Affiliation:
1Rheumatology Research Unit, Institute of Molecular Medicine João Lobo Antunes, Faculty of Medicine of the University of Lisbon, Lisbon Academic Medical Centre, 1649-028 Lisbon, Portugal
Email: ines.correia@medicina.ulisboa.pt
ORCID: https://orcid.org/0000-0002-9405-8208
Affiliation:
1Rheumatology Research Unit, Institute of Molecular Medicine João Lobo Antunes, Faculty of Medicine of the University of Lisbon, Lisbon Academic Medical Centre, 1649-028 Lisbon, Portugal
2Pediatric Rheumatology Unit, Santa Maria University Hospital, North Lisbon University Hospital Centre, Lisbon Academic Medical Centre, 1649-028 Lisbon, Portugal
ORCID: https://orcid.org/0000-0002-7174-8302
Affiliation:
3Nutrition Laboratory, Faculty of Medicine of the University of Lisbon, Lisbon Academic Medical Centre, 1649-028 Lisbon, Portugal
ORCID: https://orcid.org/0000-0002-8730-6517
Affiliation:
1Rheumatology Research Unit, Institute of Molecular Medicine João Lobo Antunes, Faculty of Medicine of the University of Lisbon, Lisbon Academic Medical Centre, 1649-028 Lisbon, Portugal
4Serviço de Reumatologia e Doenças Ósseas Metabólicas, Santa Maria University Hospital, North Lisbon University Hospital Centre, Lisbon Academic Medical Centre, 1649-028 Lisbon, Portugal
ORCID: https://orcid.org/0000-0003-1432-3671
Explor Med. 2022;3:540–560 DOI: https://doi.org/10.37349/emed.2022.00112
Received: May 31, 2022 Accepted: July 27, 2022 Published: December 26, 2022
Academic Editor: Alexander Swidsinski, Universitätsmedizin Berlin, Germany
Systemic lupus erythematosus (SLE) is a chronic, immune-mediated disease associated with significant morbidity and mortality. New evidence suggests that diet, gut microbiota, intestinal permeability, and endotoxemia may modulate chronic inflammation and disease activity in SLE. This review focus on what is known about the gut microbiota in lupus mouse models and SLE patients and the possible mechanisms that connect the gut microbiota with SLE. It included 29 studies (12 animal studies, 15 human studies, and 2 included data on both), with variable results regarding alpha and beta-diversity and gut microbiota composition between lupus-mouse models and SLE patients. Ruminococcus (R.) gnavus was significantly increased in lupus nephritis (LN) in one study, but this was not corroborated by others. Despite the different results, mechanistic lupus mouse model studies have shown that gut microbiota can modulate disease activity. Interestingly, pathobiont translocation in monocolonized and autoimmune-prone mice induced autoantibodies and caused mortality, which could be prevented by a vaccine targeting the pathobiont. Moreover, studies on fecal transplants and diet on different lupus mouse models showed an effect on disease activity. In SLE patients, a higher adherence to the Mediterranean diet was associated with lower disease activity, which may be explained by the connection between diet and gut microbiota. Although gut dysbiosis has been observed in SLE patients and lupus mouse models, it remains to clarify if it is a cause or a consequence of the disease or its treatments. Further studies with larger and well-characterized populations will undoubtedly contribute to deciphering the role of gut microbiota in SLE development, progression, and outcome.
Systemic lupus erythematosus (SLE or lupus) is a prototypical, systemic, immune-mediated disease with an uneven prevalence among gender, ethnic, and age groups. It has multiple clinical manifestations, affecting virtually every organ. An interaction between genetic, epigenetic, and environmental factors contributes to SLE pathogenesis [1], resulting in dysregulation of innate and adaptative immune responses [2]. Despite the improvements in the available treatments, SLE remains a disease with high morbidity and mortality, greatly influencing patients’ quality of life and productivity [3]. In the United States of America, SLE is the seventh cause of non-traumatic death in women between 15 and 24 years old, ahead of diabetes mellitus, human immunodeficiency virus (HIV), or septicemia [3]. In African-American and Hispanic women in the same age group, the SLE burden is even higher, being the fifth cause of death [3]. Furthermore, organ damage occurs in 32% to 50% of the patients, associated with the disease itself and the drugs used for its treatment, mainly corticosteroids [4]. Understanding SLE pathogenesis and finding new treatment strategies is, therefore, imperative.
Next-generation sequencing technology allowed a deeper knowledge of the gut microbiota and its contribution to health and disease [5]. The gut microbiota is composed of a collection of bacteria, archaea, viruses, protists, and fungi [6]. The bacterial phyla Firmicutes and Bacteroidetes represent over 90% of the gut microbiota, while Actinobacteria, Spirochaetes, and Proteobacteria, are represented to a lesser extent [7]. While gut homeostasis is associated with health, alterations in the composition and function of the gut microbiota (dysbiosis), driven by external and host-related factors, have been associated with several diseases [8], including inflammatory bowel disease [5], malignancies [9], and immune-mediated diseases [8].
Dysbiosis is characterized by one or more of the following parameters: overgrowth of pathobionts; loss or decrease in the number of commensals by the death of the microorganisms or reduction in its proliferation, and decrease in the gut microbiota diversity (alpha diversity) [8]. Dysbiosis is associated with diet and other lifestyle factors [6] and genetic factors [10].
In SLE patients and lupus-mouse models, gut microbiota dysbiosis has been identified, suggesting it may participate in lupus pathogenesis. Moreover, SLE patients have increased circulating lipopolysaccharides (LPS) [11], demonstrating that higher gut permeability may facilitate the translocation of gut microorganisms and their metabolites to the circulation, influencing the systemic landscape. In fact, the gut microbiota can modulate immune responses by upregulating toll-like receptor (TLR) expression on antigen-presenting cells, altering T-cell subsets [11], and facilitating post-translational modifications of luminal proteins that will activate stochastically generated autoreactive T and B cells [12]. Moreover, in a lupus mouse model, antibiotics and a vaccine against a pathobiont improved disease manifestations and prevented death [13], showing that gut bacteria can be involved in SLE outcomes. Nevertheless, the mechanisms by which the gut microbiota participate in SLE pathogenesis are unclear. Overall, the current hypothesis is that diet, gut microbiota, and intestinal permeability modulate endotoxemia and, consequently, the chronic activation of the immune system seen in SLE. This review looks at the current evidence related to the gut microbiota in lupus mouse models and in SLE patients, with the ultimate goal of providing a comprehensive overview of the recent advances in this field.
A literature search in MEDLINE and SCOPUS was conducted to identify papers written in English and published in the last ten years (until the 13th of April 2022). The following search terms were used: “SLE”[Title/Abstract] OR “lupus”[Title/Abstract] AND “gut microbiota”[Title/Abstract] OR “gut mycobiota”[Title/Abstract] OR “gut virome”[Title/Abstract] OR “intestinal microbiota”[Title/Abstract] OR “gut dysbiosis”[Title/Abstract] OR “intestinal dysbiosis”[Title/Abstract] OR “gut microbiome”[Title/Abstract] OR “actinobacteria”[Title/Abstract] OR “Bacteroidetes”[Title/Abstract] OR “firmicutes”[Title/Abstract] OR “mollicutes”[Title/Abstract] OR “patescibacteria”[Title/Abstract] OR “proteobacteria”[Title/Abstract] OR “tenericutes”[Title/Abstract] OR “verrucomicrobia”[Title/Abstract] OR “alistipes”[Title/Abstract] OR “akkermancia”[Title/Abstract] OR “bacteroides”[Title/Abstract] OR “Bifidobacterium”[Title/Abstract] OR “bilophila”[Title/Abstract] OR “blautia”[Title/Abstract] OR “clostridium”[Title/Abstract] OR “coprobacter”[Title/Abstract] OR “dehalobacterium”[Title/Abstract] OR “dialister”[Title/Abstract] OR “dysgonomonas”[Title/Abstract] OR “desulfovibrio”[Title/Abstract] OR “dorea”[Title/Abstract] OR “Escherichia”[Title/Abstract] OR “eggerthella”[Title/Abstract] OR “enterococcus”[Title/Abstract] OR “erysipelatoclostridium”[Title/Abstract] OR “eubacterium”[Title/Abstract] OR “ezakiella”[Title/Abstract] OR “faecalibacterium”[Title/Abstract] OR “flavonifractor”[Title/Abstract] OR “gemmiger”[Title/Abstract] OR “hungatella”[Title/Abstract] OR “incertae sedis”[Title/Abstract] OR “klebsiella”[Title/Abstract] OR “lachnoclostridium”[Title/Abstract] OR “lachnospira”[Title/Abstract] OR “lactobacillus”[Title/Abstract] OR “lactobacilli”[Title/Abstract] OR “Lactococcus”[Title/Abstract] OR “megasphaera”[Title/Abstract] OR “morganella”[Title/Abstract] OR “odoribacter”[Title/Abstract] OR “oscillospira”[Title/Abstract] OR “oxalobacter”[Title/Abstract] OR “paraprevotella”[Title/Abstract] OR “parabacteroides”[Title/Abstract] OR “porphyromonas”[Title/Abstract] OR “prevotella”[Title/Abstract] OR “pseudobutyrivibrio”[Title/Abstract] OR “pseudomonas”[Title/Abstract] OR “rothia”[Title/Abstract] OR “rhodococcus”[Title/Abstract] OR “roseburia”[Title/Abstract] OR “rudaea”[Title/Abstract] OR “ruminococcaceae”[Title/Abstract] OR “ruminococcus”[Title/Abstract] OR “stenotrophomonas”[Title/Abstract] OR “streptococcus”[Title/Abstract] OR “turicibacter”[Title/Abstract] OR “veillonella”[Title/Abstract]. Firstly, the titles and abstracts of the papers found were reviewed, and then the full texts were evaluated. All studies in which the gut microbiota was assessed in SLE or lupus-mouse models were included. Reviews and papers with interventions to modulate the gut microbiota without a previous characterization were not included. The details of the papers included in this review are presented in Tables 1 (lupus mouse models) and 2 (human studies).
Studies in lupus mouse models and controls
| Authors & Reference | Mouse model | Age | Sequencing | Alpha and beta-diversity | F/B ratio | Functional pathways | Other findings |
|---|---|---|---|---|---|---|---|
| Wang et al. [14] | MRL+/+ MRL/lpr C57BL/6 (control) | 6–16 weeks | 16S rDNA: V4 regionIllumina Miseq | Alpha-diversity: lower in MRL/lpr 6 week-old mice but not in 18-week-old.Without difference in MRL+/+ mice.Beta-diversity: differential bacterial community composition in MRL+/+ and MRL/lpr mice at 6 weeks and 18 weeks. | Lower in 6 week-old MRL/lpr mice vs. age-matched MRL+/+ mice. | NA | In 18-week vs. 6-week MRL+/+ mice: increase in Bacteroidetes and Verrucomicrobia phyla; Akkermansiaceae and Rikenellaceae families; Akkermansia, Alistipes, Blautia, and Ruminiclostridium genus, and Akkermansia muciniphila species.Reduction in Firmicutes and Tenericutes phyla, Anaeroplasmataceae and Peptostreptococcaceae families, Anaeroplasma and Romboutsia genus, and Clostridium clostridiiforme and Faecalibacterium prausnitzii species.No statistically significant alterations were found between 18-week and 6-week MRL/lpr mice.6-week-old MRL/lpr mice vs. MRL+/+ mice: decreased Peptostreptococcaceae and Lactobacillaceae, increase in Rikenellaceae; decreased Muribaculaceae and Anaeroplasmataceae. |
| Chen et al. [41] | MRL/lpr MRL/MpJ (control) | 17 weeks | Illumina HiSeq | Alpha-diversity: without significant alterations, but decreased number of species.Beta-diversity: significantly different from controls. Higher inter-group distribution in lupus mice than controls. | NA | Positively related to increase in MRL/lpr and SLE patients: biosynthesis of L-arginine and L-ornithine, tryptophan, menaquinol, palmitoleate, and oleate, phosphatidylglycerol. | Increased R. torques and genera Blautia in SLE patients and lupus mice.Decreased genera Desulfovibrio in MRL/lpr. |
| Mu et al. [15] | MRL/lprMRL/lpr with no antibiotics (control) | 9–16 weeks | 16S rRNA: V4 regionIllumina MiSeq | Alpha-diversity: no alteration with a mix of antibiotics given after disease onset but reduced when only vancomycin was given. | NA | Treatment of MRL/lpr mice with vancomycin up-regulated peptidoglycan biosynthesis and transcriptional factors pathways, and down-regulated glyoxylate, decarboxylate, and histidine metabolism, and phenylalanine, tyrosine, tryptophan, and lipopolysaccharide biosynthesis. | MRL/lpr: Marked depletion of Lactobacilli, and increase of Clostridial species.Antibiotics treatment initiated after disease onset improved lupus-like symptoms; post-disease onset reduced bacterial load and did not decrease bacterial diversity.Antibiotic reshaped the composition of the gut microbiota, increasing the relative abundance of Lactobacillus spp., and decreasing Lachnospiraceae.A mix of antibiotics and vancomycin led to Lactobacillus spp. enrichment. |
| Zhang et al. [16] | MRL/MpJMRL/lprB6/lprC56BL/6 (control) | 6–14 weeks | 16S rRNA: V4 regionIllumina MiSeq | Alpha-diversity: higher in lupus-prone mice.Beta-diversity: phylogenetic difference. | NA | Down-regulated in lupus mouse models: DNA replication, DNA repair, protein synthesis, galactose and glycolysis metabolism.Up-regulated in lupus mouse models: bacterial chemotaxis, flagellar assembly, sporulation, glyoxylate and decarboxylate metabolism, ABC transporters, amino-acid metabolism. | MRL/lpr: reduction in Lactobacillaceae (family), increase in Lachnospiraceae (family).The abundance of Lachnospiraceae positively correlated with lymphadenopathy and renal pathology.Treatment with retinoic acid attenuated lupus-like symptoms while increasing the Lactobacillaceae. |
| Chen et al. [17] | NZB/W F1 induced by HCMVpp65422–439NZB/W F1 treated with PBS (control) | 12–24 weeks | 16S rRNA | Alpha-diversity: higher than controls.Beta-diversity: differences in microbial community composition. | Increased in lupus mice vs. controls | Differentially expressed in lupus vs. controls: cell motility, lysosomes, flagellar assembly, cytoskeleton proteins, bacterial motility proteins, bacterial chemotaxis. | Increase in lupus mouse models: families Saccharimonadaceae, Marinifiaceae, and Desulfovibrionaceae.Candidatus Saccharimonas showed significant positive correlations with the creatinine level, anti-dsDNA IgG titer, and glomerulonephritis severity. All four lupus-like effects were positively correlated with Odoribacter, Desulfovibrio, and Roseburia. |
| Johnson et al. [18] | (SWRxNZB)F1 (SNF1)C57BL/6 (control) | 4–24 weeks | 16S rRNA: V3–V4 regionIllumina Miseq | Alpha-diversity: lower in 24-week SNF1 females than males.Alpha-diversity: significantly different in 16-week mice.Beta-diversity clustering of gut microbiota of 16-week males and females but not at 4 weeks. | NA | Up-regulated in female 16-week mice lupus model: glycol degradation, glycosphingolipid biosynthesis pathways, nitrogen, amino sugar, nucleotide sugar, galactose, starch, and sucrose metabolism pathways.Down-regulated in female 16-week mice lupus model: secondary metabolite synthesis and degradation, fatty acid, glycerophospholipid, and lysine metabolism. | The gut microbiota composition of lupus-prone SNF1 mice is different at adult ages but not at juvenile ages.Females at 16 weeks: higher abundance of Bacteroidetes and Parabacteroides genus members, and lower Dysgonomonas genus members of Bacteroidetes phylum.Significant age-dependent differences in the abundance of multiple microbial communities in 16-week mice. |
| Choi et al. [19] | TCC57BL/6 (control) | 6 weeks to 6 months | 16S rDNA: V4–V5 regionsIllumina Miseq | Alpha-diversity: no alterations. | NA | Up-regulated in TC mice: Tryptophan metabolism, microbial metabolism derived from shikimate pathway, purine and pyrimidine metabolism, protein digestion and absorption, biosynthesis of antibiotics, biosynthesis of amino acids, nucleoside pathways. | TC mice: marked alterations in Prevotellaceae, Paraprevotella, and Lactobacillus.Different bacterial taxa composition between young and adult TC and controls. |
| Luo et al. [42] | NZB/W F1 treated with dexamethasone NZB/W F1 (control) | 10–33 weeks | 16S rRNA: V4 regionIllumina Miseq | Alpha-diversity: increased bacterial diversity with disease progression in NZB/W F1 mice.Beta-diversity: difference between pre-disease and diseased time points. | NA | NA | Genera Clostridium, Dehalobacterium, Lactobacillus, Oscillospira, Dorea, Bilophila, and AF12, and an unnamed genus within the family Ruminococcaceae, increased from the pre-disease stage to the diseased stage.Decrease of Akkermansia muciniphila and a species within the genus Anaerostipes from the pre-disease stage to the diseased stage.The relative abundance of Lactobacillaceae increased significantly from the pre-disease stage to the post-disease stage, and treatment with dexamethasone was able to significantly decrease its abundance. |
| Ma et al. [20] | TCC57BL/6 (control) | 34–36 weeks | 16S rRNA genes: V4 regionIonS5™XL | Alpha-diversity: decreased richness and diversity in TC mice.Beta-diversity: differences between mouse-models.Fecal transplantation to GF mice: the recipient mice maintained the same tendency of diversity as donors. | NA | NA | TC mice showed a dysbiotic gut microbiota when compared to controls.Increased Firmicutes and Actinobacteria phylum in TC mice.Fecal transplantation induced a gut mucosal immune response, higher with TC than with feces of control mice.The expression of Irf7 and Csk genes was significantly higher in germ-free mice that received TC fecal transplant than in germ-free mice that received fecal transplant from controls.Fecal transplant with feces of TC mice triggered alterations in gene expression in the colon. |
| Mu et al. [21] | MRL/lprMRL/MpJ (control) | 3–14 weeks | 16S rRNAIllumina Miseq | NA | NA | NA | Sugestive positive correlation between a higher abundance of Lactobacillales and improved lupus-like symptoms.Before disease onset, Lactobacillus treatment significantly increased the survival of female lpr mice. L. reuteri and an uncultured Lactobacillus sp. accounted for most of the observed effects.Lactobacillus spp. in the gut can attenuate kidney inflammation in lupus-prone mice in a sex hormone-dependent manner when given before disease onset. |
| Valiente et al. [22] | NZM2410C57BL/6 | 10–30 weeks | 16S rRNA: V4 regionIllumina Miseq | Alpha-diversity: higher in SFB+ mice than in SFB– mice (not statistically significant). | Trend towards a reduced F/B ratio at 30-week-old SFB+ mice without statistical significance. | NA | SFB+ mice showed increased gut permeability and a decrease in gut barrier integrity.SFB+: increased Prevotellaceae, Lactobacillaceae, and Clostridiaceae, and decreased Ruminococcaceae and Lachnospiraceae.30-week-old mice had significant enrichment of R. torques spp. when compared to 15-week-old SFB+ and SFB– mice. |
| Cabana-Puig et al. [23] | MRL/lpr(2 different colonies) | 3–15 weeks | 16S rRNA: V4 regionIllumina Miseq | Alpha-diversity: different between colonies of MRL/lpr mice. | NA | NA | Housing/environmental differences justify alterations in the gut microbiota of two different colonies of MRL/lpr mice.Some phenotypic differences may be explained by the alterations in the gut microbiota. |
| Hong et al. [24] | BXD2C57BL/6 (control) | 6 months or 12 months | 16S rRNA: V4 regionIllumina Miseq | NA | NA | NA | Lupus phenotype is not influenced by the gut microbiota composition.The gut microbiota composition favors the maintenance of immune dysregulation and disease progression in older mice. |
| Shirakashi et al. [25] | B6SKGB6SKG germ-free (control) | 3–12 months | 16S rRNA: V1–V2 regionsIllumina Miseq | Alpha-diversity: moderate decrease. | NA | NA | In B6SKG mice, dysbiosis may occur due to altered Tfh and germinal center development in Peyer’s patches, and IgA production.An altered T cell receptor signaling, which contributes to the alteration of the gut microbiota, favors autoimmunity. |
B6SKG: SKG mice with a B6 background; Csk: tyrosine-protein kinase; dsDNA: double-stranded DNA; F/B ratio: Firmicutes/Bacteroidetes ratio; HC: healthy controls; IgG: immunoglobulin G; Irf7: interferon Regulatory Factor 7; MRL: Murphy Roths large; NA: not applicable; NZB/W: New Zealand black x New Zealand white; PBS: phosphate-buffered saline solution; R.: Ruminococcus; rDNA: ribosomal DNA; rRNA: ribosomal RNA; SFB–: not inoculated with segmented filamentous bacteria; SFB+: inoculated with segmented filamentous bacteria; TC: triple congenic (B6.Sle1.Sle2.Sle3); Tfh: T follicular helper cells
The search resulted in 52,334 records. Three papers were found by consulting the references of the screened papers. After duplicate removal, 52,454 records were screened. After excluding the studies that did not meet the inclusion criteria, 81 papers were assessed, fifty-two of which were excluded for the following reasons: 1 without access to the full text, 27 reviews or editorial articles, and 24 did not include SLE microbiota characterization before any intervention.
A total of 29 studies were included in this review, twelve of which used mouse models [14–25], fifteen included SLE patients [26–40], and two both [41, 42].
The studies with SLE patients comprised 597 patients (29 males, representing 5% of the total sample) and 1,563 HC (548 males, representing 35% of the total sample). Additionally, a GWAS involved 7,219 SLE cases and 15,991 HC [29]. Ten studies included Chinese populations [26, 27, 30–33, 39–41], one Egyptian [28], two American [37, 42], one Spanish [34], one Japanese [35], and one Dutch [38]. The mean age of the participants ranged between 12.4 and 49.2 years old for SLE patients and 10.6 and 46.9 years old for HC. Only one study exclusively included children [31].
The authors only analysed bacterial variations when assessing the gut microbiota alterations, thus not assessing other microorganisms, such as viruses and fungi.
The different sequencing techniques and regions used in each study are mentioned in Tables 1 and 2.
The papers included in this review used diverse lupus-mouse models and controls and showed different results. Lupus characteristics, such as the production of autoantibodies, the timing of disease onset, the organs involved, and the lupus relative gender prevalence, are factors considered when choosing one specific mouse model [43]. NZB/NZWF1 and NZM2410 mice develop autoantibodies, splenomegaly, and immune complex glomerulonephritis [43]. The NZM2410 model shows a faster disease development, with 50% mortality at six months, while in NZB/NZWF1, there is a 50% mortality at nine months in female mice [43]. This mouse model lacks anti-Smith (Sm)/ribonucleoproteins (RNP) antibodies. However, the renal lesions are similar to those seen in SLE patients [44], and it has the same female bias recognized in humans [42]. MRL/lpr is a well-studied model that has the lpr spontaneous mutation of the FAS cell surface death receptor (FAS) [45]. It develops systemic auto-immunity, glomerulonephritis [45], arthritis, cerebritis, skin rash and vasculitis, antinuclear antibodies (ANA), anti-dsDNA, anti-Sm, anti-Ro, anti-La antibodies [43], and shows rapid and aggressive disease onset [14]. On the other hand, the development of autoantibodies in MRL+/+ mice is slow, and glomerulonephritis only occurs in the second year of life [14]. It has a similar autoantibody profile and glomerulonephritis to humans [46]. MRL/Mp mice do not carry the lpr mutation and develop symptoms later, usually after 18 weeks [16], with less severity and lower incidence [47]. SNF1 is a spontaneous lupus mouse model with detectable dsDNA autoantibodies at 16 weeks and severe nephritis at 22 weeks that shows gender bias [18, 43]. TC or B6.Sle1.Sle2.Sle3 mice develop severe systemic autoimmunity and fatal glomerulonephritis [19, 45] in 100% of the cases by nine months [45]. BXD2 mice develop autoantibodies, arthritis, glomerulonephritis, and splenomegaly [24]. Mouse models have been essential in improving the knowledge about SLE. However, human SLE characteristics are difficult to mimic in mice. In humans, SLE is a highly heterogeneous disease in which the pathogenesis is influenced by several factors beyond genetics, making it challenging to develop a mouse model that fully represents this disease [43].
Alpha-diversity is an indicator of microbial species diversity, which considers the variety of species (richness) and the evenness of species abundance [48]. Variations in this parameter have been observed in several diseases [49]. Most of the studies in mice included in this review assessed alpha-diversity, but the results were heterogeneous. In six-week-old MRL/lpr mice [14], a lower alpha-diversity was found, while in MRL+/+ mice, the decrease was not significant [14]. On the other hand, in NZB/NZWF1 mice, the diversity of the microbiota increased after SLE onset, approximately at 20 weeks of age, as well as in NZB/W F1 mice induced by HCMVpp65422–439, a viral peptide of human cytomegalovirus [17], suggesting that the disease progression influences the microbiota diversity [42].
Differences in the microbial community composition (beta-diversity) were also found by several authors [14, 16, 18–20, 42]. One study only identified changes in the beta-diversity of 16-week-old SNF1 mice housed littermates but not in 4-week-old mice [18], showing, once again, that disease progression affects the gut microbiota.
The F/B ratio has been considered a marker of gut homeostasis. Thus a higher or lower ratio has been considered a dysbiosis marker associated with several diseases [50]. Nevertheless, it should be considered together with other markers of gut microbiota diversity and composition [49]. The heterogeneous results between studies highlight the variation in the gut microbiota of lupus mouse models. An increased F/B ratio was found by some authors [17], but others found an opposite trend, with a decreased ratio in 6-week-old MRL/lpr mice [14] or a trend toward a decreased ratio in 30-week-old segmented filamentous bacteria (SFB) positive mice without statistical significance [22].
Within the phyla Firmicutes and Bacteroidetes, several bacteria may contribute to an altered F/B ratio. In 6-week-old MRL/lpr mice, the lower F/B ratio was attributed to a decreased abundance in Peptostreptococcaceae and Lactobacillaceae (phylum Firmicutes) and increased abundance in Rikenellaceae (phylum Bacteroidetes) [14]. However, there was no correlation with advanced lupus activity, suggesting that a lower ratio may contribute to early disease onset [14].
The gut microbiota dysbiosis may exacerbate SLE manifestations rather than trigger the disease [19, 24, 25], and the translocation of pathobionts is one of the possible mechanisms involved [22, 37, 51]. In SNF1 mice, the depletion of the gut microbiota with an antibiotic resulted in decreased expression of pro-inflammatory cytokines while not decreasing immune regulatory ones, such as interleukin 10 (IL-10) [18]. Moreover, after antibiotic use, only 40% of the female mice developed severe nephritis, while, without antibiotics, 80% of the mice developed this manifestation [18]. Interestingly, the microbiota depletion in male mice did not alter the disease incidence, immune phenotype, disease stage, or renal involvement [18]. Moreover, the gut microbiota composition in male and female littermates was significantly different only at adult ages [18]. Furthermore, castrated male SNF1 mice had different microbial communities at the genus level than non-castrated mice and showed higher lupus incidence and earlier onset (although not statistically significant) [18]. This work highlights that the gut microbiota in SLE may have a different influence in females and in males, which is reinforced by the results obtained with fecal transfer from male to female SNF1 mice, showing a slower lupus progression, lower proteinuria, and a modest decrease in anti-dsDNA and anti-nucleohistone antibodies than female mice that did not receive the fecal transplant [18].
Other authors also identified different variations in the microbial communities of female and male mice. Increased levels of Lachnospiraceae [16], Bacteroidetes, and Parabacteroides, and decreased Dysgonomonas [18] were found in females, while higher levels of Bifidobacterium were found in males [16].
Besides gender bias, the disease’s duration and characteristics may also contribute to the identified variability in the gut microbiota. In female and castrated male lpr mice, fecal transplant with Lactobacillus spp., particularly an uncultured Lactobacillus spp. and L. reuteri, starting before the disease onset, improved renal function and survival [21]. These alterations were associated with the increased expression of tight-junction proteins, meaning an improved gut barrier function and a decreased endotoxemia induced by Lactobacillus spp. treatment [21]. The improvement of the gut barrier integrity, and consequent decrease in bacterial translocation and activation of immune responses, was corroborated by the reduction in T cell migration to the gut lamina propria [21]. Moreover, Lactobacillus spp. also increased the systemic production of IL-10, meaning that these bacteria may have a systemic anti-inflammatory effect [21]. Furthermore, treatment with Lactobacillus spp. increased the influx of CD8+ T cells and forkhead box P3 (Foxp3)+ T reg cells to the kidney while decreasing T helper 17 (Th17) cells, attenuating LN, and improving the Treg-Th17 balance in castrated mice (that showed a similar response to female mice), but not in non-castrated ones. These results suggest that androgens attenuate Lactobacillus spp. effects, and in this mouse model, the control of LN induced by the gut microbiota was sex hormone-dependent [21].
In another study of two strains of lupus mouse models with different disease characteristics, distinct bacterial communities were identified [14]. Six-week-old MRL/lpr mice had a lower abundance of Muribaculaceae and Anaeroplasmataceae than 6-week-old MRL+/+ mice, while in 18-week-old MRL+/+ mice, there was an enrichment in Akkermansiaceae [14]. The authors suggested that this change may contribute to immune-mediated responses preceding the disease progression, favoring altered gut permeability and mucosal immune activation [14]. Lactobacillaceae showed a decreasing trend between 6-week-old and 18-week-old MRL/lpr mice, meaning that the inflammatory responses of the gut mucosa may have originated in a prolonged decrease in Lactobacillaceae [14]. Other authors found a similar reduction in Lactobacillaceae in the same mouse model [16]. Akkermansiaceae was significantly increased in 18-week-old MRL+/+ mice (more aggressive disease) compared with 6-week-old, but lower in MRL/lpr mice, suggesting that these bacteria can have different roles in different lupus phenotypes [14]. Other authors have also found that Akkermansiaceae and R. were negatively correlated with the serum levels of complement component 3 (C3) [33].
The bacterial composition of the gut microbiota varies between mouse studies. In 30-week-old NZM2410 mice inoculated with SFB, higher Prevotellaceae and Rikenellaceae and reduced Lachnospiraceae were present in severe glomerulonephritis [22]. In NZB/W F1 mice, Odoribacter, Desulfovibrio, and Roseburia genera were positively correlated with lupus-like effects, and the creatinine level, anti-dsDNA IgG titer, and glomerulonephritis were positively correlated with Candidatus Saccharimonas. These four genera are linked to the regulation of the host immune system, and immune-mediated diseases [17], demonstrating that the gut microbiota alterations are associated with lupus-like symptoms in this mouse model. Moreover, the authors also identified increased levels of the proinflammatory cytokines IL-6 and IL-17A related to the observed alterations in the gut microbiota [17].
In another study Bacteroidetes, Succinivibrio, Bilophila, and Parabateroides were positively correlated with IL-17, tumor necrosis factor (TNF)-related weak inducer of apoptosis (TWEAK), IL-2 receptor (IL-2R), IL-21, IL-35, IL-10, and interferon (IFN)-γ, while Dialister and Gemmiger were negatively associated with IL-17, IL-2R, and IL-3 [26], meaning that interfering with the production of these inflammatory cytokines, may be one of the mechanisms by which the gut microbiota influences SLE pathogenesis [37].
Finally, the influence of genetic variations on the composition and modulation of the gut microbiota is a new focus of research [52]. Several polymorphisms have been associated with the diversity of the gut microbiota, and some with specific bacteria, such as the well-described LCT gene association with Bifidobacterium [53]. Recently, in a transgenic mouse model, defective T cell receptor (TCR) signaling altered the gut microbiota and promoted systemic autoimmunity by driving Th17 cell differentiation [25]. Once again, this study showed the connection between genome, microbiota, and autoimmunity.
The connection between gut microbiota and lupus phenotype was further reinforced by the observation that a colony of MRL/lpr mice with a milder disease phenotype had an altered gut microbiota profile [23].
Further alterations in the gut microbiota in lupus mouse models can be found in Table S1.
The differences between the gut microbiota of lupus-mouse models and SLE patients are striking and are also observed among different SLE patient populations, making it challenging to establish consensual conclusions about SLEs’ gut microbiota characteristics. Considering that in any population, several internal and external factors impact the gut microbiota [54] and that in SLE patients both the disease and treatments seem to influence its composition and diversity [55], different results should be expected.
Similarly to the results in mouse studies, in humans the alpha-diversity alterations were also heterogeneous, varying between lower alpha-diversity [20, 26, 32, 33, 35, 37, 39, 41, 42], increased alpha-diversity [30] or no alterations in this marker compared with HC [16, 19, 27, 31, 34]. No differences were found between the newly onset patients and other patients, nor between patients with and without LN [35]. The same authors did not find differences in alpha-diversity between patients with high and low SLEDAI scores [35]. However, other authors found a trend toward an inverse correlation between alpha-diversity and higher disease activity (SLEDAI ≥ 8) [37]. When assessing differences between active and in remission patients, one study did not find global differences but detected an increase in Bifidobacterium [32]. Some authors identified that, despite decreasing disease activity, SLE treatment further decreased the gut bacterial diversity [41].
The beta-diversity was altered between SLE patients and controls [16, 27, 30, 32, 35, 37–39, 41], but not in all studies [31]. Although no association was found with medication [32], some authors found that after PPI use, there was an approximation between the microbiota profile of SLE patients with HC [39]. Beta-diversity also varied between patients with low and high disease activity [37, 38], although not consensually [35], and before and after disease onset [22, 42].
Five studies found a decrease in the F/B ratio in SLE patients compared to HC [26–28, 31, 38], while others only found a decreasing trend [22, 32], two did not find differences in this variable [33, 42] and one detected an increase [27]. An inverse correlation between disease activity and F/B ratio was found in some SLE patients [28]. Paradoxically, in active disease an increased F/B ratio was also reported [17, 26].
Several authors identified gut microbiota changes in SLE [14, 17, 32, 38, 39], but the bacteria involved varied between studies. One explanation may be the disease duration since variations in bacterial composition have been observed with disease progression [16, 42] and severity. Some authors found differences in the gut bacterial composition of patients with higher and lower disease activity [28, 33, 37], but not all [26], and some found a correlation between certain bacteria and disease activity, such as Streptococcus anginosus and Veillonella [32, 37], Acholeplasma, Capnocytophaga and Leptotrichia [33]. Some bacteria were associated with SLE risk (Bacilli, Eggerthella, and Lactobacillales), while others were found to be protective (Coprobacter, Bacillales, and Lachnospira) [29]. Interestingly, Lactobacillales were increased in active SLE patients but not in patients in remission [32].
R. gnavus has been recognized as a possible pathobiont contributing to renal involvement in SLE patients, as a 5-fold increase in this species, and a decrease in bacteria with beneficial roles, such as B. uniformis, were identified in this population [41]. Azzouz and collaborators [37], in 2019, mentioned that SLE patients showed decreased bacterial diversity, which was inversely correlated with disease activity and altered dynamics between species. When assessing the bacterial abundance, R. gnavus, a Gram-positive bacteria, was significantly increased and correlated with lupus disease activity [41]. Moreover, when LN was present, the increase in R. gnavus was even higher [41]. Other authors reported similar findings, encountering an increased abundance of R. gnavus in SLE patients with high disease activity [37]. SLE patients, especially with LN, showed increased gut permeability, leading to increased IgG anti-rat glioma model (RG) 2 strain antibodies, which correlated with disease activity [41]. R. gnavus seems, therefore, to contribute to renal damage [41]. These results were not replicated in other studies. Strikingly, some SLE patients with active disease showed a decreased abundance of R. gnavus compared to patients in remission [32]. Differences between populations related to genetics and diet and other lifestyle factors should be considered to explain these differences. Further studies assessing the variability of R. gnavus will help clarify these observations.
An association between gut microbiota composition and immune factors seems to exist [31]. Certain strains of B. fragilis, which are increased in some SLE patients [41], produce a toxin that binds to a colonic epithelial cell receptor and alters gut permeability [56]. However, in murine models, B. fragilis have beneficial effects, preventing colitis [56]. The immunomodulatory characteristics of this bacterial species are due to polysaccharide A (PSA) that can induce IL-10 secretion, by Treg cells, reducing gut inflammation [57]. It can also have a protective effect against pathogenic infections and systemic immune-mediated diseases [58]. Despite these different results, the gut microbiota and the host immune system certainly crosstalk, shaping the chronic activation of the immune system.
Akkermansia muciniphila showed immunogenic and inflammatory potential in a group of SLE patients with increased levels of this bacterium in the gut. In these patients, a bacterial peptide, which mimics an extracellular part of the human FAS, can bind IgG and positively correlates with serologic markers of inflammation [41]. The genus Lactobacillus was lower in SLE patients than in HC [28], although Lactobacillus abundance has not been steady through the different studies [32, 33], which may be due to the different species included [28].
Phylum Bacteroidetes [26, 38] and Proteobacteria [30], and genus B. and Alistipes were found to be increased in SLE patients [38], although there are inconsistent results between studies [33]. Bacteroidetes species are glycan-degrading and short-chain fatty acid-producing bacteria, usually considered beneficial for the host. However, simultaneously B. theta has been considered a potential gut pathobiont, enhancing lupus-like disease [38], and Proteobacteria, a marker of dysbiosis [31].
Despite the variations between the bacterial communities of SLE patients and lupus-mouse models, some shared alterations were also found. R. torques and Blautia genera were increased, and the Desulfovibrio genera were decreased in SLE patients and MRL/lpr mice. However, the role of the mucin degrader R. torques in SLE warrants a better understanding [41].
Streptococcus anginosus and Streptococcus intermedius, two species belonging to the oral and gut microbiota, were increased in SLE metagenome [35], reinforcing the concept of an oral-gut interaction. Nevertheless, the gut bacterial composition is not always consistent with the oral bacterial composition [33].
Although some studies included both male and female participants, the differences in the gut microbiota composition between genders were not assessed, possibly due to the low number of male participants, considering that SLE is mainly a female disease. Further changes in the gut microbiota can be found in Table S1.
Despite the heterogeneous results of the studies included in this review, it seems undeniable that gut microbiota participates in SLE pathogenesis [20]. Although the precise mechanisms remain to be fully known, some aspects connecting SLE and the gut microbiota have been elucidated.
Gut mucosa immunity is key to host homeostasis [59]. Its important role is corroborated by studies in germ-free mice in which, without the gut microorganisms, there was a lower production of cells involved in defense and inflammation, such as intraepithelial lymphocytes, as well as a decrease in Treg cells [59]. This finding shows that the production of bacterial components is essential for maintaining gut mucosal immunity.
On the other hand, in female lupus-prone mice, depleting the gut microbiota suppressed the pro-inflammatory gut immune phenotype and decreased disease incidence and progression, demonstrating that there is an involvement of the gut microbiota in SLE, possibly through its interactions with the gut mucosa [18]. Furthermore, other authors reported that, although the gut microbiota did not seem to contribute to the development of lupus phenotype in BXD2 and SKG mice, especially when older, it favors the maintenance of immune dysregulation, influencing the disease progression [24, 25].
Additionally, the gut microbiota participates in the host defense by producing antimicrobial components that will prevent colonization by pathobionts [60], which means that alterations in the gut microbiota composition may lead to an increase in pathogenic bacteria that can induce immune responses.
The gut microbiota of TC lupus-prone mice (B6.Sle1.Sle2.Sle3) stimulated the production of autoantibodies and activated immune cells when transferred into germ-free B6 mice [19]. Fecal transfer to B6 mice induced autoimmune phenotypes only when the TC donor mice were older and exhibited autoimmunity [19]. Metabolomic screening identified increased tryptophan metabolites in the feces of TC mice, including kynurenine, which were decreased with antibiotics [19]. In this model, the diet had a clear role in autoimmunity. Low dietary tryptophan prevented autoimmune pathology in TC mice, whereas high dietary tryptophan exacerbated disease [19]. Reducing dietary tryptophan changed the gut microbiota patterns in lupus-prone TC mice and B6 mice [19]. Furthermore, fecal transfer from TC mice fed a high tryptophan diet but not a low tryptophan diet induced autoimmune phenotypes in germ-free B6 mice [19].
In another mouse model, fecal transfer from lupus mice also induced anti-dsDNA antibodies and led to increased stimulation of B cells and decreased Treg cells, highlighting that feces and, consequently, the gut microbiota can induce gut mucosal immune responses [20]. However, the influence of the gut microbiota extends beyond the gut mucosa, impacting peripheral immune responses and gene expression of type I interferon-related genes and susceptibility genes for SLE in mice, such as Irf7 and Csk [20].
Intracellular tight junctions (TJ) are proteins that form a barrier between the apical and the basolateral sides of the gut, regulating the diffusion of molecules between cells (paracellular pathway) [61]. Zonulin, is the best described regulator of intestinal permeability through TJ modulation [62]. Interestingly, its release is triggered, among other factors, by dysbiosis, including small intestinal bacterial overgrowth, which strengthens the connection between the gut microbiota, and gut permeability. Zonulins’ release leads to the opening of the paracellular pathway and consequent passage of gut microorganisms and their antigens, resulting in the activation of the host immune system, targeting tissues and organs, and ultimately favoring the occurrence of immune-mediated diseases in genetically predisposed individuals [61]. In line with this observation, an altered gut barrier integrity was identified in 30-week-old NZM2410 mice inoculated with SFB, associated with increased renal involvement [22]. Moreover, LPS-producing bacteria can enter circulation and have systemic effects by triggering immune-mediated responses, which was seen with “YLYDGRIFI” peptide of Odoribacter splanchnicus that increased IFN-γ and IL-17A secretion in peripheral blood mononuclear cells of anti-Sm positive SLE patients [41]. Increased levels of LPS were identified in SLE patients [11] and were associated with SLE progression [15], showing that Gram-negative bacteria may favor SLE pathogenesis [15].
To summarize, the gut microbiota can shape intestinal mucosal immunity and gut permeability by adjusting zonulin levels. These mechanisms will affect endotoxemia and will have an important role in the chronic activation of the immune system. Some of the mechanisms through which the gut microbiota can participate in SLE pathogenesis, are highlighted in Figure 1.
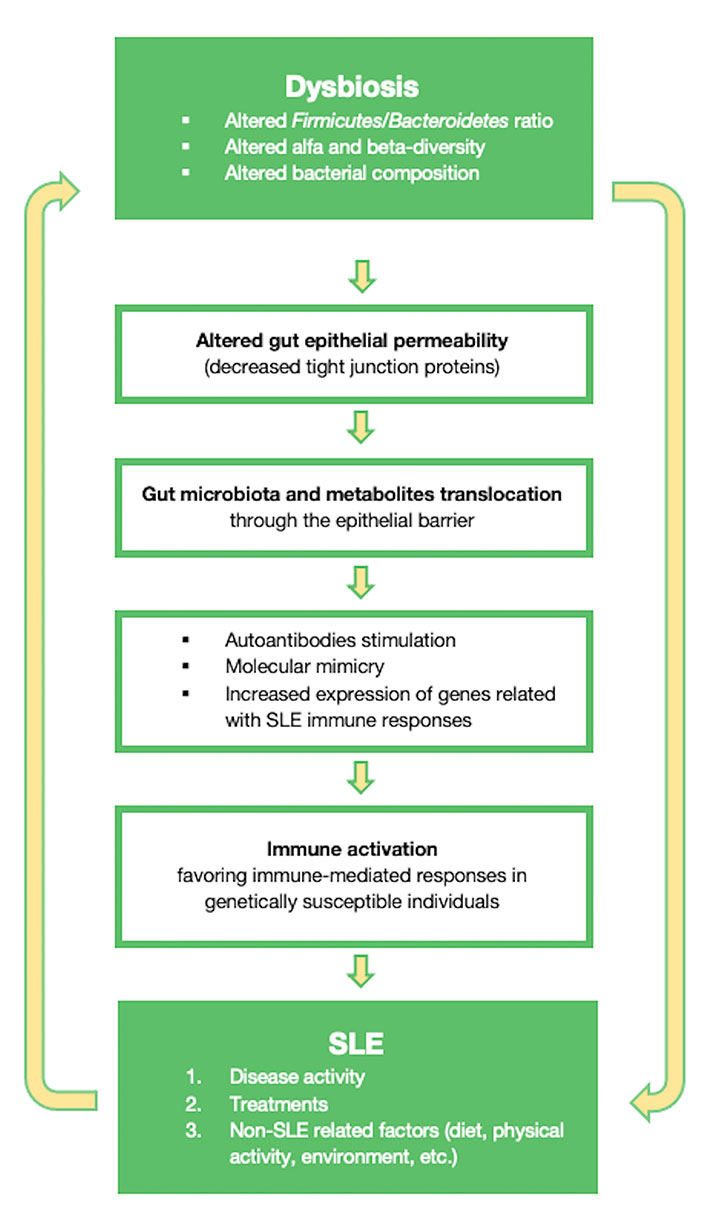
Diagram showing some of the mechanisms through which the gut microbiota can influence SLE
Some interventions have shown promising results in improving SLE disease activity by modulating the gut microbiota. Treatment with a probiotic of Lactobacillus fermentum CECT5716 (LC40) altered the gut microbiota composition of NZB/NZWF1 mice [63]. This probiotic decreased the abundance of Blautia and Lachnospira and endotoxemia by improving the gut barrier function [63]. Furthermore, LC40 also reduced anti-dsDNA and splenomegaly as well as the pro-inflammatory cytokines TNF-alpha and IL-1β and increased the expression of IL-18 [63], meaning it can be a promising tool in SLE management.
Vitamin D is well known for its effect on immunity, namely inhibiting Th17 and Th1 responses, favoring the development of regulatory T cells, reducing the activation and function of B cells and monocytes, and reducing pro-inflammatory cytokine production [64, 65]. Additionally, vitamin D can influence the gut microbiota composition [64, 66], probably as an indirect consequence of the hosts’ response to vitamin D status [66]. In fact, polymorphisms in the encoding vitamin D receptor (VDR) gene significantly influenced the gut microbiota beta-diversity and influenced Parabacteroides abundance, in a GWAS with a cohort of 2029 individuals [67]. Vitamin D also seems able to influence the gut epithelial integrity, particularly under stress conditions, either by regulating mucus production [66], or modulating TJ proteins, improving gut permeability [64]. These findings are of particular interest in SLE context, as ultraviolet-light exposure can trigger flares, and patients are frequently advised to avoid sun exposure, the main source of vitamin D [65].
Diet is one of the main factors influencing the gut microbiota composition through the life cycle, a reason why it may be an important ally in SLE disease control. Resistant starch is a non-digestible fiber fermentable by gut bacteria [68]. Resistant starch fermentation by the gut bacteria results in the production of short-chain fatty acids, which are relevant metabolites for host immunity [68]. In a TLR 7-dependent lupus mouse model, the overabundance and translocation of L.reuteri were reduced by the increase in dietary resistant starch, which led to higher production of short-chain fatty acids [68]. This intervention also resulted in an increase in survival and a decrease in organ involvement [68].
The Mediterranean diet is a dietary pattern associated with several beneficial health outcomes, such as lower incidence of inflammatory, immune-mediated, and metabolic diseases [69]. SLE patients with higher adherence to the Mediterranean diet showed lower BMI and fat mass, better cardiovascular and inflammatory profiles, as well as lower disease activity [69]. These results show that diet, particularly the Mediterranean diet, influences disease activity [69], which may occur through the modulation of the gut microbiota [70].
Considering that the gut microbiota can be influenced by several factors, including SLE treatments [41], it is relevant to include this parameter when assessing the gut microbiota variation.
SLE treatments may also influence the gut microbiota diversity [26, 39]. MRL/lpr mice treated with vancomycin after disease onset but not before [15, 71] showed improvement in lupus-like symptoms and decreased bacterial diversity [15], increasing Lactobacillus spp., and decreasing Lachnospiraceae [15]. The first has been linked to better outcomes [16]. Nonetheless, vancomycin does not have the same impact on all Lactobacillus species, as it decreases L. animalis, and L. intestinalis, while not changing L. frumenti [71]. Moreover, vancomycin treatment decreased fecal short-chain fatty acids, probably by reducing Clostridia [71]. Also, it decreased intestinal permeability, Gram-negative bacteria, and consequently, LPS levels [15]. The same beneficial effects were obtained with a mix of antibiotics but without decreasing the bacterial diversity [15].
Besides the alteration in bacterial composition, treatments can also alter the metabolic pathways associated with SLE. In MRL/lpr mice, treatment with vancomycin altered functional pathways just four days after its initiation [15]. This antibiotic downregulated genes related to lipid A biosynthesis, an endotoxic component of LPS, meaning that it can downregulate Gram-negative bacteria abundance by reducing LPS in circulation [15]. Other authors made similar observations regarding a Gram-positive gut commensal in NZB/NZWF1 mice [51]. In this work, higher gut permeability led to the translocation of Enterococcus gallinarum (E. gallinarum) to mesenteric lymph nodes, mesenteric veins, and liver at 16 weeks and spleen at 18 weeks. A reduction in the translocation of E. gallinarum, and a concomitant reduction in mortality, were achieved using vancomycin, ampicillin, and a vaccine against the pathobiont [51]. Moreover, vancomycin countered the upregulation of Th17 cells, which are involved in SLE pathogenesis [51]. E. gallinarum also down-regulated relevant molecules involved in maintaining the gut barrier integrity while up-regulating molecules involved in inflammation [51]. Once again, this study highlighted the tight connection between gut microbiota, intestinal barrier permeability, and inflammation.
Differences between treated and non-treated patients seem to exist [41, 42], such as with glucocorticoids [26] and antibiotics [15]. Some SLE treatments can further decrease the patients’ gut microbiota diversity [41].
Glucocorticoids’ increased Lactobacillus and Bifidobacterium [26], which is extremely relevant as Lactobacillus can be reduced in SLE patients, and Bifidobacterium was inversely correlated with disease activity [32]. Both Lactobacillus and Bifidobacterium produce short-chain fatty acids from dietary fiber fermentation, which have anti-inflammatory effects on epithelial and immune cells [26], improving the gut barrier function through this pathway. In fact, L. reuteri and L. rhamnosus strains are reported to improve the gut barrier function [15] and induce Treg cells [16]. Glucocorticoid therapy may also favor the reduction of F/B ratio and alter the gut microbiota structure [26].
PPI have been associated with an increase of oral microbiota bacteria in the gut [38], suggesting a possible translocation of these microorganisms and in fact, some oral bacterial species, such as A. massiliensis, S. satelles, Clostridium sp. ATCC BAA-442, B. fragilis, and C. leptum were more related to the gut bacterial composition of SLE patients than controls [41], meaning that they may contribute to the alteration of the gut microbiota by facilitating the translocation of oral microorganisms to the gut. In another study, patients with SLE not taking PPI showed different alpha-diversity than HC. However, when this medication was used, the alpha and beta-diversity of the patients were not significantly different from HC [39]. In addition, the PPI altered the gut microbiota composition, increasing the abundance of the genus Desulfovibrio, Oxalobacter, Roseburia, Streptococcus, and Lactobacillus and decreasing the abundance of Veillonella, Escherichia, Pseudomonas, Stenotrophomonas, and Morganella when compared with patients not taking this medication. In summary, PPI brought the microbiota of SLE patients closer to the one of HC [39].
This review has some limitations. The search was limited to two databases and articles written in English. The studies included in this review were very heterogeneous in population, models, and methods, limiting broad conclusions. Moreover, the studies that included SLE patients were limited to a few countries, which is particularly relevant in the context of the gut microbiota, as geographic and cultural habits have a relevant impact on its composition. Nevertheless, we provided a perspective of the main findings regarding the gut microbiota in SLE, contributing to a better understanding of what is currently known in this field and helping to define the current research agenda.
Diverse and, in some cases, contradictory results were found in the studies included in this review, possibly due to the heterogeneity of the lupus-mouse models and populations, the latter regarding ethnicity, disease activity, chronicity, medication, sample size, and gender ratio. Nevertheless, most studies found gut microbiota dysbiosis in SLE, but it remains to clarify if it is a cause or a consequence of the disease or its treatments. The impact of specific bacterial strains is also unclear, and the specific mechanism through which the gut microbiota participates in SLE needs to be further studied. Even so, it seems clear that the gut microbiota has a role in the pathogenesis of SLE, either directly or through its metabolic functions. Thus, finding new treatment approaches targeting the gut microbiota may lead to beneficial outcomes.
Further studies with larger and well-characterized populations will undoubtedly contribute to clarifying some of the remaining questions and deciphering the role of gut microbiota in SLE development, progression, and outcomes.
B.: Bacteroides
BMI: body mass index
dsDNA: double-stranded DNA
F/B ratio: Firmicutes/Bacteroidetes ratio
GWAS: genome-wide association study
HC: healthy controls
IgG: immunoglobulin G
IL-10: interleukin 10
LN: lupus nephritis
LPS: lipopolysaccharides
MRL: Murphy Roths large
NZB/W: New Zealand black x New Zealand white
PPI: proton pump inhibitors
R.: Ruminococcus
SFB: segmented filamentous bacteria
SLE: systemic lupus erythematosus
SLEDAI: systemic lupus erythematosus disease activity index
TC: triple congenic (B6.Sle1.Sle2.Sle3)
Th17: T helper 17
TJ: tight junctions
The supplementary material for this article is available at: https://www.explorationpub.com/uploads/Article/file/1001112_sup_1.pdf.
IAC: Conceptualization, Writing—original draft. PCR: Conceptualization, Writing—original draft. CSG: Writing—review & editing. JEF: Writing—review & editing. All authors contributed to the final version of the manuscript, read, and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2022.
Copyright: © The Author(s) 2022. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
