Affiliation:
1Biotechnology in Health and Environmental Sciences Research Laboratory, Natural Sciences College, Autonomous Guerrero State University, Chilpancingo, Guerrero 39070, Mexico
Email: rockdrig@yahoo.com.mx
ORCID: https://orcid.org/0000-0002-4695-7129
Affiliation:
1Biotechnology in Health and Environmental Sciences Research Laboratory, Natural Sciences College, Autonomous Guerrero State University, Chilpancingo, Guerrero 39070, Mexico
ORCID: https://orcid.org/0000-0002-7275-7120
Affiliation:
2College of Medicine, Autonomous Guerrero State University, Av. José Francisco Ruiz Massieu S/N, Colonia Insurgentes, Acapulco, Guerrero 39300, Mexico
3Genomol Laboratory, Costera Miguel Aleman, Acapulco, Guerrero 39640, Mexico
ORCID: https://orcid.org/0000-0002-1027-8028
Affiliation:
1Biotechnology in Health and Environmental Sciences Research Laboratory, Natural Sciences College, Autonomous Guerrero State University, Chilpancingo, Guerrero 39070, Mexico
ORCID: https://orcid.org/0000-0002-1038-9975
Explor Immunol. 2023;3:1–16 DOI: https://doi.org/10.37349/ei.2023.00085
Received: September 05, 2022 Accepted: November 23, 2022 Published: February 24, 2023
Academic Editor: Pierre-Antoine Gourraud, Public Health Université de Nantes, France
Immunoinformatics is an emerging area focused on development and applications of methods used to facilitate vaccine development. There is a growing interest in the field of vaccinology centered on the new omic science named ‘vaccinomics’. However, this approach has not succeeded to provide a solution against major infections affecting both animals and humans, since tick vaccines are still being developed based on conventional biochemical or immunological methods to dissect the molecular structure of the pathogen, looking for a candidate antigen. The availability of complete genomes and the novel advanced technologies, such as data mining, bioinformatics, microarrays, and proteomics, have revolutionized the approach to vaccine development and provided a new impulse to tick research. The aim of this review is to explore how modern vaccinology will contribute to the discovery of new candidate antigens and to understand the research process to improve existing vaccines. Under this concept, the omic age of ticks will make it possible to design vaccines starting from a prediction based on the in silico analysis of gene sequences obtained by data mining using computer algorithms, without the need to keep the pathogen growing in vitro. This new genome-based approach has been named “reverse vaccinology 3.0” or “vaccinomics 1.0” and can be applied to ticks.
Parasitic diseases are an important problem in a changing world in which borders no longer exist, due to free trade agreements and free market policies between countries. The free circulation of pathogens affects animal health as well as food safety [1]. Parasites can be internal (endoparasites) or external (ectoparasites), this time we will be focused on the cattle tick Rhipicephalus (Boophilus) microplus, an external parasite, and the growing problems of multiple acaricide resistance around the world. Looking for a solution, different research groups have centered their interest on an omic-based research, looking for new candidates for vaccine development, to prevent tick infestations caused by tick strains resistant to multiple ixodicides [2].
The search and discovery of new candidates for vaccine development have been the result of the discovery of new technologies and the emergence of new fields of knowledge in parallel, such as the bioinformatics and functional genomics [3]. In the specific case of the cattle tick R. microplus, the integration of these fields of knowledge has made it possible to innovate and rediscover new lines of research based on reverse genetics and vaccinology approaches, which has facilitated the use of the genomes of various organisms in order to find new alternatives for the development of vaccines and diagnostic systems [4].
The word ‘vaccination’ was coined by Edward Jenner in 1796 to describe the injection of smallpox vaccine [5, 6]. Louis Pasteur developed the concept through his innovative work in microbiology. Now, vaccination is the administration of an antigenic component, applied to stimulate the immune system of an individual to stimulate the adaptive immunity against a disease. Vaccines can ameliorate, or prevent the effects of an infection and are generally accepted to be the most effective method to prevent infectious diseases [7], since the efficacy of vaccination has been extensively studied and verified [8–10]. However, the field of vaccinology remains empirical in many aspects, since, vaccine development, vaccine immunogenicity, and vaccine efficacy, have been historically driven by two empiric models, the “isolate-inactivate-inject” and the public health paradigm of “the same dose for everyone for every disease”. These two models were the predominant thinking during the pre-genomics era with regard to vaccine-preventable infectious diseases [11]. Preventable diseases represent a health threat worldwide, which can be avoided by protective and long-lasting vaccination. Vaccines prevent three million deaths every year by providing protection against relevant pathogens, affecting vulnerable populations including newborns, pregnant women, and elderly individuals as well as those patients affected by chronic or compromising medical conditions [12].
Vaccination as an effective prophylactic treatment has been demonstrated for over 200 years for bacterial and viral diseases. However, eukaryotic pathogens continue causing some severe diseases in both humans and animals, as they continually spread into non-endemic countries. Combined with the increasing number of reports of drug resistance and the lack of effective treatment programs, the impact that these organisms will have on life quality remains a global challenge [13]. From the earliest traditional variolation procedures to the state-of-the-art technologies currently employed, many protective candidates have been successfully developed in order to be used in medical and veterinary disciplines. Despite the successes of these technologies in the discovery of subunit vaccines against prokaryote pathogens, vaccines directed against metazoan parasites, have not been successfully developed. With currently developed omic technologies and metadata for eukaryotic parasites, molecular target discovery for vaccine development can be accelerated [13].
Biochemical, serological, and microbiological methods have been used to identify the molecular components, useful for the development of vaccines against parasites. Although this approach has been successful in many cases, it is time-consuming and fails when the pathogen cannot be cultured in vitro or when the sequence of the candidate antigen is variable. Currently used bioinformatic algorithms allow us to retrieve gene sequences from public databases, predict antigens and their physicochemical properties, three-dimensional structure, and allergenicity, regardless of their abundance and immunogenicity, furthermore, there is no need to culture the pathogen in vitro [14]. This facilitates vaccine development using non-conventional antigens or linear B or T cell epitopes predicted in silico. Since the process of vaccine discovery starts in silico by using the genomic information deposited in databases rather than the pathogen itself, many vaccines impossible to develop will become a reality. This novel process has been named “reverse vaccinology”, and is based on the increasing availability of public genome sequence databases (Figure 1). Its use has led to the development and application of technologies to vaccine discovery, including comparative genomics, transcriptomics, proteomics, immunomics, and structural genomics. Antigen candidates identified from a pathogen genome or proteome, can then be expressed as a recombinant protein and tested in an appropriate in vitro or in vivo model to predict and assess immunogenicity and protection. In the omic era, vaccines have had a significant impact on public health, and vaccinology is taking advantage of new technologies to develop vaccines that have not succeeded, since most of the existing vaccines were developed based on traditional vaccinology methods, which depended on the empirical detection of few candidates at a time, depending on the known characteristics of the pathogen [15]. However, the ability to sequence a pathogen’s genome provides access to its entire antigenic repertoire. Genomics has catalyzed a shift from a traditional vaccinology approach to a sequence-based “reverse vaccinology” development, using high-throughput in silico screening of public whole-genome databases in combination with bioinformatics algorithms, resulting in a great tool to identify protein-coding genes with most of the attributes of a good vaccine candidate (Figure 1) [15].
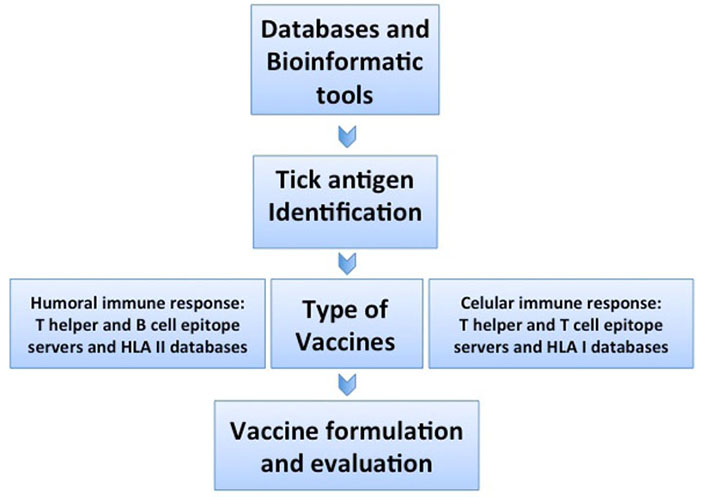
Reverse vaccinology pipeline starts using public databases to retrieve gene sequences of interest that can be processed in silico, in order to identify genes that encode proteins containing immunogenic linear B or T cell epitopes with the attributes of a good tick vaccine target. HLA: human leukocyte antigen
The aim of this review is to explore how modern vaccinology contributes to the discovery of new candidate antigens and to understand the research process to improve existing vaccines. These new concepts will revolutionize veterinary vaccinology, from development and manufacturing to administration, and how this process can be used to improve animal health, protect the environment, and reduce food contamination resulted from the intensive use of acaricides for tick control. Perhaps most exciting will be the increasingly complex high-throughput data generated by the application of systems biology and vaccinology approaches.
A vaccine is a biological product that can induce a protective response against a disease, infection, or a subsequent exposure to a pathogen by stimulation of the immune system to produce neutralizing specific antibodies. However, a wide range of dangerous pathogens are out of control, via conventional vaccination strategies. Recent advances in molecular biology, immunology, genetics, biochemistry, and bioinformatics have provided new alternative routes to vaccine development (Figures 1 and 2).
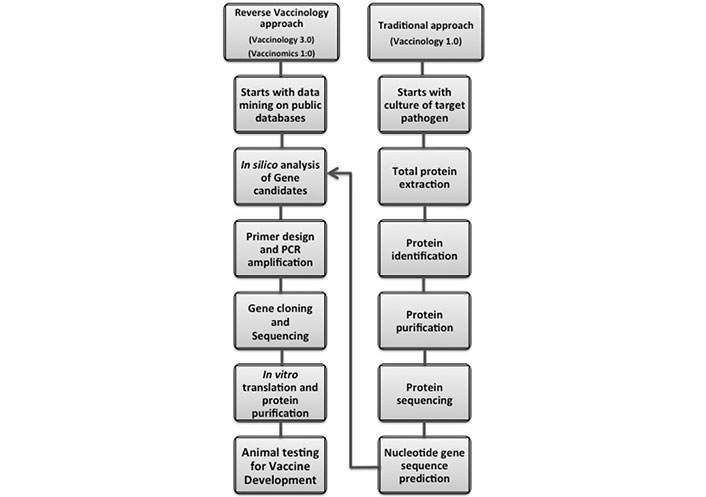
Pipeline showing how recent advances in molecular biology, immunology, genetics, systems biology, and bioinformatics have shortened the alternative routes to vaccine development, facilitating the transition from traditional to the reverse vaccinology. PCR: polymerase chain reaction
Those innovative new concepts on vaccine development are currently urgent issues in a changing world, in which the immigration and globalization phenomena have led us to the emergence and reemergence of infectious diseases at the animal-human interface [16], shaping the future of a new vaccinology concept and moving forward, into the personalized vaccine design and administration, as well as in the precision medicine field. Precision medicine approaches are based on pharmacogenomics and have been successfully implemented, in order to predict more efficient treatments and prevention strategies based on the genetic background of the patient. This approach has already been proposed for vaccines, since it has great potential to address public concerns about vaccine safety and to promote increased public confidence, higher vaccination rates, and fewer serious adverse events in genetically predisposed individuals [17].
Vaccine development following the empiric Pasteurian paradigm (isolate, inactivate, and inject), also called “vaccinology 1.0”, requires the identification inactivation and injection of the disease-causing pathogen or its disease-mediating entity (e.g., a toxin) in order to elicit a protective immune response [18–20]. This conventional and empiric method was developed before the omics revolution but enabled the discovery and development of effective vaccines against pathogens such as influenza, tetanus, diphtheria, pertussis, smallpox, measles, mumps, and rubella.
Vaccinology 1.0 was officially born in 1796 thanks to the contribution of Edward Jenner. The typical vaccine produced under the concept of vaccinology 1.0 is the anti-rabies virus vaccine, the first human vaccine manufactured in the laboratory in 1885 [21]. Other “first generation” vaccines are: the Bacillus Calmette-Guerin, pertussis, polio, and smallpox [22]. Basically, as we already mentioned, vaccinology 1.0, consisted of isolation, cultivation, inactivation, and injection of total or partial components of infectious agents.
Vaccinology 2.0 was based on the use of the purified pathogen cell components. The transition from vaccinology 1.0 to vaccinology 2.0 was possible due to great technological advancements, including genetics, protein engineering, recombinant DNA [23], polysaccharide/carbohydrate chemistry, and combinatorial chemistry [24] among others. Examples of “second generation” vaccines include vaccines against tetanus, diphteria, anthrax, pneumoniae, influenza, hepatitis B, and Lyme disease [22]. Entering the twenty-first century, vaccinology has shifted from a classical discipline based on the Pasteurian principle of “isolate, inactivate, and inject” to a new paradigm, characterized by a rational and scientific design [25].
Vaccinology 3.0 started from the pathogen genomic sequences or from the repertoire of protective human antibodies (reverse vaccinology 2.0) [26, 27]. This shift was possible thanks to new and different dimensions of omics data, such as high volume, velocity, and a wide variety of data [28]. New cutting-edge high-throughput technologies, enabled genomic and post-genomics emerging disciplines such as: transcriptomics, proteomics, metabolomics, cytomics, immunomics, secretomics, surfomics, or interactomics (Table 1) [19, 29, 30].
Some omic terms definitions frequently used in this review
| Relevant disciplines in vaccinology | Definition |
|---|---|
| Genomics | Genome-wide investigation of genes |
| Proteomics | Proteome-wide investigation of proteins |
| Transcriptomics | Transcriptome-wide investigation of transcribed genes |
| Metabolomics | Metabolome-wide investigation of metabolites |
| Cytomics | Cytome-wide investigation of biochemical events at a single cell level |
| Immunogenomics | Immunogenome-wide investigation of immunological relevant genes |
| Immunoproteomics | Immunoproteome-wide investigation of immunologically relevant proteins |
| Immunometabolomics | Immunometabolome-wide investigation of immunologically relevant metabolites |
| Interactomics | Interactome-wide investigation of interactions among proteins and/or other molecules or components |
| Secretomics | Secretome-wide investigation of all secreted proteins of a given cell, tissue, |
| or organism | |
| Exoproteomics | Exoproteome-wide investigation of proteins in the extra-cellular proximity of a biological system |
| Surfomics | Surfome-wide investigation of surface proteins and other components, such as surface-exposed moieties |
| Immunomics | Immunome-wide investigation of the immune system regulation, and response to a given pathogen |
| Adversomics | Adversome-wide investigation of potential vaccine-related adverse effect |
| Vaccinomics | Comprehensive integration of previously described omics disciplines, for advancing vaccine discovery and development, as well as personalized vaccinology |
Post-genomic specialties interaction has converged in a sort of bioinformatic data-driven holistic investigation currently known as “vaccinomics”, defined by Poland and collaborators as the “integration of immunogenetics and immunogenomics with systems biology and immune profiling” [31]. The immunoinformatics and computational biology played a central role in the evolution from classical vaccinology to vaccinomics, since the access to public databases required the use of powerful bioinformatic algorithms to achieve the highest prediction accuracy of three-dimensional protein structure, protein folding rates, stability of proteins based on mutations, mapping epitopes, and protein localization, as well as some other features prediction methods and measures of prediction performance, desirable for vaccine development (Figure 2 and Table 2).
Search tools, bioinformatic web server names, and URLs necessary to predict subcellular localization, protein structure, immunogenic characteristics, and some other desirable features for vaccine development
The popularity of new emerging disciplines for the identification of new vaccine candidates, by using immunomics or immunoinformatics, started in 2007 and they were composed of a combination of reverse vaccinology with computational epitope mapping (Table 2) [32]. The post-genomic era brought new research approaches in different areas of human and veterinary medicine. One of these new approaches is the emerging discipline currently called ‘vaccinomics’, which is based on the search for genes deposited in databases generated by research groups or scientists, dedicated to deciphering the genomes of diverse organisms (Figure 3) [3].
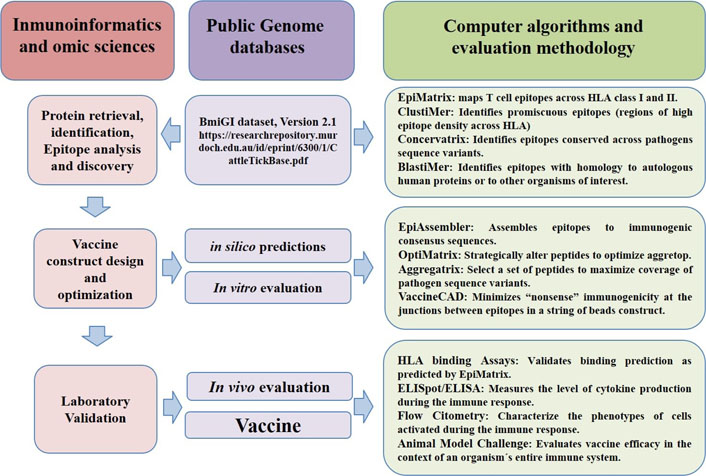
Analysis of public genome databases with immunoinformatics methodologies, omic sciences and computer algorithms have shortened the road to tick vaccines discovery, going from protein retrieval identification and in silico analysis to vaccine design and animal model challenge validation. ELISA: enzyme-linked immunosorbent assay; ELISpot: enzyme-linked immunosorbent spot
Vaccinomics is based on the use of cutting edge high-dimensional experimental and novel bioinformatics approaches to the development of next-generation vaccines. Vaccinomics is based on the use of cutting edge high-dimensional experimental and novel bioinformatics approaches to the development of next-generation vaccines. Vaccinomics will allow us to move faster, beyond the empiric Pasteurian approach, “isolate, inactivate, and inject”, characterizing past vaccine development efforts, and toward a more detailed molecular understanding of biological processes involved in developing an anti-tick vaccine-induced immunity (Figure 2). This enhanced knowledge will then be applied to overcome the obstacles to the creation of an effective anti-tick vaccine or to protect cattle against pathogens, with the greatest current impact on public health [33].
The discovery of new vaccine candidates to control tick infestations and transmission of pathogen infections requires the development of effective screening platforms and algorithms that allow the analysis and validation of data produced by systems biology approaches [34]. Tick vaccines could be used to vaccinate human and animal populations, to reduce host exposure to infected ticks and transmission of pathogens affecting human and animal health worldwide. The search for new vaccine candidates in the post-genomic era invariably begins with the in silico genome research of the organism of interest or specific genes, through the use of specialized bioinformatic algorithms (Figures 1 and 2, Table 2).
These algorithms can be assigned from the putative protein function to the prediction of useful characteristics of antigenic sequences for vaccine development [35]. These gene sequences can be later analyzed or amplified by in vitro synthesis of nucleic acids, more commonly known as PCR for its acronym in English, polymerase chain reaction. PCR amplification products from nucleic acid templates [genomic DNA or complementary DNA] from the organism of interest can be cloned and expressed in vectors whose characteristics may vary depending on the interest of the study or research purpose [36].
This approach denotes the benefits of the process seen from the pragmatic point of view of the system as a whole, since the maintenance of the living organism is not required up to this point. The strategy is already a saving in terms of time and work invested in the process (Figure 2), although its success still depends on the accuracy of the algorithms used to predict the candidate antigens immunogenic properties [37].
This is always a problem since some proteins, even when they share structural and biological characteristics, they may have no obvious similarity in sequence and for this reason, the discovery of truly new antigens can be frustrated since the antigenicity of a sequence can be encoded by such a subtle way, that it may not be identified by the alignment of the sequence alone, due to this absence of similarity with the antigens of known origin [38]. These problems are being corrected by the development of bioinformatics, which, like genomics, has recently had impressive advances.
Ticks and mosquitoes are considered to be envolved worldwide as vectors of animal and human diseases and vaccines are a friendly and cost-effective alternative to prevent tick infestations and transmission of tick-borne diseases. These premises stress the need to improve tick vaccines, in order to prevent tick-borne diseases, and therefore tick vaccinology will be significantly accelerated by using vaccinomic approaches, starting from the study of the tick-host-pathogen interactions to the characterization and validation of vaccine formulations.
The intensive use of chemical control of ticks around the world has led to major drawbacks, the selection of acaricide resistance genes and the appearance of resistant tick phenotypes within populations in addition to the food and environmental contamination by acaricide residues. The problem of acaricide resistance has motivated the interest in tick vaccine development, as an alternative to prevent the parasitism caused by these arthropods and at the same time to block the transmission of diseases to their hosts. Ticks, unlike other ectoparasites, feed on blood slowly, remaining attached to the host for several days, this biological behavior puts them in contact with the immune response components, generating a natural interface between the intestinal epithelium of the tick and the host’s antibodies [39].
The use of anti-tick vaccines is currently seen as an alternative method to control ticks that promises to be viable, not only to mitigate the acaricide resistance, but to prevent tick infestations, tick-borne diseases and therefore the improvement of animal health, environmental pollution, and reduction of contaminants on animal products for human consumption, derived from livestock operations [2, 40]. Historically, the study of useful tick proteins to be developed as immunogens starts with the identification of limiting factors that interrupt vital functions, such as tick attachment, tick feeding, water balance, blood digestion, and the physiology of tick-host interphase.
Vaccines developed during the pre-genomic era (also called vaccinology 1.0) were based on the use of dead, live, or attenuated organisms, or the use of one or more purified or semi-purified proteins obtained from total extracts of the organisms of interest [41]. To develop this type of vaccine, a critical step is necessary, the identification of the proteins of interest and the elimination of others that are not useful. In this case, an antigen is recognized as a protein capable to induce immune responses and provide protection, against subsequent challenges against the microorganism, or parasite in question [42, 43].
The empirical approach for the development of subunit vaccines includes several steps: a) the cultivation of the parasite, microorganism, or pest to be controlled, b) the analysis and purification of its components, c) the identification of the antigen that possesses the immunogenic characteristics required for the final product development, and d) the subsequent challenge by the infectious agent or parasite against which the vaccine has been made, in order to evaluate the immunogenic characteristics of this technology in an appropriate animal model (Figure 3) [44, 45]. This methodology has inherent difficulties in the purification process and identification of the fractions with the optimal antigenic characteristics for vaccine development, as well as the availability of the macro or microorganism to be controlled by this biotechnological tool, since the production of the vaccine is severely limited when the target organism cannot be easily cultured [46]. There are other drawbacks associated with the biology of the target organism, since in some cases the most abundant proteins are not necessarily immunoprotective, or it may be the case that the antigens expressed during the in vivo infection or infestation, are not the same as those expressed during in vitro culture, the latter may not be the case of the R. microplus tick [47].
Commercially available vaccines against cattle fever ticks, including TickGARD (Hoechst Animal Health, Australia), TickGARDPLUS (Intervet Australia, Australia), and Gavac® (Heber Biotec, Havana, Cuba), are based on different recombinant allele forms of the concealed midgut antigen Bm86, developed in the early 1990’s, to prevent cattle tick infestations and subsequently registered in Australia and Latin American countries respectively [38, 48]. These vaccines showed the advantages of being a cost-effective and environmentally friendly alternative with a dual effect to reduce tick infestations and transmission of disease-causing pathogens [49]. The protective mechanism characterized for Bm86 tick vaccines (and the homolog Bm95 gene), is based on its capability to induce the production of antigen-specific antibodies in immunized cattle that subsequently affect the biological function of the targeted antigen located on the surface of the intestinal tick cells, impairing tick feeding and blood digestion in the immunized hosts. The application of the Bm86/Bm95-based vaccines also showed a reduction in the number of engorged female ticks, their weight, and reproductive capacity, although the greatest vaccine effect was the reduction of larval infestations in subsequent generations [48].
The Bm86 antigen, derived from tick intestines, has been the most successful tick antigenic component used to formulate a vaccine [48]. It is a protein anchored to the membrane surface of intestinal cells, which has been analyzed, cloned, expressed, and developed as the classic example of a commercial vaccine. As we already mentioned, both commercial tick vaccines, TickGARD [50], and Gavac® [51], have been derived from the Bm86 homologous genes from Australian and Cuban tick strains respectively. Bm86 gene was used in Argentina with the name of Bm95 [52], although it is actually recognized as an allelic variant of the same gene, it was used to solve the differences in immunity observed in R. microplus populations declared susceptible and resistant to immunization with this antigen [53]. Based on these observations, different degrees of susceptibility to vaccination associated with variations in the Bm86 gene sequence were published [54, 55]. The results of the experiments using the Bm95 gene suggested that it was a universal antigen that solved the problem of populations susceptible and resistant to vaccination with the original antigen, however, what is clear is that the real difference, was the use of a local native gene, since the alignment of the Bm95 gene sequence with the Bm86 sequences shows that it is a variant of the same gene. Pointing out the importance of using native antigens to improve the efficiency of tick vaccines, especially when vaccines are based on a single gene sequence.
The identification of the antigen Bm86/Bm95 on intestinal cells of the tick R. microplus is the first example of antigens used for tick control. The response to vaccination with these antigens produces antibodies that cause lysis of intestinal cells, resulting in a reduced survival, a decrease in body weight, and fertility of the engorged females [56, 57].
The advancement of omic sciences has allowed the incorporation of massive techniques for the analysis and identification of useful proteins, by immunization assays of expression libraries (ELI) [58]. Recently, gene silencing as a functional genomic tool, which in addition helps us to predict the putative function of the proteins encoded by the genes of interest, allows us to observe the effect of gene silencing in ticks. This allows us to make inferences about the importance of the protein function in the arthropod’s physiology, such is the case of the “subolesin” gene which was discovered in genetic material of Ixodes scapularis as a protective antigen and is currently known to be an evolutionary conserved gene that has also been identified in Aedes albopictus [59]. Mosquitoes and ticks subolesin sequences are known from multiple alignments to maintain conserved epitopes, and vaccination experiments have shown that immunization with the orthologous mosquito and tick subolesin proteins significantly reduces infestation and tick weight of Ixodes scapularis in the same way. For this reason, there are preliminary results that point out the development of vaccines designed for the control of multiple arthropod vectors using the orthologous proteins of subolesin, although the use of multiple antigens is suggested to produce an effective vaccine [59].
Genomic-based studies have been conducted in different tissues and organs of different tick species, trying to enhance the understanding of tick-host interaction, and as an important scientific methodology to assist the identification of novel candidate antigens for tick vaccine development. The sialotranscriptome of Amblyomma americanum [60–62], Dermacentor variabilis [62], Ixodes scapularis [63, 64], Ixodes ricinus [65], Dermacentor andersoni [66], and R. appendiculatus [67] have been determined. R. microplus transcriptomes of larvae [68–72], engorged female gut [73], ovary [74], and synganglia [75] have been also recently reported. The full-length coding sequences of numerous proteins have been described based on these genomic approaches and their roles during host-parasite interaction.
Mulenga et al. [76] in 1999, identified a 29 kDa protein by immunological mapping of antigens from salivary glands of Haemaphysalis longicornis. Immunization of rabbits with the recombinant protein p29 conferred a 40% and 56% reduction in engorgement and mortality of larvae and nymphs respectively. The p64 antigen is a 15 kDa protein that was identified in the three-host tick Rhipicephalus appendiculatus, the putative function of this protein is related to cement and its role in tick adherence and feeding. The effect of immunization with this protein in guinea pigs infested with nymphs and adults was to decrease the infestation of 48% and 70% respectively [77].
Among the list of antigens, there are some candidate genes that participate in the host-parasite tick-feeding interface and have currently been rediscovered from the analysis of proteomic maps of saliva [78]. The putative biological functions of some of these sialome derived proteins have been related to: a) suppression of the host antibody response, via an immunoglobulin binding-protein, b) inhibition of host hemostatic responses, via a thrombin inhibitor, c) the possible destruction of host extracellular matrix, necessary for the formation of the feeding pool via a metalloprotease, and d) attachment of the tick to its host via a glycine rich cement protein [79]. Experimental immunization trials were carried out using a breed of Holstein calves, highly susceptible to tick infestations. A group of calves were vaccinated with the four tested antigens, and a significant reduction of tick infestation and an efficacy of 73.25% was found when they were experimentally exposed to larval tick bites, by infestation with a R. microplus tick strain. These results showed that a reverse vaccinology pipeline guided by different levels of anti-tick immunity will be a powerful strategy for the identification of promising antigens that can boost host immunity during natural infestations, and that salivary exposed antigens [79] or any other potential candidate, can be used to improve current anti-tick vaccines, or produce a new one by using vaccinomic approaches.
The most important limiting factor in the development of tick vaccines is the identification of antigens, since they have traditionally been identified by evaluating proteins derived from crude extracts used to immunize animals that are subsequently subjected to massive experimental infestations with ticks. The purification of a protein from a total extract is a long and laborious process, therefore, at present, the use of databases containing genomes facilitates the reverse process of determining the putative function of the genes of interest and to observe the effect of immunization with proteins considered important for its function and survival in vaccination experiments against ticks (Figure 2) [80].
There are two basic immunological concepts that must be considered for the identification of an effective tick protective antigen, a) a sufficient amount of the produced antibodies must have access to the tick target antigen and b) the formation of the complex antigen-antibody should be able to disrupt the normal function of the tick target protein [81]. These basic concepts are necessary to the best selection of genes encoding the tick protective antigens. To fulfill these criteria, a protective antigen must be exposed to host antibodies, and their interaction should disrupt an important protein function.
The general pipeline starts with sequence protein retrieval, the protective antigen identification (subcellular localization and secretion proteins), B cell epitope prediction, vaccine construct design, PCR amplification, gene cloning and laboratory validation (Figure 3).
The protective antigen identification is the starting point of the experimental strategy to develop a vaccinomic pipeline. The algorithm can include the generation of a proteomic or transcriptomic dataset or start from public genomic databases, containing genome sequences for datamining. There are some desirable characteristics of protective antigens that can be predicted by using different algorithms such as WoLF PSORT, CELLO, Protein Prowler, TargetP, SLP-Local and PredSL, used for analysis of genes coding for secreted proteins. Gene ontology (GO) analysis of cellular components, molecular function, and biological process, is conducted using the complementary DNA annotation system software (The Office of Technology Information Systems, National Institute of Allergy and Infection Diseases), AmiGO and UniProt, using non-redundant sequence databases and tick specific sequence databases, in order to obtain the GO annotations for selected candidate tick protective antigens. To characterize the subcellular localization of proteins encoded by selected gene candidates, it is necessary to analyze the proteomic or transcriptomic dataset or the non redundant sequence dataset obtained from public tick databases, in order to generate a dataset of candidate antigens to be analyzed for antigenicity, and to balance the variations in epitope length, as well as the physico-chemical properties of antigens. The most commonly used linear epitope predictors are: BepiPred, ABCpred, and Bcepred, (Table 2). Once, the linear B cell epitope aminoacid sequence has been predicted, synthetic or recombinant peptides can be used to validate the vaccine preparation in the laboratory. Coding sequences containing the linear B cell epitopes can be amplified by reverse transcription-PCR and expressed in E. coli, to be used in a recombinant peptide vaccine preparation.
It is important to point out that multiantigen vaccines have been proposed as an alternative, based on the premise that many antigens will provide better protection, although in practice there are many experimental examples demonstrating that this is not entirely true, since “the results have been limited and contradictory, that is why it is suggested that a greater attention should be paid to the efficacy of multiantigenic vaccines, since they must have a greater experimental support than they have so far deserved” [42]. At this time, the use of bioinformatics and omic sciences will allow us to use public databases with biotechnological purposes, in order to identify new candidates that can be redesigned, for the improvement of existing vaccines, through vaccinomic approaches. The development of tick vaccines will establish the difference of an excellent integrated control system in the near future.
Research progresses in omic sciences and emergent disciplines will have a direct impact on the discovery of new candidate targets for the development of recombinant vaccines resulted from vaccinomic approaches.
Functional genomics and reverse vaccinology are emerging omic disciplines, whose applications in the discovery of new candidate genes for vaccine development have recently made great contributions, for instance, the RNA interference (RNAi) technology used to silence genes allowed us to know, how genes silencing affects the expression or function of the gene of interest. In this way, the gene encoding the subolesin protein was discovered because of the associated effect of gene silencing on tick reproduction, that is why it is currently being studied to be used as an alternative for tick control.
The emerging field of vaccinomics is a promising field in terms of the search for new antigenic molecules that allow us to advance faster on the route to vaccine development, since the availability of technology to sequence complete genomes of important parasites has had an impressive advance, in parallel with the increasing amount of information deposited in public genome databases.
The search for new candidate proteins to develop recombinant vaccines has been the results of the application of new technologies and the emergence of newly developed disciplines, such as bioinformatics, molecular genetics, systems biology, functional genomics, gene editing, as well as their interactions. The integration of these emerging fields of knowledge will allow scientists to innovate and rediscover new experimental routes based on data mining strategies in combination with reverse genetics and vacunomics approaches, making possible the use of public genome databases in order to identify new candidates for tick vaccine development and diagnostic systems.
Rough work, difficult as it may be, has already made great advances, the list of genes cloned and analyzed is already large, and experiments have shown that genes like Bm86, and a growing number of orthologous genes can be used to control ticks. The future in the field of vaccine development is getting shorter, and the scope of the modern technology in the field of the omic sciences is becoming wider, thus the landing of knowledge regarding the tick vaccines development used for tick control, It is very close to bearing fruit due to the growing list of new antigens discovered, although the tick control still represents a challenge for the scientific community in Mexico and all over the world.
PCR: polymerase chain reaction
We greatly appreciate the support received from Dr. Consuelo Almazán-García for her constructive comments made on the original draft of this manuscript.
RRC: Conceptualization, Formal analysis, Writing—original draft. DIDG: Formal analysis, Writing—original draft, Writing—review & editing. SLS: Formal analysis, Writing—original draft, Writing—review & editing. FRD: Resources, Writing—review & editing, Visualization.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
