Affiliation:
UMR5095, CNRS, Université de Bordeaux, 33077 Bordeaux, France
Email: manon@ibgc.cnrs.fr
ORCID: https://orcid.org/0000-0002-5792-1084
Explor Target Antitumor Ther. 2022;3:128–148 DOI: https://doi.org/10.37349/etat.2022.00076
Received: December 07, 2021 Accepted: February 14, 2022 Published: April 02, 2022
Academic Editor: Donatella Del Bufalo, Regina Elena National Cancer Institute, Italy
The article belongs to the special issue The Role of Bcl-2 Family Proteins in Cancer Progression and Their Relevance to Cancer Therapy
The budding yeast Saccharomyces cerevisiae, a favorite model in biology, does not contain any protein of the Bcl-2 family. From initial experiments with two-hybrid systems to the heterologous expression of human Bcl-2 family members, and the characterization of several forms of yeast programmed cell death, it has however always been a powerful tool to gain information on the mechanisms of apoptosis in general and on Bcl-2 family in particular. This is a short survey of 25 years of experiments that have provided, and at times initiated, insights into the molecular mechanisms underlying the function of Bcl-2 family members.
Apoptosis is the main form of programmed cell death (PCD) in animals. Beyond its crucial functions during development, it plays a central role in the maintenance of tissue homeostasis. As a matter of fact, alterations of the apoptotic process are one of the early characteristics of tumor cells. Furthermore, the efficiency of anti-cancer therapies depends on the apoptotic response of the cells. Hence, defects in the mechanisms of apoptosis are responsible for the failure of these treatments.
Classical anticancer therapies, such as radiotherapy or anti-proliferation chemotherapies, target DNA maintenance and replication. DNA alterations trigger the expression of transcription factor p53, which acts as a tumor suppressor by promoting both cell cycle arrest and apoptosis. A major target of p53 is the Bcl-2 family. The first member of this family, Bcl-2, was identified in 1984 in B-cell lymphomas (hence its name) where its over-expression is the main cause of increased survival [1]. Since then, homologs of Bcl-2 have been identified on the basis of homologies in 4 domains, called BH1 to BH4 [2]. Some of these proteins, like Bcl-2, are anti-apoptotic (Bcl-xL, Mcl-1, A1/Bfl-1, et al.) while others are pro-apoptotic (Bax, Bak, Bok). Other proteins contain only the BH3 domain and are regulators of both anti-apoptotic and pro-apoptotic proteins. Among them, the protein Bid is a close homolog of Bcl-2, while the others have only the BH3 domain in common (e.g., Bim, Bad, Puma). It is noteworthy that, later, proteins having a distantly related BH3 domain have been identified both in mammals and other organisms, where they have functions that are not directly related to apoptosis [3]. However, under certain conditions, these proteins may interact with Bcl-2 family members: for example, the autophagy effector Beclin-1/Atg6 was initially identified as an interactor to Bcl-2 (hence its name) [4, 5].
The main role of Bcl-2 family members is to modulate the permeability of the outer mitochondrial membrane (OMM): pro-apoptotic proteins Bax and Bak are able to form large pores, that promote the release of proteins from the intermembrane space (IMS) towards the cytosol or the nucleus [6, 7]. At least 5 released proteins playing a role in apoptosis have been identified: apoptosis inducing factor (AIF) [8], cytochrome c [9], smac/diablo [10], Omi/HtrA2 [11], and endonuclease G [12]. Released cytochrome c interacts with the protein APAF-1 to form the apoptosome, that initiates the cleavage/activation of procaspase 9 to caspase 9. Once active, caspase 9 cleaves and activates other caspases, namely caspases 3 and 7, that cleaves different substrates. This process has been named the “caspases cascade” [13–15]. Caspases can be inhibited by death regulators, such as the IAPs, that promote caspase degradation by the proteasome. IAPs are themselves inhibited by smac/diablo and Omi/HtrA2 that act by “trapping” them, thus preventing their interaction with caspases. AIF and endonuclease G do not modulate caspases, but act downstream, directly on nuclear DNA degradation. The anti-apoptotic protein of the Bcl-2 family is generally considered to act early in the process by preventing the localization and/or activation of Bax and Bak on the OMM.
This rapid descriptive summary of apoptotic events triggered by anticancer treatments shows that mitochondria are at the heart of the process. It is sometimes written that OMM permeabilization is a “point of no return” of apoptosis. This might be somewhat exaggerated, since caspases activation may still be blocked by IAPs. Nevertheless, the action of the Bcl-2 family on mitochondria is a major step of the apoptotic process, and its deregulation is a major cause of antitumor treatments failure [16–18].
The budding yeast Saccharomyces cerevisiae is a favorite model in biology. It can be grown rapidly and easily in every laboratory with minimal equipment. It displays all the basic functions of a eukaryotic cell, with a large set of proteins that are greatly conserved in mammals: it is estimated that 30% of human genes involved in pathologies have a homolog in yeast. Genetic studies are greatly facilitated by its compact genome, with almost no intron (only 4% of yeast genes are spliced after transcription) and the genuine ability of yeast to achieve homologous recombination: yeast can then be “humanized”, by replacing a gene with its human ortholog. It is also easy to maintain non-yeast genes on replicative plasmids that are maintained when yeast divides, or to integrate them at a pre-selected positions in the genome. Last but not least, yeast is very useful for mitochondrial studies: indeed, when mitochondria are deficient, yeast cells are still able to grow owing to alcoholic fermentation, thus providing a way to isolate and study altered mitochondria. For these different reasons, yeast has been used by different investigators to identify new potential functions and targets involved in apoptosis regulation that might be further tested in mammalian cells. In this short survey, we will show several examples of different types of experiments that have been done over the years.
The two-hybrid method has been developed in 1989 to test the interaction between any couple of proteins expressed in yeast [19]. It is based on the reconstitution of the transcription factor GAL4 of which the DNA-binding domain and the activation domain are fused to the two proteins of interest. The interaction of the proteins of interest brings the domain together, and the activity of the reconstituted transcription factor can be followed by the capacity of the cells to grow on galactose, or by a colorimetric method with a GAL1-lacZ fusion. The method has been improved over the years, with other transcription factors such as LexA [20], and is now a classic element of the molecular biology toolbox.
As early as 1993, a two-hybrid was used to show interactions between Bcl-2 and the protein R-ras p23, which was next confirmed by co-immunoprecipitation [21]. Soon after, the interaction between Bcl-2 family members was evidenced by the two-hybrid approach [22]. The method was then used to investigate which domains and residues of these proteins were involved in the interactions [23, 24]. However, by serendipity, these experiments also established that Bax was able to hamper yeast growth while anti-apoptotic proteins Bcl-2, Bcl-xL, and Mcl-1 prevented this inhibition. Consequently, the investigators quickly moved to the utilization of yeast to identify factors that could modulate the effects of Bcl-2 family members on yeast growth and viability (see below).
Nevertheless, the two-hybrid system remained a method of choice to identify new interactants of known apoptosis regulators. Owing to the specific interactions between BH domains, many BH3-containing proteins were identified through their interaction with Bcl-2 by two-hybrid methods. The most important is the major cell death regulator Bad, identified as a Bcl-2 interactant [25]. The BH3-containing protein BNIP3 was found to interact with Bcl-2 and Bcl-xL and also with the C.elegans Bcl-2 homolog Ced-9 [26]. The BH3-containing protein MAP-1, later renamed MOAP-1, was identified as a Bax interactant [27]. More recently, the protein Bcl2L12 was identified through its interaction with both Bcl-2 and Bcl-xL [28]. Other BH3-containing proteins were identified through their interaction with apoptosis regulators that are not Bcl-2 family members, such as NBK/Bik, which was identified as an interactant to the viral protein E1B 19K [29]. Although it is not directly involved in apoptosis, but rather in autophagy, the BH3-containing protein Nix was identified in a two-hybrid screen against a neurotrophin receptor involved in Bax-dependent apoptosis [30].
Other proteins, unrelated to Bcl-2 family members but still able to interact with them, were also identified by two-hybrid, such as the protein Btf [31] and the endophilin-related protein SH3GLB [32]. The two-hybrid system can also be combined with other methods of interaction measurements, such as co-immunoprecipitation, to refine the characterization of hits [33].
Outside from the Bcl-2 family, two-hybrid allowed to identify players of the caspases cascade, such as a new substrate of caspase-7 [34], an inhibitor of APAF-1-driven caspase-9 activation [35], or a regulator of XIAP [36]. Note that, conversely, Bcl-2 family members have also been found as hits in two-hybrid screens with other cancer-related proteins as baits [37].
The general approach used in the studies cited above was to choose an adequate bait (anti- or pro-apoptotic Bcl-2 family member, or any apoptosis effector/regulator of interest) and to screen a cDNA library of possible interactants from healthy or tumor mammalian cells. The main limitations of this approach are the “false positive” responses. Indeed, two-hybrid might be so sensitive that non-specific interactions might be detected. This namely happens in the case of proteins having a hydrophobic α-helix that may interact with any other hydrophobic α-helix. This led to the later-questioned observation that Bax could physically interact with the mitochondrial inner membrane adenine nucleotides transporter (ANT) [38]. To avoid this bias, most investigators choose to work with Bcl-2 family members deprived of their C-terminal hydrophobic α-helix. However, this might have contributed to underestimating the function of this helix in the regulation of Bcl-2 family members, by considering it only as a membrane anchor and a burden for this type of assays.
Since the initial report that the expression of Bax constructs inhibited yeast growth and that this inhibition could be prevented by the co-expression of Bcl-2 or Bcl-xL constructs [22], the investigators found in this unexpected observation a powerful way to identify new regulators of apoptosis. The inhibition of growth was simply measured by drop-tests or replica-plating on a Bax-inducing medium (such as a medium containing galactose when Bax was expressed under the control of the GAL1/10 promoter) and the restoration of growth was measured on the same medium when Bcl-2 or Bcl-xL were expressed under the same GAL1/10 promoter or a constitutive promoter, such as ADH1. This growth/no-growth phenotype led to some confusion in early papers where the absence of growth was interpreted in terms of cell death [22] while other authors discriminated against the absence of growth and actual cell death [39]. It is now widely accepted that the two phenotypes should be evaluated independently through adequate methods, the more accurate being plating efficiency [40]. Both absorbance or fluorescent molecular probes aiming at differentiating between growing cells, not growing cells, and dead cells are also widely used, but care must be taken that the outcome signal reflects the status of the cells.
The power of studies in yeast can be illustrated through the Bax-induced release of cytochrome c. The demonstration that cytochrome c was released from mitochondria [41] and that cytosolic cytochrome c was required for the apoptotic program [9] was published in 1996. One year later, the demonstration that anti-apoptotic Bcl-2 [42] and Bcl-xL [43] could block cytochrome c release, thus preventing apoptosis, was published. However, the first demonstration that pro-apoptotic Bax directly induced the release of cytochrome c was obtained through the expression of Bax in yeast [44], even before a similar demonstration was done in mammalian cells [45].
Although it does not accurately reflect cell death, the clarity and the practicality of the no-growth phenotype induced by Bax expression in yeast opened the way to the identification of suppressors. The first obvious suppressors were the anti-apoptotic members of the Bcl-2 family [22, 46–49]. But this has been extended to other mammalian proteins, through the identification of suppressors of Bax (or Bak)—induced growth impairment [50–58] (Table 1 and Figure 1).
An overview of proteins that have been shown to modulate Bax-induced cell death in yeast
| Function | Activation: overexpression stimulates Bax effects and/or deletion inhibits Bax effects | Inhibition: overexpression inhibits Bax effects and/or deletion stimulates Bax effects | |||
|---|---|---|---|---|---|
| Mammalian proteins | Yeast proteins | Mammalian proteins | Yeast proteins | Plant and viral proteins | |
| Bcl-2 family and related | tBid [171, 172]Puma [173]Noxa, Bik [174] | Bcl-2, Bcl-xL [22, 39, 44, 46, 175]Bfl-1/A1 [176] | see below for Ybh3 | M11 [177]DPV022 [178] | |
| Bax-inhibitor | BI-1 [50] | BI-1/Bxi1/Ybh3 [59] (not tested with Bax in [89, 90]) | BI-1 [59, 61, 179, 180] | ||
| Traffic | Tom22 [135, 137] | Tom22 [135, 139]TOM complex [136]Mdm34 [148] | Vps34 [54] Vamp3 [181] | Sec22 [182] | AtVAMP [63] |
| Autophagy and protein quality control | Uth1 [73, 183]Yme1 [184] Naa20 [185] | Atg5, Atg7 [73] | Sentrin [186] | ||
| Protein aggregates | PrP [187,188]α-synuclein [189] | ||||
| Protein- kinases | AKT [128]PKCα [190] | AKT [128] | Sch9 [191] | ||
| Stress responses | Hsp60 [192]Hog1 [193] | Trx1 [194]Fth1 [195] | Tsa1 [194]Rgi1 [195] | tQM [196]LePHGPx [64] AtEBP [62, 65] BI-GST [60, 197]sAPX [198] | |
| Energetic metabolism | Qcr7, Cyc3, Cox4, Cox7, Atp4, Pet9 [161, 199]Data not confirmed in [155] | COX6A1 [200] | OYE2 [69, 201]Por1 [199] | GAPDH [202] | |
| Lipid metabolism | SMS1 [203] | Hfd1 [184]Fah1 [204] | fah1, fah2 [204]BcLCB(2) [205] | ||
| Others | hRPL9 [206]Septin7 [70]Tsc22 [55] | Cdc10 [70] | Omp2b [67] | ||
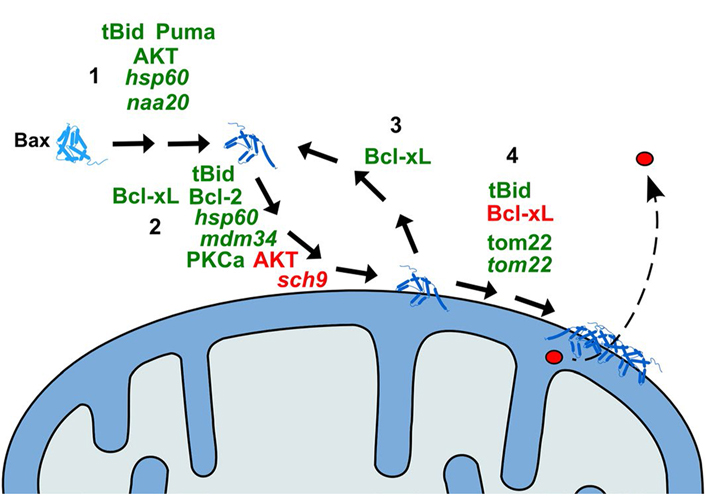
Schematic representation of the action of modulators of Bax effect on yeast mitochondria. 1: Activation of Bax (conformational change); 2: translocation of Bax to the MOM; 3: retrotranslocation of Bax from the MOM; 4: oligomerization of Bax and formation of the pore. Only the proteins of which the direct effect on Bax has been demonstrated are represented. Activators of the relevant step are in green; inhibitors are in red. Names in italic are yeast proteins
Conversely, it is theoretically possible to identify suppressors of the anti-apoptotic function of Bcl-2 or Bcl-xL. However, this is more difficult since it implies the use of a negative screening (i.e., finding clones that lose their ability to grow when Bcl-2 or Bcl-xL is co-expressed with Bax) by replica plating. To our knowledge, no successful large-scale assay has been published to date.
Because a large number of functions and regulations are conserved between yeast and higher eukaryotes, the identification of Bax suppressors (or Bcl-2/Bcl-xL suppressors) is not limited to mammalian genes. The screening of libraries from invertebrates [59] and plants [59–67] allowed to identify proteins relevant to cell death that might have not been previously identified in mammals, with the noticeable exception of the protein Bax inhibitor-1 that has been found in every organism tested [68]. cDNA from the yeast itself has been used and several genes involved in yeast resistance to Bax have been identified this way [69, 70].
It is necessary to point out, however, that these experiments may display a major bias, related to the way Bax prevents yeast growth or even induces yeast cell death. As we will see in the next chapter, moderate oxidative stress induces a form of yeast PCD that exhibits several similarities to mammalian apoptosis. It has been shown that Bax expression in yeast actually induces moderate oxidative stress [71, 72]. Furthermore, Bax-induced yeast growth inhibition might not be related to PCD but rather to autophagy [73]. This might indicate that the protective effect of over-expressed mammalian, plant, or yeast genes over Bax effects in yeast, might not be fully related to Bax itself but rather to downstream events specific to yeast. It is therefore crucial to complete the identification of Bax suppressors by experiments in the original organism before being conclusive. Yeast is such a powerful tool that we, investigators, should not forget that it is only a tool.
The first evidence that yeast displays a form of PCD having common features with apoptosis was obtained in a mutant of the cell cycle protein cdc28 [74] and, rapidly after, it was observed that Bax expression in yeast induced similar features [75]. It was then observed that (moderate) oxidative stress was a major regulator of yeast PCD [76]. It followed that any alteration producing moderate oxidative stress (including Bax expression, as we cited above) could induce yeast PCD having some similarities to mammalian apoptosis. This included acetic acid stress [77], high salt concentration [78], α-mating pheromone [79], amiodarone [80], aging [81], defects in N-glycosylation [82], among others (see [83] for a recent exhaustive survey).
The large collection of available yeast deletion mutants has allowed identifying proteins involved in this form of cell death. The main interest is that several of these proteins have homologs in mammals. For example, the deletion of the gene SRO7/SOP1 induced a loss of viability, with several markers of apoptosis, when the cells were grown in the presence of high NaCl concentration and, interestingly, this protein is the homolog of the Drosophila l(2)gl tumor suppressor [84]. HIR1, a co-repressor of histone transcription, acts as a suppressor of yeast cell death induced by various stress, such as mRNA stabilization and acetic acid treatment [85]. The human homolog of HIR1, HIRA, had been previously associated with different apoptosis alterations during development [86]. Studies in the fission yeast Schizosaccharomyces pombe have shown that calnexin is involved in apoptotic cell death induced by endoplasmic reticulum (ER) stress [87]. A genetic screen identified HMG1/2, the homolog of the human protein HMGB1, as a suppressor of both yeast and human calnexin-induced apoptosis, showing that this death pathway is largely conserved between fission yeast and human cells. Like for Bax-induced cell death, a large number of proteins having direct or indirect effects on oxidative stress have been identified as suppressors, such as PGK1 [88].
A particular interest has been focused on the protein called Ybh3, identified as a BH3-containing protein (hence its name, for yeast BH3) [89]. Beyond the discussion on whether or not the BH3 domain of this protein is a genuine one [3], it was particularly intriguing that this protein, that favors cell death, has been simultaneously identified as an anti-apoptotic protein [90], and is actually a homolog of the widely characterized Bax inhibitor identified in plants and animals [91, 92]. The identification as pro-apoptotic protein was done in cells submitted to moderate oxidative stress, while the identification as anti-apoptotic proteins was done on cells submitted to ER stress. It is noteworthy that several Bcl-2 family members may have opposite functions, depending on different modifications. A shorter splicing variant of Bcl-xL, named Bcl-xS, was identified at the same time and was shown to be pro-apoptotic [93]. Caspase-3-mediated cleavage of Bcl-2 was shown to generate a ΔN34 variant that permeabilized mitochondria to cytochrome c [94]. Conversely, the phosphorylation of Bax on Ser184 not only prevented Bax mitochondrial localization (see the discussion below) but could, under certain conditions, convert Bax into an anti-apoptotic protein through its binding to BH3-only proteins [95]. There is no indication, to date, that such processes occur for yeast Ybh3, but this might be a trail to explore its apparent dual function.
Yeast PCD also involves proteins that have been identified as homologs of known mammalian apoptosis regulators. The most obvious is cytochrome c, a universal and highly conserved mobile electron transporter [96]. In non-apoptotic eukaryotic cells, it is localized in the mitochondrial IMS but remains in close proximity of the inner membrane, more specifically of respiratory complexes III and IV. When released on the cytosol during apoptosis, cytochrome c interacts with APAF-1 to form the apoptosome, in a process that depends on the conserved K72 residue [97]. No yeast homolog of APAF-1 has been identified to date and the role of cytochrome c in yeast cell death, if any, remains unclear. It can be speculated that cytosolic cytochrome c contributes to the modulation of the intracellular redox status [98] but this has never been clearly supported.
Yeast does not contain caspase but expresses a metacaspase, called Yca1. Its deletion delays or decreases physiological cell death induced by many stimuli [99], but not by Bax [73]. The actual role of this enzyme in yeast cell death has been and still is a matter of debate [100]. Other homologs of apoptogenic factors, such as yeast AIF [101], endonuclease G [102], and Nma111/Omi/HtRA2 [103] have been identified as regulators of several physiological yeast death pathways. Yeast AIF is not involved in Bax-induced death [73] and the two others have not been investigated to date.
Beyond the study of apoptotic mechanisms senso strictu, yeast can be a powerful tool to study the connections between PCD and the regulation of growth and survival processes such as cell cycle [104], autophagy [104, 105], RNA stability [106], and, more generally, metabolism [107–109]. These processes are generally better conserved than PCD, between yeast and mammals. The information gained from studies in yeast, either from “physiological” yeast cell death or from “ectopic” cell death induced by mammalian proteins can therefore bring useful information on cross-talks between PCD and these processes in mammalian cells.
However, extrapolations from yeast to mammals should be done with great care. For example, two major autophagy regulators in mammalian cells have a BH3 domain: Beclin-1, which is involved in the initiation of the formation of autophagosomes [110], and Bcl2L13, a bona fide Bcl-2 family member, that is the mitochondrial receptor of mitochondria-targeted autophagy [111]. Besides their role in autophagy, these two proteins are able to regulate (and be regulated by) Bcl-2 family members [112, 113]. Their yeast homologs, Atg6 and Atg32, do not have a BH3 domain. Two speculations can be made: (i) autophagy/apoptosis cross-talks that exist in mammals do not exist in yeast or (ii) autophagy/apoptosis cross-talks in yeast do not require any BH3 domain (because there are no genuine Bcl-2 family members in yeast).
These examples underline that yeast PCD should be considered as a biological process that deserves to be studied somewhat independently from mammalian apoptosis. Both processes clearly share some similarities, some signaling pathways are undoubtedly homologous to the point that human proteins can compensate for the absence of yeast proteins, but the two processes of yeast PCD and mammalian cell death have evolved in parallel but not identically [40]. The obvious rationale behind this distinct evolution is probably the fact that yeast is a unicellular organism. It might exhibit some cooperative features [114]. An outstanding study further demonstrated the role of yeast PCD in the organization of yeast colonies [115, 116]. But it remains that yeast PCD does not have the same raison d'être as mammalian PCD and it must be kept in mind that they are not the copy of each other.
The initial observation that Bax constructs were able to inhibit yeast growth and that this inhibition was prevented by Bcl-2 and Bcl-xL [39, 117] opened the way to the utilization of yeast as a tool to investigate further the mechanistic events underlying the action of the Bcl-2 family members on mitochondria. Indeed, yeast provided a “living test tube” to investigate the function of a limited number of proteins, thus simplifying the interaction network. As we already noted, Bax expression in the yeast provided the first demonstration that the protein was directly responsible for cytochrome c release [44]. Furthermore, Bax expression in yeast complemented studies on mammalian mitochondria to show that Bax alone was able to form a large pore in the OMM, having a size compatible with the release of cytochrome c [118].
Hence, our group and several others have identified domains and residues involved in the intricate process of Bax mitochondrial relocation and activation. The first question was about the actual role of the C-terminal hydrophobic α-helix of Bax as a membrane anchor. This was not a trivial question since, contrary to Bcl-xL, purified Bax was not able to bind to isolated mammalian mitochondria. Furthermore, the replacement of the C-terminus of Bax by the C-terminus of Bcl-xL generated a chimera Bax-CxL that was able to bind to isolated mitochondria, while the reverse construction did not [119]. When expressed in yeast, truncated Bax was able to permeabilize the OMM to cytochrome c but lost its sensitivity to the inhibition by Bcl-xL [46]. The chimera Bax-CxL was still localized to mitochondria both in mammalian and yeast cells, but lost its pro-apoptotic properties, showing that the mitochondrial localization of Bax is not sufficient to promote the permeabilization [47, 120]. Substitutions of residues in the Bax C-terminus confirmed that its hydrophobic nature was not crucial for Bax mitochondrial localization, but that its movement out from the hydrophobic groove formed by the BH domains was a crucial step in its activation, that could be mimicked by the single substitution P168A in yeast [121], in mammalian cells [122] and in liposomes [123].
Part of the regulation of this movement relies on the possible phosphorylation of residue S184. It was demonstrated that the deletion of this residue generated a membrane anchor much more “efficient” than the genuine hydrophobic α-helix [124]. The identification of S184 as a target of several protein kinases, including the survival kinase AKT [125] raised the hypothesis that the phosphorylation of this residue could control the movement of the C-terminal helix. The substitution of S184 by non-phosphorylatable (A, V) or phosphomimetic (D, E) residues was in accordance with the hypothesis that S184 phosphorylation limited Bax mitochondrial localization, both in mammalian cells [125] and yeast cells [126]. However, the actual effects on Bax function seem to remain somewhat contradictory [95, 127–129], as a possible consequence of a differential effect of anti-apoptotic proteins on non-phosphorylated or phosphorylated Bax [127, 130].
Another interesting issue that was addressed in yeast, was the role of the N-terminus of Bax (Figure 2; Table 2). Among natural Bax variants found in tumors, an N-terminally truncated mutant called BaxΨ was found in low-grade glioblastoma [131], and this variant was found to be more active than full-length Bax in mammalian cells, in yeast, and in reconstituted systems, leading to the conclusion that the N-terminal end of Bax was a negative regulator of its activity [132, 133]. It was next observed that the domain that follows immediately, corresponding to helix α1 contained residues that were crucial for Bax interaction with mitochondria, suggesting the existence of a mitochondrial Bax receptor [134] that was further identified as Tom22 both in mammalian and yeast cells [135–137]. It is noteworthy that this regulation was lost on isolated mitochondria [138], which might be related to the fact that Tom22 does not act as a bona fide mitochondrial receptor to Bax, but rather as a regulator for its insertion under a regulatable conformation [137, 139].
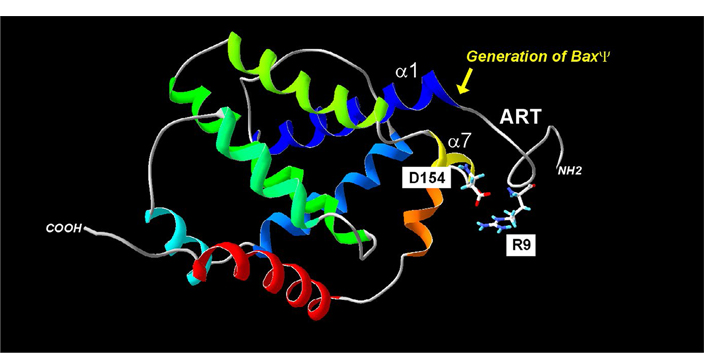
Regulation of Bax mitochondria localization by the ART. The absence of the ART (20 N-terminal residues) generates the BaxΨ variant, which is spontaneously mitochondrial and active. Within the ART, the R9 residue may interact with the residue D154 of helix α7. Point mutations in the helix α1 decrease the mitochondrial localization of both wild-type Bax (BaxWT) and BaxΨ. Together, these data suggest that a movement of ART away from the core of the protein may contribute to the exposure of helix α1 to mitochondrial partners
| Sequence | Protein | Binding to mitochondria | Cytochrome c release | ||
|---|---|---|---|---|---|
| Yeast | Mammals | Yeast | Mammals | ||
| (1) Role of ART and Proline residues at positions 8 and 13 | |||||
| -P8RGGGP13- | full length Bax | +/− | +/− | +/− | +/− |
| BaxΨ/ΔART | +++ | +++ | +++ | +++ | |
| -G8RGGGG13- | ART mutant | +++ | n.d. | +++ | n.d. |
| -V8RGGGV13- | ART mutant | n.d. | +++ | n.d. | +++ |
| -V8RGGGP13- | ART mutant | n.d. | +/− | n.d. | +/− |
| -P8RGGGV13- | ART mutant | n.d. | +++ | n.d. | +++ |
| (2) Role of helix α1 | |||||
| -A24LLL27- | BaxΨ/ΔART | +++ | +++ | +++ | +++ |
| BaxΨ/ΔARTΔHα1 | n.d. | − | n.d. | − | |
| -R24LLL27- | BaxΨ/ΔARTHα1 mutant | + | − | + | − |
| -A24LGV27- | BaxΨ/ΔARTHα1 mutant | +++ | − | +++ | − |
| (3) Interaction between ART and BH2 (helix α7/8) | |||||
| -P8RGGGP13--I152QDQG156- | full length human Bax | +/− | +/− | +/− | +/− |
| -P8EGGGP13--I152QDQG156- | ART mutant | +++ | n.d. | +++ | n.d. |
| -P8RGGGP13--I152QKQG156- | BH2 mutant | +++ | n.d. | +++ | n.d. |
| -P8EGGGP13--I152QKQG156- | ART mutantBH2 mutant | − | n.d. | − | n.d. |
(1) The substitution of prolines 8 and 13, like the complete deletion of the 20 N-terminal residues, increase Bax binding and activity both in yeast and mammals; (2) substitutions in helix α1 decrease the binding of BaxΨ in mammals. The decrease is partly visible in yeast; (3) individual point mutations inverting charges in ART and in BH2 induce stimulation of Bax in yeast, but a double charge change revert to the wild-type behavior of Bax, suggesting the existence of an interaction between charged residues in these two domains, stabilizing the inactive conformation. Bold residues indicate mutations. −: inactive; n.d.: not determined; +/−: poorly active; +: active; +++: strongly active
The ART domain contains a positively charged residue, R9, which, given the high mobility of ART can be in relatively close proximity to a negatively charged residue, D154, in the helix α7 (~7Å). The introduction of two negative charges (E9/D154) or two positive charges (R9/K154) generates mutants that have a greater mitochondrial localization (and a greater activity) than BaxWT (R9/D154) or the reverse mutant (E9/K154), suggesting that the interaction between these two residues regulated negatively Bax mitochondrial localization and activity [121].
Strikingly, a recent study showed that Bak, the other pro-apoptotic member of the Bcl-2 family, also supports a negative regulation of its insertion/activation process by a domain located on the N-terminal moiety of the protein [140]. Although the domain and mode of regulation are different, this negative regulation seems to be a conserved feature between the two proteins.
The role of mitochondrial receptors in the localization of Bcl-2 family members, and the utilization of yeast to investigate this role, are not limited to Bax. Yeast mitochondria deficient for Tom20 are less able to bind Bcl-2 [141]. The same study pointed at the critical role of positive charges flanking the C-terminal α-helix of Bcl-2 in its mitochondria addressing, a property that was later confirmed in mammalian mitochondria [142]. More recently, the role of Tom20 was specified, both in mammalian and yeast cells, showing that Tom20 facilitated the ER to mitochondria transfer of Bcl-2 after apoptosis has been initiated and Bax has been activated [143]. The role of Tom70 as a receptor for the BH3-only protein Bim has also been suggested through experiments in yeast, even though the actual role of this interaction remains unclear [144].
Yeast is also a powerful tool to investigate the role of contacts between ER and mitochondria, corresponding to domains called mitochondria-associated membrane (MAM). Indeed, contrary to mammalian cells in which the occurrence of MAM depends on different complexes [145, 146], yeast MAM depends on the well-characterized ER-mitochondrial encounter structure (ERMES) [147]. The deletion of one component of ERMES, the protein mdm34, has been shown to limit the mitochondrial localization of Bax [148] and, as a consequence, of Bcl-2, when both proteins are co-expressed [144].
A most remarkable characteristic of Bax mitochondrial localization is its reversibility, which has been identified by demonstrating that Bcl-xL could retrotranslocate Bax from mitochondria to the cytosol [149]. It was next demonstrated that the C-terminal end of Bcl-xL was required for retrotranslocation [150]. The retrotranslocation of Bax by Bcl-xL was also observed in yeast, where it was further shown that Bcl-xL also stimulated translocation, by a process that did not depend on an intact C-terminal end of Bcl-xL, thus explaining why truncated Bcl-xL greatly increased Bax mitochondrial content [151, 152].
A large number of mitochondrial proteins are conserved between yeast and mammals, and yeast has extensively been used as a simplified model to confirm or rebut suspected mitochondrial regulations of Bcl-2 family members addressing and activating. Voltage-dependent anion-selective channel (VDAC), the channel responsible for the permeability of the OMM to metabolites, has been proposed to be involved in Bax- induced cytochrome c release [153]. However, yeast cells depleted for VDAC isoforms did not respond to Bax differently from wild-type yeast cells [48, 154, 155], and this was later confirmed in mammalian cells [156]. Here again, experiments in simple yeast cells have been more reliable than experiments in more intricate mammalian cells.
Another example of conserved proteins between yeast and mammals is given by chaperones, such as the Hsp70 family that is universally present in prokaryotes and eukaryotes [157, 158]. The BH3-only protein Bim has been shown to interact with mammalian Hsp70 through its BH3 domain [159]. The heterologous expression of Bim in yeast stimulated cell growth and increased the protection against heat shock [160], thus providing a simple model to study the role of Bim as a co-chaperone.
The yeast model is not without inconvenience, however. The ability of yeast to grow without functional mitochondria can generate confusion. For example, it has been observed that a deletion of subunit 4 of mitochondrial FoF1-ATP synthase prevented the effect of Bax on yeast growth, leading to the conclusion that this complex was somehow involved in the Bax effect [161]. However, yeast mutants in ATP synthase are prone to generate cells losing a large portion (when not all) of their mitochondrial DNA, thus lacking all of the respiratory complexes. Yeast cells lacking mitochondrial DNA had been previously shown to survive Bax expression (although their growth rate was affected) likely because of different sensitivity to oxidative stress [38] and this could explain the resistance of the ΔAtp4 mutant.
Besides mammalian proteins, yeast has also been used as a complementary tool to further characterize the function of newly identified Bcl-2 family members from other organisms such as zebrafish [162] or Trichoplax [163].
Due to the clarity of the growth/no growth phenotypes linked to the expression of Bcl-2 family members in yeast, it was obviously tempting to use yeast as a rapid “pre-screen” to identify molecules of interest in targeting apoptosis, especially that known molecules had a significant effect on yeast [164]. Since it is easier to identify a drug that suppresses the “no growth” phenotype than the opposite, such screening is expected to be more efficient to identify anti-apoptotic molecules, of potential interest in treating degenerative diseases, than to identify pro-apoptotic molecules, of potential interest in treating proliferative diseases [165].
Several screenings have been successful in identifying caspases activators [166] or p53/mdm2 pathway activators [167, 168]. However, to our knowledge, no successful screening has been published for the Bcl-2 family. The reason for this failure might rely on the fact that the Bax-induced phenotype of growth inhibition/death is complex, involving yeast PCD [75], oxidative stress response [71, 72], and autophagy [73].
This is, however, not without a solution. Owing to genetic manipulation and the plasticity of yeast, it might be possible to limit the “side responses” of yeast by working in an autophagy-deficient or stress response-deficient genetic context. Also, yeast can be used to further characterize the effects of an already known molecule in a simpler model, which allows for confirmation of the relevance of a target. As an example, the BH3-mimetic ABT-737, a well-established inhibitor of both Bcl-2 and Bcl-xL binding to Bax, had been suggested to promote a further activation of Bax because the binding of the anti-apoptotic proteins caused a conformational change of the pro-apoptotic protein, thus promoting its full activation once the binding is challenged by the inhibitor [169]. This “pre-activation” process of Bax by Bcl-xL causing a greater efficiency of ABT-737 was next confirmed in yeast, showing that this process is dependent only on the two proteins, without the involvement of a third partner [151, 152].
With the outstanding development of genetic tools in mammalian cells, one might think that yeast has lost a large part of its attractivity to study processes such as apoptosis. However, beyond the basic manipulation of genes, which is greatly facilitated by its compact genome and the nearly infinite collection of mutants available in the world, yeast keeps many interests: its ability to grow even though mitochondria are altered, the possibility to study a limited number of proteins and its outstanding capacity to overcome adverse conditions by activating or inhibiting enzymes or whole pathways far away from the initial alteration, thus providing unsuspected possible regulations. Still a vivid tool for geneticists, yeast is sometimes snubbed by some cell biologists and biochemists, who label yeast studies as “old science”. Considering the number of Nobel Prizes awarded to yeast investigators [170], they are likely wrong.
AIF: apoptosis inducing factor
ER: endoplasmic reticulum
MAM: mitochondria-associated membrane
OMM: outer mitochondrial membrane
PCD: programmed cell death
The work done in the lab of the author has been supported by the CNRS, the Université de Bordeaux, and the Ligue Régionale contre le Cancer. The author thanks all his present and former students and colleagues for these studies, namely Nadine Camougrand, Muriel Priault, Hubert Arokium, Ingrid Bhatia-Kiššová, Thibaud Renault, Dario Trindade, Lilit Simonyan and Akandé Rouchidane Eyitayo.
The author contributed solely to the work.
The author declares no conflict of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2022.
Copyright: © The Author(s) 2022. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Donatella Del Bufalo ... Simona D’Aguanno
Prasad Sulkshane, Tanuja Teni
George Shapovalov ... Natalia Prevarskaya
Rui Caetano Oliveira ... José Casanova
