic target of rapamycin
mTORC1: mechanistic target of rapamycin complex 1
OGD: oxygen glucose deprivation
SIRT1: sirtuin 1
TET1: ten-eleven translocation 1
TIPARP: TCDD inducible poly[ADP-ribose] polymerase
TJs: tight junctions
YH and LG: Investigation, Writing—original draft, Writing—review & editing. BH: Conceptualization, Validation, Writing—review & editing. HY: Conceptualization, Validation, Writing—review & editing, Supervision. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
This work was supported by the
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
