Affiliation:
Engineering Research Center of Cell & Therapeutic Antibody (MOE), School of Pharmacy, Shanghai Jiaotong University, Shanghai 200240, China
Email: mabuyong@sjtu.edu.cn
ORCID: https://orcid.org/0000-0002-7383-719X
Explor Immunol. 2021;1:27–36 DOI: https://doi.org/10.37349/ei.2021.00004
Received: January 15, 2021 Accepted: February 23, 2021 Published: April 30, 2021
Academic Editor: Dominique J Charron, Hôpital Saint-Louis, France; Reem Al-Daccak, Hôpital Saint-Louis, France
Aim: Fragment crystallizable (Fc) glycans modulate Fc conformations and functions, and glycan may also regulate antigen recognition. In the antibody drug development, glycosylation patterns affect antibody drug characteristics and quality control. In order to provide a global feature of N-glycan interactions in response to antigen and Fc receptor bindings, the interactions among Fc N-glycans and N-glycans’ interaction with Fc CH2 and CH3 domains have been studied.
Methods: Molecular dynamics simulations were used to generate conformation ensembles of free antibody, antibody-antigen complex, antibody-human Fc-gamma-receptor-I (hFcγRI) and antibody-antigen-hFcγRI, the hydrogen bonds and radial distance distribution involving N-glycans carbohydrate chains have been analyzed.
Results: Two important interaction patterns have been observed. The first is the strong but non-specific interactions between two carbohydrate chains in free antibody. Secondly, it has been found that N-glycans carbohydrate chains can directly interact with CH3 domain in free antibody, and that the distance distribution between carbohydrate chains and CH3 domain clearly differentiate the free antibody, antibody-antigen complex, antibody-hFcγRI complex, and final antibody-antigen-hFcγRI complex.
Conclusions: N-glycans partially acts as allosteric sensor and respond to antigen and hFcγRI binding.
Three structural elements [fragment antigen-binding (Fab), fragment crystallizable (Fc), and glycan] synergistically determine antibody activity. The specificity and affinity of antibody-antigen recognition is mainly decided by the variable domains, especially complementarity-determining regions (CDRs). The recognition process involves conformational transitions mediated by its inherent flexibility. The Fc domain contains CH2 and CH3 domains. The CH2 domain mainly interacts with Fc receptors (FcRs) on the cell surface and plays pivotal roles in immunological responses. The Fab and Fc domains are connected by a flexible unstructured hinge region. Fc chains have conserved glycosylation sites at Asn297 (N-glycan) for immunoglobulin G (IgG)1, each is conjugated to a core oligosaccharide, differing from a complete biantennary dodecasaccharide (di-sialilated, core fucosylated form) to the minimal pentasaccharide (mannose terminating, not fucosylated). Fc glycans modulate Fc conformations and functions [1, 2]. The glycans help maintain the quaternary structure and Fc stability [3], and thus Fc-FcR recognition [4–7]. Deglycosylation of G0F in IgG1 resulted in a 40-fold loss in Fc-gamma-receptor-I (FcγRI) binding [4]. Glycan may also regulate antigen recognition, and it has been reported that core fucosylation of IgG B cell receptor is required for antigen recognition and antibody production [8]. In antibody-drug development, glycosylation patterns affect antibody-drug characteristics and quality [9].
The noncovalent interactions of multiple Fc domain residues with the N-glycan are necessary for optimal recognition of FcγRI [10]. Single amino acid mutations of these Fc residues affect glycan processing [10–12]. In the apo form, terminal carbohydrate N-glycans residues are flexible: α(1–3)Man-linked branch is usually unconstrained, while the α(1–6)Man-linked branch has two states, free and bound to nearby Fc domain polypeptides [13, 14]. N-glycans dynamics are crucial in FcR interactions and enzymatic glycan remodeling [13]. The composition of N-glycans can modulate the binding affinity of IgG1 Fc to FcγRs [15–18]. Lee et al. [19] reported that the C’E loop and the CH2-CH3 orientation are dynamic, and changes in N-glycan composition optimize the interface with the Fc receptor. X-ray crystallography and nuclear magnetic resonance (NMR) data indicated that the two arms of N-glycan are either in the bound state (attached to the Fc) [20] or in the free state (detached to the Fc) [13]. N-glycans may directly interact with the human FcγRI (hFcγRI) D2 domain [4], but do not show direct contact with hFcγRI in a high resolution structure [21].
Computational approaches have been used to understand the effects of glycan on various antibody properties. Sakae et al. [22] used replica exchange molecular dynamics (REMD) simulations to study core fucosylation of IgG1-Fc N-glycans, and they found that increased conformational fluctuation of the N-glycan at Asn162 of sFcγRIIIa upon fucosylation of IgG1-Fc. Combination of molecular dynamics (MD) simulations and small-angle X-ray scattering and nuclear magnetic resonance spectroscopy suggested that the N-glycans restrict the motional freedom of CH2 and endow quaternary-structure plasticity through multiple intramolecular interaction networks [23]. Our previous study has verified that antigen binding allosterically promotes Fc receptor recognition [24]. The analysis of extensive MD simulations found that the energy landscape of antibody may play decisive role in coordinating antibody conformation change. The above previous studies focused on the interaction of N-glycan with residues on Fc CH2 domain. In order to provide a global feature of N-glycan interactions in response to antigen and Fc receptor bindings, here we extend our investigation to interactions among Fc N-glycans and with Fc CH3 domain.
Starting free antibody conformation is adapted from the Protein Data Bank (PDB) structure ID: 1IGT [25]. The antigen is β-amyloid (Aβ) peptide and its binding with antibody is adapted from solanzumab crystal structure PDB ID: 4XXD [26]. Antibody-FcγRI complex is adapted from PDB ID: 4W4O [21]. As described earlier [24], initial antibody random conformations were generated by adjusting three sets of torsion angles. 231C-232N-232CA-232C, 232N-232CA-232C-233N, and 232CA-232C-233N-233CA (numbering in 1IGT), each step with 60° rotation. During the conformation randomization, the Fc domain was fixed and the Fab domains move freely, leading to 216 conformations. Excluding conformations with closed Fab domain or with Fc domain clashes, 12 conformations were selected as the starting points for the simulations. Twelve independent MD simulations of antibody with 12 different initial conformations have been performed. A total of 160,000 structures were generated and 1,600 conformers were used for antibody and antibody-antigen complex analysis, respectively. In the complexes between hFcγRI and antibody, 4 out of 12 representative conformations are selected to avoid clashes between Fabs and hFcγRI. After four independent MD simulations of antibody-hFcγRI complexes, 77,500 conformations were saved, and 1,550 conformers were used for antibody-hFcγRI and antibody-antigen-hFcγRI complexes, respectively. Visual molecular dynamics (VMD) program was used to calculate radial pair distribution and root-mean-square deviation (RMSD) trajectory analysis.
In the MD simulations, the systems were solvated by transferable intermolecular potential 3 point (TIP3P) water molecules, and sodium and chlorides were added to neutralize the system and to achieve a total concentration of ~ 150 mM. The systems were energy minimized for 5,000 conjugate gradient steps, where the protein was fixed and water molecules and counterions could move, followed by additional 5,000 conjugate gradient steps, where all atoms move. In the equilibration stage, each system was gradually relaxed by a series of dynamic cycles, in which the harmonic restraints on proteins were gradually removed to optimize the protein-water interactions. All MD simulations were performed using the using the NAnoscale Molecular Dynamics (NAMD) software [27] with CHARMM36 force field [28]. All simulations are carried out in the isothermal-isobaric (NPT) ensemble with a temperature of 36.85°C and a pressure of 0.1 mPa. MD trajectories were saved by every 2 ps for analysis.
It is interesting to see how glycan conformation changes in the crystal structures of a full antibody (PDB code: 1IGT) and Fc-hFcγRI complex (PDB code: 4W4O). Both structures have the G1F glycan sequence (the top panel of Figure 1), with fucose connected to core. The 1IGT is an intact, anti-canine lymphoma monoclonal antibody (Mab231) structure [25], with an Fc fragment having a nearly exact, local 2-fold symmetry. The glycan in the 1IGT is known to be very similar to those of the free human Fc fragment. In the Fc-hFcγRI complex (PDB code: 4W4O) structure [21], Fc undergoes rigid-body displacement of the CH2 domains, but not the CH3 domains, upon engagement with the receptor [21]. After superimposing the backbone of the Fc domain protein chain, it can be seen that glycan conformations in the superimposed right chain are very similar.
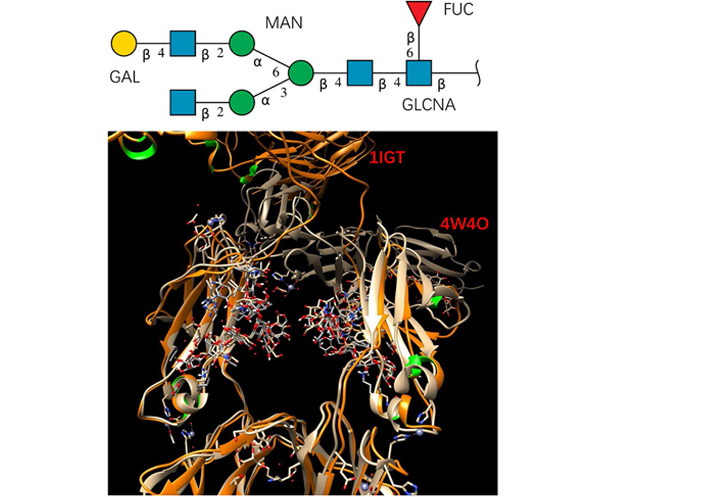
The studied glycan has stable conformation in free antibody and in the Fc-hFcγRI complex. Top panel: glycan sequence. Bottom panel: superimposing the crystal structures of a full antibody (PDB code: 1IGT) and Fc-hFcγRI complex (PDB code: 4W4O). Proteins are represented by ribbons, and the glycans are shown as sticks. GAL: Galactose; MAN: mannose; FUC: fucose
The glycan conformations in free antibody in MD simulations are much more flexible than in the static crystal structures. For each antibody system (free antibody, antibody-antigen complex, antibody-hFcγRI complex, and antibody-antigen-hFcγRI complex), 150 conformers are evenly selected from MD trajectories and superimposed. As can be seen in Figure 2, except second N-acetylglucosamine and the branch point mannose, other residues are flying around. It can be seen from the RMSD of both carbohydrate chains in the 150 conformations (Table 1) that all these conformers have relatively small average RMSDs, but with different distribution in response to antigen and Fc receptor bindings. Overall, the free antibody has the largest carbohydrate chain RMSDs (2.2 Å and 2.1 Å). With antigen binding, average carbohydrate chain RMSDs decrease (1.8 Å and 2.0 Å ), and the standard deviation also narrowed from 0.8 Å /0.6 Å to 0.6 Å /0.5 Å. Without antigen binding, antibody-hFcγRI complex has uneven carbohydrate chain response. One chain has the same average RMSD (2.2 Å) with a larger standard deviation (0.9 Å), and the second chain has a smaller average RMSD (1.9 Å) and standard deviation (0.5 Å). The carbohydrate chains in the antibody-antigen-hFcγRI complex are the most stable, and its average carbohydrate chain RMSDs are 1.6 Å and 2.0 Å, and the standard deviations are 0.6 Å /0.5 Å. Comparing free antibody and antibody-antigen-hFcγRI complex the decrease of average RMSD from 2.1 Å to 1.6 Å is significant in both percentage (25%) and absolute value (0.5 Å). The influence of N-glycan flexibilities is not symmetrical, with one of them is more stabilized. It can also be seen from Figure 2C and D that residue fluctuations of core fucose and N-acetylglucosamine also decreased in the antibody-antigen-hFcγRI complex. It is interesting to see that the terminal residues in the carbohydrate chains still have large conformation flexibility in the antibody-antigen-hFcγRI complex, probably with effects of partially decreasing entropy penalty in FcR binding.
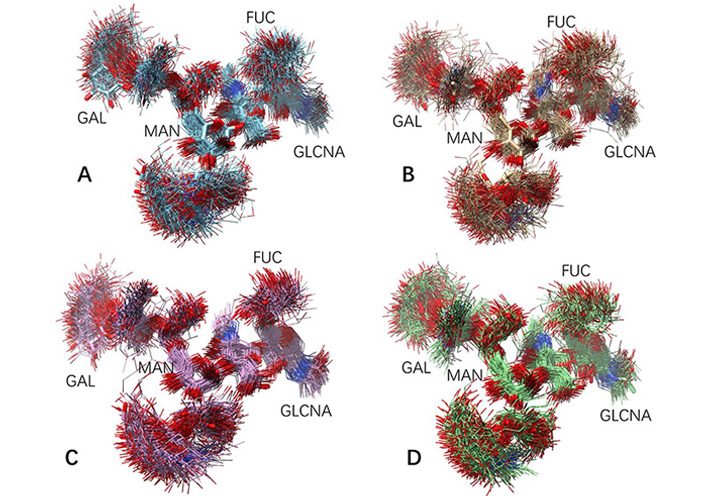
Fluctuation of glycan conformation in free antibody and in the antibody-antigen-hFcγRI complex. One hundred and fifty conformers are evenly selected from MD trajectories and superimposed. (A) and (B) Two carbohydrate chains in free antibody; (C) and (D) two carbohydrate chains in the antibody-antigen-hFcγRI complex. The G1F carbohydrate chain sequence is in Figure 1, and the key residues are labeled here
RMSD (Å) of glycan conformations in different antibody binding states
| System | Glycan | Ave | Sd | Min | Max |
|---|---|---|---|---|---|
| Antibody | Chain A | 2.2 | 0.8 | 0.6 | 4.6 |
| Chain B | 2.1 | 0.6 | 0.8 | 4.2 | |
| Antibody-antigen | Chain A | 1.8 | 0.6 | 0.6 | 4.4 |
| Chain B | 2.0 | 0.5 | 0.7 | 4.2 | |
| Antibody-hFcγRI | Chain A | 2.2 | 0.9 | 0.6 | 5.0 |
| Chain B | 1.9 | 0.5 | 0.9 | 3.8 | |
| Chain A | 1.6 | 0.5 | 0.6 | 5.2 | |
| Chain B | 2.0 | 0.6 | 0.8 | 3.9 |
Ave: average RMSD; Sd: standard deviation; Min: minimum RMSD; Max: maximum RMSD
The conformation fluctuations of core N-acetylglucosamine, even though a small one, may cause large spatial changes of carbohydrate chains. This simple hinge-like motion, in addition to terminal residues conformation change, may cause large variations of interactions between two carbohydrate chains and their interaction with protein residues. In the crystal structure of free antibody (1IGT), the shortest distance between two hydroxyl groups in two mannoses in two glycan arms is 4.35 Å, implying possible hydrogen bonds when antibody residues fluctuate. Indeed, we observed large number of nonspecific hydrogen bonds between two carbohydrate chains for antibodies before FcR binding (Figure 3).
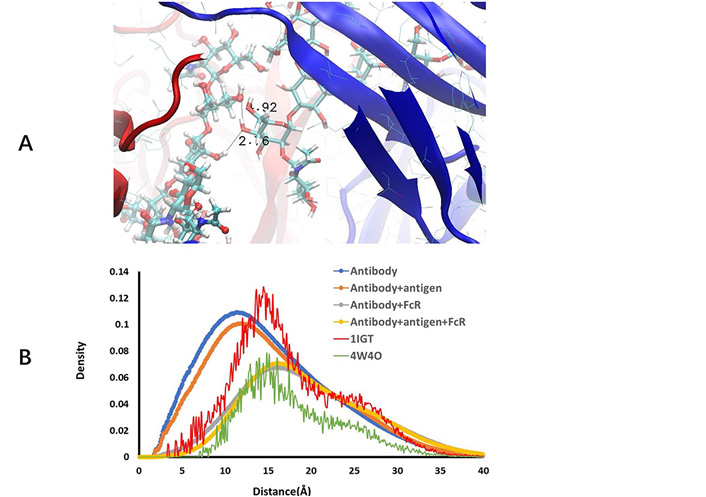
Free antibody has strong but non-specific hydrogen bonds between two carbohydrate chains. (A) Illustration of hydrogen bonds between two carbohydrate residues; (B) radial distance distribution between atoms in two carbohydrate chains in free antibody, antibody-antigen complex, antibody-hFcγRI complex, antibody-antigen-hFcγRI complex, and two crystal structures (1IGT and 4W4O)
As can be seen in Figure 3A, depicts a snapshot of hydrogen bonds between two carbohydrate chains. We compare the radial distance distribution between atoms in two carbohydrate chains in free antibody, antibody-antigen complex, antibody-hFcγRI complex, antibody-antigen-hFcγRI complex, and two crystal structures (1IGT and 4W4O; in Figure 3B). Using the free antibody crystal structure 1IGT as reference, one can see that the two carbohydrate chains separate further in the Fc-hFcγRI complex crystal structure 4W4O, mainly due to the opening of CH2 domains when binding to hFcγRI. However, the contractions of distances between two carbohydrate chains in MD simulations are unexpected. In both free antibody and antibody-antigen complex, not only the hydrogen bonds form between two carbohydrate chains, but also the overall distances between the atoms in the two carbohydrate chains are much shorter than those in the crystal structure. Antigen binding increases the separation a little bit, but still shorter than in the static state in the crystal structure. After binding to hFcγRI, the radial distance distributions approach the level observed in the crystal structure of Fc-hFcγRI complex (4W4O). The order of increasing the distance separation is free antibody < antibody-antigen < antibody-hFcγRI < antibody-antigen-hFcγRI complex.
Examination of hydrogen bonds between carbohydrate chains and protein revealed that overall hydrogen bonds frequencies increase in the order of free antibody < antibody-antigen < antibody-hFcγRI < antibody-antigen-hFcγRI complex (Supplementary material Table 1). Charged amino acids, Asp, Glu, followed by Lys and Arg dominate the interaction with carbohydrates. Polar and hydrophobic residues may use backbone to interact with carbohydrates. We found that a few amino acids in CH3 domain have small percentage hydrogen bonds with fucose in the free antibody (Figure 4A), but sharply decreased as long as antigen binds to antibody (Figure 4B). It is unexpected that core fucose can reach as far as CH3 domain. We studied the distance distributions of fucose to all amino acids (Figure 4C) but found no characteristic feature. However, when including all carbohydrates in consideration, the distance distribution of carbohydrates to CH3 domain residues revealed clear stepwise separation distribution in the order of free antibody < antibody-antigen < antibody-hFcγRI < antibody-antigen-hFcγRI complex (Figure 4D). The clear separation of curves in Figure 4D indicates the importance of glycan in allosteric response to antigen and FcR binding.
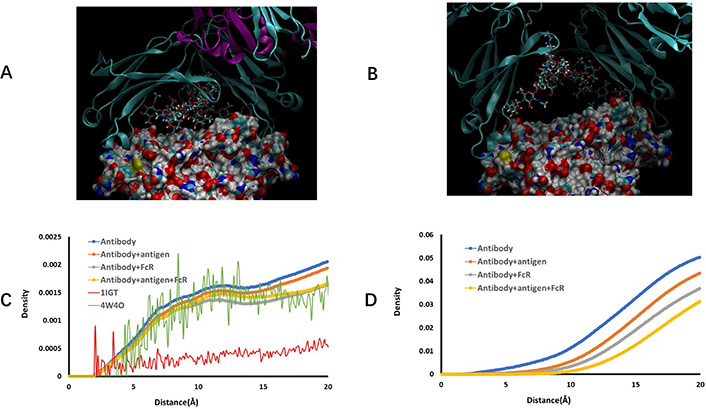
Two carbohydrate chains interact with Fc CH3 domain residues in free antibody conformation ensemble. (A) and (B) Snapshots of carbohydrate-CH3 domain interaction in free antibody and antibody-antigen complex, respectively. CH2 domain proteins chains are represented by ribbons, and CH3 domains are shown as solid surface. carbohydrate residues are depicted as stick bonds; (C) distance distribution of fucose residue from antibody amino acid residues; (D) distance distribution of carbohydrate residues from CH3 domain amino acid residues
Recent studies found that antibodies with identical variable domains, but different constant domains, may have significantly different affinities to Aβ [29]. Engineering CH and CL in Trastuzumab and Pertuzumab recombinant models also affect antigen-binding [30]. A previous antibody structural analysis found that distant CH1-1 loops undergoing significant fluctuations upon antigen binding [31]. These results suggest allosteric effects during antibody-antigen recognition, with both the variable and constant domains playing a role [32–37].
Many works have shown that there is a two-way communication between the Fc and Fv domains; that is, modifications of Fc can influence the variable domain antigen recognition. Various experiments, for example circular dichroism [38], NMR [39], and crystallography [40] have shown that upon antigen binding, the C domains can affect the V region paratope conformation, Simulations and experiments showed that modification of the constant domain influences binding affinity [41–43] and specificity [44, 45] of the antibody-antigen interaction. For example, Immunoglobulin A (IgA) Fc mutations reduced Her2 binding [46]. Antibody effectors can be antigen specific [47], indicating the intrinsic connection between antigen recognition and Fc receptor binding. There is substantial evidence for configurational allostery due to glycosylation and sequence variations [48].
In our study, it is interesting to see that N-glycans move with flexibility and overall conformation integrity with small overall RMSD. Thus, N-glycans can interact with various parts in Fc region with similar conformations.
The terminal residues in the carbohydrate chains still have large conformation flexibility in the antibody-antigen-hFcγRI complex. This kind of residual conformation fluctuations partially decrease entropy penalty in FcR binding. In the experiment to study Fc-hFcγRI binding, it has been found that the values of the thermodynamic parameters were consistent with the stabilization of the complex with respect to the unbound state, a process driven by the formation of numerous non-covalent interactions and opposed by the loss of entropy. It is expected that the magnitude of the entropy/enthalpy compensation was even greater for the full-length antibody. Given this large enthalpy lose in antibody-antigen-hFcγRI complex, any residue fluctuation that does not decrease binding site interaction enthalpy will help the overall free energy change in the binding processes.
In conclusion, we showed that the N-glycans partially acts as allosteric sensor and respond to antigen and hFcγRI binding. This is achieved largely by conformation fluctuation of connecting and terminal carbohydrate residues. Two important interaction patterns have been observed. The first is the strong but non-specific interactions between two carbohydrate chains, and second is that the distance distribution between carbohydrate chains and CH3 domain clearly differentiate the free antibody, antibody-antigen complex, antibody-hFcγRI complex, and final antibody-antigen-hFcγRI complex.
Fab: fragment antigen-binding
Fc: fragment crystallizable
FcR: fragment crystallizable receptor
FcγRI: fragment crystallizable-gamma-receptor-I
hFcγRI: human fragment crystallizable-gamma-receptor-I
IgG: immunoglobulin G
MD: molecular dynamics PDB: Protein Data Bank
RMSD: root-mean-square deviation
The supplementary materials for this article are available at: https://www.explorationpub.com/uploads/Article/file/10034_sup_1.pdf.
BM thanks Dr. Jun Zhao for helpful discussion.
The author contributed solely to the work.
The author declares that there are no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
BM thanks Shanghai Jiaotong University for supporting this work. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2021.
Copyright: © The Author(s) 2021. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
