Affiliation:
1Laboratory BVBGR-LR11ES31, Biotechnopole Sidi Thabet, University Manouba, ISBST, Ariana 2020, Tunisia
2Association Tunisienne de Lutte contre le Cancer (ATCC), Tunis, Tunisia
Email: soumayakouidhi@gmail.com; soumaya.kouidhi@isbst.uma.tn
ORCID: https://orcid.org/0000-0002-3987-8617
Affiliation:
1Laboratory BVBGR-LR11ES31, Biotechnopole Sidi Thabet, University Manouba, ISBST, Ariana 2020, Tunisia
3Department of Biologu, Faculty of Sciences of Tunis, University of Tunis El Manar, Tunis 1068, Tunisia
ORCID: https://orcid.org/0000-0003-2953-2193
Affiliation:
2Association Tunisienne de Lutte contre le Cancer (ATCC), Tunis, Tunisia
5Service d’Oncologie Médicale, Hôpital Salah-Azaïz, Tunis 1006, Tunisia
Affiliation:
6Department of Pathology, Abderrahman Mami Hospital, University of Tunis El Manar, Ariana 2080, Tunisia
Affiliation:
1Laboratory BVBGR-LR11ES31, Biotechnopole Sidi Thabet, University Manouba, ISBST, Ariana 2020, Tunisia
ORCID: https://orcid.org/0000-0001-5642-8805
Affiliation:
1Laboratory BVBGR-LR11ES31, Biotechnopole Sidi Thabet, University Manouba, ISBST, Ariana 2020, Tunisia
ORCID: https://orcid.org/0000-0001-7310-3842
Affiliation:
7Laboratory of Genetics, Immunology and Human Pathology, Department of Biology, Faculty of Sciences of Tunis, University of Tunis El Manar, Tunis 1068, Tunisia
Explor Target Antitumor Ther. 2023;4:240–265 DOI: https://doi.org/10.37349/etat.2023.00132
Received: September 04, 2022 Accepted: January 04, 2023 Published: April 26, 2023
Academic Editor: Fathia Mami-Chouaib, Université Paris-Saclay, France
The article belongs to the special issue Cancer Immunotherapy and Tumor Microenvironment
It is now well-acknowledged that microbiota has a profound influence on both human health and illness. The gut microbiota has recently come to light as a crucial element that influences cancer through a variety of mechanisms. The connections between the microbiome and cancer therapy are further highlighted by a number of preclinical and clinical evidence, suggesting that these complicated interactions may vary by cancer type, treatment, or even by tumor stage. The paradoxical relationship between gut microbiota and cancer therapies is that in some cancers, the gut microbiota may be necessary to maintain therapeutic efficacy, whereas, in other cancers, gut microbiota depletion significantly increases efficacy. Actually, mounting research has shown that the gut microbiota plays a crucial role in regulating the host immune response and boosting the efficacy of anticancer medications like chemotherapy and immunotherapy. Therefore, gut microbiota modulation, which aims to restore gut microbial balance, is a viable technique for cancer prevention and therapy given the expanding understanding of how the gut microbiome regulates treatment response and contributes to carcinogenesis. This review will provide an outline of the gut microbiota’s role in health and disease, along with a summary of the most recent research on how it may influence the effectiveness of various anticancer medicines and affect the growth of cancer. This study will next cover the newly developed microbiota-targeting strategies including prebiotics, probiotics, and fecal microbiota transplantation (FMT) to enhance anticancer therapy effectiveness, given its significance.
Although precision medicine has made enormous strides in the treatment of cancer, and disease-free survival has shown clinical improvements, the required success rate has not yet been reached. Cancer patients still face resistance to therapy that limits reaching optimal cures, with high tumor recurrence [1]. Several mechanisms of resistance caused by chemotherapy and immunotherapy have been intensively described [1]. These side effects were related to genetic and epigenetic alterations or DNA damage repair, modifications, deregulation of cell death, unfavorable immune responses, and complex interactions within the tumor microenvironment (TME) [2, 3]. Interestingly, emergent mechanisms of cancer therapy resistance have been recently proposed. Emergent proof from preclinical and clinical studies has focused on the key role of gut microbiota in anticancer remedy response via modulating drug efficacy, abolishing the anticancer effect, and interceding toxicity [4]. Accumulating evidence has discovered that an altered intestine microbiome is related to resistance to chemotherapy or immune checkpoint inhibitors (ICIs) [5–7]. Therefore, it is not surprising that the latest investigations spotlight the possibility of modulating the gut microbiota to overcome the failure of remedy and poor cases issues, increase the efficacy of cancer treatment, and repair the original healthy microbiota [8–10].
Hence, a deep understanding of the interplay between microbiota and cancer treatment could help to implement new strategies for cancer prevention and patients’ stratification for more efficacy and less complication due to therapy.
In this review, authors first provide the scientific evidence highlighting the critical role of the gut microbiome in cancer establishing the links between gut microorganisms and cancer growth, immune responses, as well as the efficacy of anticancer treatment mainly chemotherapy and immunotherapy. This review will next go over the latest microbiome-targeting strategies, including probiotics and prebiotics, antibiotics, and fecal microbiota transplantation (FMT), which may improve the effectiveness of cancer treatments.
From birth, many bacteria that are primarily found on the skin, mouth cavity, vagina, and stomach coexist with humans [11]. Nevertheless, the gastrointestinal (GI) tract as well as distal organs including the brain, liver, and pancreas are all impacted by the gut microbiota, which has become an important factor in human health [12, 13]. Since the introduction of culture-independent techniques like omics, we have gained a greater understanding of the structure and function of the human gut microbiota [14–16]. The adult human gut microbiota incorporates 1013–1014 microorganisms/mL of luminal content, with an estimated biomass of about 1.5 kg [17]. Bacteria predominate microbial communities, as is well known. However, a more recent sharp increase in studies examining the involvement of viral and fungal components of the microbiome suggests that viruses, archaea, and fungus may potentially play crucial roles in preserving gut homeostasis [18]. Though it has been hypothesized that the virome may control the microbiome and affect bacterial complexity and population dynamics. It is likely that the most common bacteria are eliminated by their phages [19]. Thus, phages have a significant impact on the bacterial communities in the intestine [19]. However, because of their reduced abundance and the lack of specialized techniques and curated reference databases for their identification and classification, the viral and fungal biomes have not been well defined up to this point [20].
The microbiota’s structure remains constant throughout adulthood despite changes brought on by environmental and developmental factors [21]. Firmicutes (60–65%), Bacteroidetes (20–25%), and Proteobacteria (5–10%) are the phyla with the highest representation [22]. Lately, the microbiota is considered as an essential organ and has been associated with health and disease status. When the intestinal ecosystem is in good microbial equilibrium, we talk about “eubiosis” which is associated with good health. Indeed, it is sometimes considered as our “forgotten organ” [23]. In eubiosis, the gut microbiota provides a range of beneficial properties to the host, and its roles in human health have been recently investigated and described in different aspects. Actually, these microbes have important roles in preserving the mucosal barrier’s integrity, delivering nutrients including vitamins, short-chain fatty acids (SCFAs), and secondary bile acids, or protecting against infections [24–26]. Even more crucially, the gut microbiota interacts with the immune system, sending messages to encourage immune cell maturation and the appropriate development of immunological functions [27]. The composition of the microbiota is susceptible to host and environmental selective factors such as alteration in the immune system, host mutations, diet, antibiotic therapy, hospitalization, and chemical exposure. Such factors lead to impaired microbiota and then we talk about dysbiosis. A shift in the gut microbiota’s composition and functions, known as dysbiosis, is defined by a decline in the proportion of commensals and symbionts and/or an increase in the proportion of pathobionts [28]. It has long been known that there is a connection between microbiota and both health and disease. In fact, as Hippocrates, the father of medicine, once said, “all disease begins in the gut”. Recently, the literature has linked dysbiosis to the development of several diseases such as obesity and metabolic disorders [29], type 2 diabetes [30], inflammatory bowel diseases [31], and cancers [32]. Zitvogel et al. [33] showed that the gut microbiota can influence oncogenesis through a variety of mechanisms, including the direct oncogenic effects of microorganisms and their products, the alteration of circulating pro-carcinogen metabolites, the stimulation of the host’s production of trophic factors, and finally, the stimulation of immune-suppressive and pro-inflammatory pathways disrupts the host’s ability to detect malignancy (Figure 1).
Cancers are diseases that can affect different organs and are due to abnormal behaviour of so-called transformed cells that have acquired this behaviour as a result of different somatic alterations of a genetic and epigenetic nature [34, 35]. Thus, starting from a normal state, a cell can undergo the action of carcinogenic agents that cause genetic abnormalities affecting the sequence of DNA and the expression of genes [36, 37]. Following this stage that generates initiated cells, the carcinogenesis occurs on this initiated already cells which will develop into a tumor under the action of the microenvironment which ensures the promotion of cancer by providing the initiated cell with proliferation signals that will perpetuate the anomalies during divisions, and thus ensure the progeny of the abnormal cell [38–40]. This progeny will accumulate in its turn new genetic abnormalities. Carcinogenesis is a dynamic process that, at each stage, selects a new cell that has undergone one or more alterations [40]. Each cancer originates from the alteration of 10 to 20 genes [41]. These alterations occur successively, each of them favoring the next. This sequence of alterations usually occurs over a very long chronological range. Deregulated genes in cancers affect signaling pathways and molecular and metabolic mechanisms leading to loss of control of cell divisions, immortality, resistance to apoptosis, and metabolic reprogramming [42–44].
The cancer progresses by giving clinical consequences: it grows within precise histological limits (we speak of cancer in situ), then exceeds them and becomes invasive. The increase in tumor size is accompanied by hypoxia which stimulates the expression of pro-angiogenic factors and inhibits the expression or production of anti-angiogenic factors, thanks to the stabilization of hypoxia-inducible factor 1-alpha (HIF1α). Tumor angiogenesis refers to the ability of tumor cells to stimulate the appearance and development of blood vessels and thus promote their own growth [45, 46]. Tumor vascularization facilitates metastases which are secondary tumor formations that derive from cancer cells that have acquired the property of disassociating [epithelial mesenchymal transition (EMT) transition] from the primary tumor and have implanted remotely through a cellular mechanism called EMT [47, 48].
Hanahan and Weinberg [49] defined the characteristics of cancer cells and tumors which share common hallmarks: self-sufficiency in growth signals, insensitivity to growth inhibitor signals, ability to avoid apoptosis, capacity for indefinite replication, induction of angiogenesis, ability to metastasize, escape from the immune system, and deregulation of cellular energy metabolism. All these characteristics are sustained by tumor cell genome instability and tumor interaction with its micro-environment particularly inflammation and nutrient and energetic sources.
The most studied examples concern the intestinal microbiota in relation to the occurrence of digestive cancers and particularly colorectal cancer (CRC). In germ-free rats that developed fewer chemically induced colorectal tumors than conventional rats, Weisburger et al. [50] presented the first study associating the gut microbiota with CRC. These findings have been verified in animals prone to CRC [51]. In humans, such a link between gut microbiota and CRC has been reported in several studies (Table 1). In a healthy gut microbiota, the dominant bacteria phyla include Firmucutes, Proteobacteria, Bacteroidetes, and Actinobacteria, with a diverse structure at the genus and species levels in gut micro-communities. To explore the role of gut microbiota in the etiology of cancer, researchers utilize metagenomic analysis that allows comparison of microbiota composition between healthy populations and those with cancer diseases, investigating cancer tissues along with matched healthy tissues.
Involvement of gut microbiota dysbiosis in CRC
| Phyla and species concerned by dysbiosis in CRC | Reference |
|---|---|
| Infection with S. bovis a risk factor for colon tumors | [52] |
| High enrichment of Fusobacteria in colorectal carcinoma tissue | [53] |
| ETBF, and Fusobacterium nucletum are highly expressed in CRC tissue | [54] |
| Firmicutes and Fusobacteria were over-represented whereas Proteobacteria was under-represented in CRC patients; Lactococcus and Fusobacterium is more abundant while Pseudomonas and Escherichia-Shigella reduced in cancerous tissues versus adjacent tissues | [55] |
| Mucosa-associated E. coli of the B2 phylogroup more prevalent in CRC tissues | [56] |
| Fusobacterium nucleatum isolated from tumor tissue and proved to be invasive in the in vitro experiments | [57] |
| B. fragilis, Enterococcus Escherichia-Shigella, Klebsiella, Streptococcus, and Peptostreptococcus is abundant in CRC patients, while Roseburia- and Lachnospiraceae-related OTUs more abundant in healthy controls | [58] |
| CRC patients have a lower microbiota diversity and Clostridia abundance, with a high abundance of Fusobacterium and Porphyromonas at the genus level | [59] |
| Bifidobacterium, Faecalibacterium, and Blautia reduction, while Fusobacterium enrichment in the CRC patients; stool samples in CRC patients enriched with Paraprevotella, Eubacterium | [60] |
| Association of Clostridioides difficile with CRC revealed by anti-tcdB antibodies in plasma, particularly the IgA level; anti-tcdB antibodies as candidate serologic markers for CRC | [61] |
| Bacteroidetes cluster 1 and Firmicutes cluster 1 decreased in CRC mucosa, whereas Bacteroidetes cluster 2, Firmicutes cluster 2, pathogen cluster, and Prevotella cluster increased in CRC mucosa; compositional alterations in the microbiota differ between distal and proximal cancers | [62] |
S. bovis: Streptococcus bovis; ETBF: enterotoxigenic Bacteroides fragilis; E. coli: Escherichia coli; F. nucleatum: Fusobacterium nucleatum; B. fragilis: Bacteroides fragilis; OTUs: operational taxonomic units; tcdB: clostridium difficile toxin B; IgA: immunoglobulin A
Contrary to gastric carcinogenesis, which seems to result from a single pathogen [Helicobacter pylori (H. pylori)] [63], the composition of the gut microbiota and more particularly the imbalance between groups of species rather than a particular species seem to be decisive in defining dysbiosis that can generate CRC cancer.
There is little agreement regarding the changes seen in this cancer, despite the fact that many studies have documented dysbiosis in patients with CRC. But several species, like S. bovis, H. pylori, B. fragilis, Enterococcus faecalis (E. faecalis), Clostridium septicum (C. septicum), Fusobacterium spp., and E. coli [64], and more recently Clostridioides difficile, have been specifically considered to have a role in CRC [61].
Overall, gut microbiota diversity decrease seems to be associated with CRC. In contrast to F. nucleatum, which has been linked to disease, Bacteroidetes fragilis acts as a protective factor in the gut by regulating inflammatory immune responses. A major question asked was whether dysbiosis is a risk factor for cancer or whether the tumor is a microenvironment that influences the composition of the microbiota. In fact, it is not to be excluded that in the context of a crosstalk, a dysbiosis makes the bed of cancer which in turn accentuates this imbalance of the microbiota that can participate in the progression of the tumor. To investigate this point, it is interesting to know if dysbiosis is associated with the presentation of cancer.
Investigations into the association of gut microbiota dysbiosis with CRC were carried out considering mostly the stage, the grade of the disease, the CpG methylation, and instability of microsatellite profiles, these latter being associated with colorectal carcinogenesis pathway. The case for differences in gut microbiota composition by stage of CRC has been provided by several researchers. It seems that there are microbial metacommunities that change according to the progression of the cancer. According to one study, it is possible to anticipate the shape and function of certain mucosal microbial communities as colorectal neoplasms develop along the adenoma-carcinoma sequence [65]. However, two man species have been particularly associated with the presentation of CRC. Some researches suggest that E. coli may contribute to the pathophysiology of CRC. A connection between the tumor-node-metastasis stage and E. coli colonization of the mucosa was specifically noted. In addition, it has been shown that pathogenic cyclomodulin-positive E. coli bacteria were more common in the mucosa of individuals with stage III/IV colon cancer than in stage I colon cancer patients. The correlation between the tumor proliferative index and E. coli colonization of the mucosa was also significant [66].
Another study that demonstrated that advanced colorectal neoplasia patients’ large intestine mucosa is colonized with more virulent strains of E. coli and that higher bacteriocin production was associated with an increasing stage of CRC [assessed according to tumor, nodes and metastases (TNM) classification], supported these findings [67]. Other species were also involved in the progression and aggressiveness of CRC. Fusobacterium and also ETBF showed important associations with CRC clinicopathological features. Fusobacterium has been particularly involved in such features. Increased levels of F. nucleatum were shown to be related to CRC progression and to CRC patient survival [68]. Fusobacterium spp. levels were significantly greater in late stage (III/IV) colorectal malignancies, and an excess of F. nucleatum has been linked to a higher risk of lymph node metastases in colon cancer [68]. Regardless of the histology, F. nucleatum was discovered in premalignant colorectal lesions, however, it was more frequently found in lesions with high CpG island methylator phenotype (CIMP) levels. Additionally, F. nucleatum increased with histological grade, indicating that it might aid in the development of colorectal neoplasia. The hypothesis of the “colorectal continuum” may be supported by the presence of F. nucleatum positive in CR tumors [69]. Furthermore, mounting data suggests that F. nucleatum may contribute to particular molecular subgroups of CRCs, including the CIMP and microsatellite instability (MSI) [70].
The pathogenesis of colon, gastric, esophageal, pancreatic, laryngeal, breast, lung, and gallbladder carcinomas has been linked to gut microbial dysbiosis. This change appears to be intimately related to the host inflammation brought on by microbial dysbiosis. We will present some here the cases of lung and breast cancers (BCs) which are the most frequent around the world.
In contrast to the control group, lung cancer patients had a much lower relative abundance of several gut microorganisms. These results imply that there may be some indirect links between lung cancer and gut microbiome [71]. Lung and intestinal flora have an impact on how lung cancer develops, is treated, and is prognosed. These factors will help develop new lung cancer prevention, detection, and treatment methods [72].
BC and gut microbial dysbiosis have been linked in several studies. As compared to healthy women, the microbiota of BC patients is different, suggesting that specific bacteria may contribute to the development of the disease. Elevated serum levels of estrogen can be modified by the gut flora. Contrarily, chemicals that resemble estrogen may encourage the growth of specific bacterial species. As a result, there may be a synergistic effect between the microbiota and both endogenous hormones and estrogen-like substances that raise the chance of developing hormone-related disorders like BC [73]. According to other research, breast tissue has a unique microbiome, with some species being elevated in the breast tissue itself as well as the gut bacteria of BC patients. More significantly, the microbiome of the breast and its associated tissues may act as potential indicators for detecting and staging BC [74]. It has been hypothesized that BC development may be influenced by changes in the mammary and intestinal microbiota. Bacteria from the phyla Proteobacteria, Firmicutes, Actinobacteria, and Lactococcus spp. are abundant in healthy breast tissue and may therefore have potential preventive effects against BC. But in malignant mammary tissues, some bacteria are more prevalent. The dysbiosis of the gut microbiota, on the other hand, has been associated with BC because some gut bacteria can affect the synthesis of advantageous metabolites and interfere with the metabolism of estrogen in the stomach. Such surprising connections between BC and gut microbiota in the breast and BC suggest potential solutions for managing BC, such as through the use of probiotics, both in terms of prevention and management [75]. It has been established that the compositional abundance of some bacterial families and the synthesis of microbial metabolites vary in BC. It might be said that microbial dysbiosis appears to be linked to the development of BC [76].
How bacterial microbiota modifications could represent novel approaches for risk assessment and, may serve as prognosis markers and/or targets for innovative therapeutic strategies in cancer? To this aim, we should better understand the pro-carcinogenic effects of gut bacteria involved in microbiota dysbiosis. Through a number of ways, the bacterial microbiota causes colorectal carcinogenesis. In this review, we explore the connections between the bacterial microbiota and colorectal carcinogenesis, concentrating on dysbiosis and the potentially cancer-causing traits of bacteria, such as genotoxicity and other virulence factors, and oxidative stress. On the other hand, gut microbiota dysbiosis may be involved in tumor promotion and progression by influencing the tumor micro-environment through inflammation, host immune defenses modulation, and bacterial-derived metabolism [64].
According to such potential effects, two main hypotheses have emerged to explain the impact of dysbiosis on CRC initiation and progression. One hypothesis holds that a dysbiotic microbial population can cause cancer by altering the microbiome as a whole, triggering inflammatory reactions, and transforming epithelial cells. The “driver-passenger” paradigm proposes that intestine “bacteria drivers”, initiate CRC by inducing epithelial DNA damage and epigenetic alterations leading to tumorigenesis. The tumor generates a microenvironment that in its turn promotes the proliferation of “passenger bacteria” that takes advantage of such tumor [77].
Instability in the genome and the proliferation of epithelial cells are two factors in the development of CRC that are brought on by these mechanisms, which also have a variety of cellular effects and affect the host’s defenses. The dysbiotic bacteria implicated in cancer initiation produce toxic molecules that can induce DNA damage leading to transformation. Cyclomodulins are bacteria toxins that can induce DNA damage, interfere with the cell cycle, and/or apoptosis. Among such toxins, cytolethal distending toxin (CDT) and colibactin can directly damage DNA and induce genomic instability. Considered true genotoxins they are able to provoke double-strand DNA breaks [78, 79].
Oxidative stress could be another mechanism involved in carcinogenesis. Highly reactive molecules called reactive oxygen species (ROS) are created from oxygen. According to research, ROS induction plays a crucial part in CRC that is connected to the microbiome. Because hydroxyl radicals are potent mutagens that may cause DNA breaks, point mutations, and protein-DNA crosslinking, enterococci species, particularly E. faecalis, can produce them [80], which can contribute to genomic instability in CRC [81, 82]. The gut microbiota also promotes host-derived production of nitric oxide and its secondary nitric oxide synthases (NOS), which can induce DNA damage. Some bacterial species such as Lactobacilli and Bifidobacteria can directly generate NOS [83]. Some enteropathogenic E. coli strains have the potential to downregulate the DNA measles, mumps, and rubella (MMR) system, demonstrating the connection between oxidative stress and DNA repair mechanisms [84]. The accumulation of mutations linked to the development of CRC can result from this downregulation of the MMR repair mechanism [85]. Viljoen et al. [54] also noted that F. nucleatum colonization was more prevalent in the colon of CRC patients who had a phenotype associated with MSI caused by MMR deficiency.
These findings support the theory that the gut bacteria and DNA repair mechanism interact to cause CRC.
The dysbiotic bacteria implicated in cancer produce toxic molecules that interfere with proliferation and apoptosis signaling pathways leading to cell transformation. Some bacteria toxins may be involved in colorectal carcinogenesis through the activation of carcinogenesis-promoting pathways. The examples of cytotoxin-associated gene A (CagA) or vacuolating cytotoxin gene A (VacA) produced by some H. pylori strains are well known [86]. Through direct binding, phosphorylation of signaling proteins, and methylation of tumor suppressor genes, CagA impacts the expression or function of critical proteins in oncogenic or tumor suppressor signaling pathways [87, 88]. VacA activates the signaling pathway of the mitogen-activated protein kinases (MAPK) p38, and extracellular signal-related kinases 1 and 2 (ERK1/2). Moreover, VacA activates a signaling pathway involving G-protein-coupled receptor kinase-interactor-1 (Git1) leading to the upregulation of the β-catenin signaling pathway involved in CRC [89].
The B. fragilis toxin (BFT) is one more illustration of a bacterial toxin that contributes to the development of CRC. In order to activate the Wnt and nuclear factor-kappa B (NF-κB) pathways, the zinc-dependent metalloprotease toxin BFT must attach to a particular colonic epithelial receptor. These outcomes result in accelerated cell division, epithelial release of pro-inflammatory mediators, and development of DNA damage [90, 91].
As widely demonstrated, the microenvironment plays a determining role in the promotion of tumors and the progression of cancer. According to the theory of the carcinogenic effect of the dysbiotic microbial community, dysbiosis could be sufficient to promote cancer, suggesting that the microbial environment of the tumor should play an important role in its progression.
Among the described mechanisms, it is necessary to consider inflammation and host immune defenses modulation, as key factors in the interactions between the gut microbiota and CRC. It is now widely accepted that people with inflammatory bowel illness have a higher chance of getting CRC. Since their microbiota underwent significant alterations throughout the development of CRC, inflammation may be a result of the host’s reaction to those changes [92, 93]. One of the microorganisms most closely linked to inflammatory bowel illness is E. coli. In individuals with inflammatory bowel illness, it has been seen that adherent and invasive E. coli abnormally colonize the gut mucosa. The bacterial control of inflammation in colorectal carcinogenesis is supported by the ability of CRC-associated E. coli to promote the expression of the pro-inflammatory gene cyclooxygenase-2 (COX-2) in macrophages [94, 95]. The gut microbiota participates in metabolic processes, and it is acknowledged that CRC development is significantly influenced by microbial-derived metabolism [96]. By controlling the production of secondary bile acids, the metabolic activation of carcinogenic substances, dietary phytochemicals, and xenobiotics, hormone metabolism, and the regulation of inflammatory pathways, these metabolic activities may influence colorectal carcinogenesis [97].
Intestinal epithelial cell differentiation and proliferation, energy storage from food sources, vitamin synthesis, ion absorption, and mucus formation are all potential outcomes of these processes [98].
The TME is constituted of all the non-tumor cells and soluble molecules surrounding the tumor. It’s classically known to include pericytes, fibroblasts, immune cells, adipocytes, vascular and lymphatic endothelial cells, and factors secreted by both tumor and non-tumor cells [99, 100]. Recently, researchers reported that the microbiome is a newly recognized component of the TME [101]. Despite the fact that the intestinal microbiome has been established as an essential biomarker of cancer development and therapeutic response, less is known about the role of the microbiome in the tumor environment [101]. Emerging evidences had reported that the local microbiota constitutes an important part of the TME across many types of cancer including laryngeal, esophageal, gastric, and CRCs, as well as primary liver cancer [102]. In order to really understand its impact on cancer, it is important to understand how the microbiome can shape the TME and interact with cancer cells.
In a study conducted by Wei et al. [103], the TME was reported to ease the growth of anaerobic and facultative anaerobic bacteria such as Clostridia, while necrotic areas of the tumor were thought to cause chemotactic compounds releasing attracting then bacterial invasion [104]. The leaky vasculature of tumoral tissues also permits bacteria to penetrate and the lack of immune cells may allow their growth [105]. Distantly, the gut microbiota was demonstrated to modulate the fate of tumors in distant organs by producing metabolites and immune signals that enter the circulation to reach tumors and become a part of their TME [106, 107]. These microbial metabolites may regulate carcinogenesis by interacting with other components of the TME and participating in the immune response or angiogenesis [108–110].
Mucosal tumors are in direct contact with bacteria and therefore are susceptible to influence from the microbiome. In the gut, CRC is exposed to various bacterial species that’s why it has been extensively studied. Interestingly, the tumor tissue microbiome is not fully consistent with the composition of the fecal microbiota [62]. In CRC tissues, Fusobacteria and Firmicutes are increased, while the abundance of taxa including Fusobacteria, Providencia, and Actinobacteria are altered between cancerous and adjacent normal tissue [111, 55]. In numerous studies, the presence of these bacteria in colonic tumor tissues is associated with histologic grade, T cell infiltration, resistance to chemotherapy, and poorer overall survival [112–114]. In colonic tumors, F. nucleatum has been reported to be enriched and it is likely to have translocated from feces in the gut into colonic epithelium, inducing proinflammatory and oncogenic pathways [115, 116]. It expresses an adhesion protein called FadA, which can bond to E-cadherin and permit the bacteria to invade CRC cells, causing β-catenin regulated transcription of the oncogenes myelocytomatosis oncogene (Myc) and Cyclin D1, and increased cancer cell proliferation [115, 116]. In mouse models, F. nucleatum has been shown to induce colorectal tumor via the microRNA (miRNA)—miR21, inhibitors of which prevented CRC cell line proliferation and invasion [116]. miRNAs have been shown different expressions between normal and tumoral colonic tissues, and have been correlated with the microbiome profile of the colon, providing another mechanism by which the gut epithelium can interact with bacteria [115].
Gut microbiota dysbiosis has been long related to decreases in butyrate production within several cancer types, therefore, decreases in butyrate may cause a leakiness of the intestinal epithelium which allows bacteria to migrate to distant tissues via the vasculature [117]. Bacteria have been found in distant tumoral tissues of the intestine such as biliary, esophageal, breast, and cervical cancer, although their significance remains unclear [118, 119]. A recent study analyzing seven human tumor types disclosed a specific microbiome composition in each, and that most bacteria were localized intracellularly within cancer and immune cells of the TME [120]. These findings point out a strong physical relationship between microorganisms and cancer cells at a local level. In agreement with this, human CRC shows the same microbiome composition with its metastatic lesions in the liver, suggesting that the microbiome is able to travel with the primary tumor cells to distant sites [120]. Recently, intratumoral CRC-associated E. coli was shown to enter the liver after gut vascular barrier disruption. Once in the liver, E. coli were reported to prime the liver microenvironment through the recruitment of innate and inflammatory immune cells to directly promote metastasis [121]. Furthermore, oncogenic bacteria ascendant from the cervix were reported able to reach and colonize the uterus and ovaries during carcinogenesis [122]. In a recent study, Klebsiella pneumoniae and some common denominators (Enterobacter cloacae, Citrobacter freundii, and Fusobacterium) were linked with pancreatic cancer as well as BC [120]. These cancer-associated microbes were found to reach the pancreas via peri-intestinal translocation through the pancreatic duct due to gut epithelial barrier damage [123]. Moreover, long-term pancreatic ductal adenocarcinoma survivors had higher intratumoral bacterial diversity and the microbiome signature was significantly different from that of short-term survivors [13]. Interestingly, three enriched genera were identified in long-term survivors (Saccharopolyspora, Pseudoxanthomonas, and Streptomyces), which had a positive correlation with the number of CD8+ T cells, suggesting their role in the antitumoral immune response [13]. Functionally, the connections between gut microbiota and intratumoral microbes of the TME are only beginning to be explored and merit further mechanistic investigation.
The human virome includes both eukaryotic and prokaryotic viruses, encompassing both enteric viruses and bacteriophage, yet, there are other types of viruses in the human virome such as archaeal viruses and virophages, which are not as deeply studied due to limited understanding of their function within the environment of the human body [124]. Viruses have been reported to hold strong direct or indirect influence over host health and disease by either directly affecting host cell behavior and structure or by altering bacterial communities [125]. Multiple studies demonstrated the implication of various eukaryotic viruses infectious to humans in carcinogenesis inside and outside the intestinal tract, principally those of the families Papillomaviridae and Herpesviridae as well as hepatitis viruses [126–128]. Infections with these viruses are often one of many contributing factors to carcinogenesis and need many years of persistent infection to develop virus-associated cancers [129]. The gut virome contributes to carcinogenesis by diverse mechanisms that vary widely between viral species. Overall, viral infections promote carcinogenesis by the following mechanisms: insertional mutation in the host genome, induction of inflammation, and modulation of signaling pathways in the infected cells by viral oncogenes [130, 131].
Papillomaviridae is a family of non-enveloped viruses with double-stranded DNA genomes [132]. The human infection member human papillomaviruses (HPV) has been associated with several types of cancer, most notably cervical cancer [127]. However, only a fraction of the approximately 150 HPV types, the so-called high-risk types such as HPV-16 and -18, are commonly associated with cancer [133]. The main mechanisms for the contribution of high-risk HPV to cervical cancer are the integration of viral DNA into the host genome and the expression of viral oncogenes. For example, the early oncogenes E6 and E7 have been shown to degrade the tumor suppressor gene p53 and the retinoblastoma protein (pRb), inter alia, causing cells to arrest in S phase, leading to genomic instability, aneuploidy, and DNA damage, and thus carcinogenesis [134]. Similar mechanisms are thought to operate in other HPV-associated cancers such as CRC [135]. In addition, E7 has been reported to induce angiogenesis [136], and E6 and E7 also deregulate cellular energy. E7 leads to the Warburg effect, a shift from standard oxidative phosphorylation-based metabolism to aerobic fermentation [137], and E6 proteins prolong activation of mammalian target of rapamycin complex 1 (mTORC1) signaling by inhibiting receptor protein tyrosine kinase signaling, making cells “blissfully ignorant” regarding their energy state [138].
Viruses of the Herpesviridae family are enveloped in double standard DNA (dsDNA). Various members such as cytomegalovirus (CMV), Epstein-Barr virus (EBV), herpes simplex virus 1/2 (HSV1/2) and HSV6 and HSV7 have been implicated in cancer formation [128, 139, 140]. The strongest association with cancer has been reported for EBV, which has been linked to several cancer types including colorectal [141], esophageal [142], and gastric cancer [142]. EBV is an oncogenic γ-1 herpesvirus associated with a variety of lymphoid malignancies, including a variety of B cell, T cell, and natural killer cell (NK cell) lymphomas and epithelial carcinomas [143]. EBV mimics B cell proliferation and survival signals, enabling it to replicate its genome while remaining latent and immunosilent in host B cells, creating a lifelong persistence that favors its spread [143]. When EBV first infects naive B cells, it adopts a growth program or Late III (Lat III) pattern, and EBV expresses Epstein-Barr nuclear antigen 1 (EBNA1)–EBNA6 as well as latent membrane protein 1 (LMP1), LMP2A, and LMP2B. This expression pattern forces infected cells to become proliferating B cells, allowing EBV episomes to replicate [143].
Recently, more and more evidences have shown that GI cancer can lead to the occurrence and development of cancer, especially GI cancer. Metagenomic analysis of stool samples from patients with CRC revealed a characteristic fecal fibroma, indicating increased abundance and diversity of intestinal fibroids [144]. Enteroviral markers that distinguish CRC from non-CRC controls include orthobuniaviruses, tunalikeviruses, phikzlikeviruses, and other viral genera [144]. Phage enrichment has been demonstrated for inoviruses and tunalikeviruses [144], reported, certain species of inoviruses can regulate the production of bacterial biofilms that contribute to colon carcinogenesis [145] and significantly reduced Enterobacteriaceae phages and crAssphage compared to healthy controls [146]. With regard to eukaryotic viruses, especially human oncogenic viruses, EBV, HPV, human polyomavirus, and CMV showed higher prevalence in CRC tissues compared with non-CRC tissues [147]. Preliminary evidences also suggest that EBV infection may contribute to the development of CRC by inducing mutations in enterocytes [148]. It has been reported that EBV-infected B lymphocytes produce microvesicles containing EBV-derived molecules that translocate to intestinal epithelial cells and subsequently initiate oncogenic transformation [149]. Polyomaviruses have been reported to produce T antigens and inactivate important regulatory tumor suppressor proteins, thereby inducing chromosomal instability and malignant transformation of colonocytes [149].
Apart from the role in carcinogenesis, recent clinical studies focusing on several types of cancer strongly suggest that gut bacteria play a key role in mediating the effects and outcomes of the host response to anti-tumor drugs, especially chemotherapy and immunotherapy [150]. Microbiota modulates therapeutic effectiveness, eliminates the anticancer effect, or mediates drug toxicity, all of which have a significant impact on how cells react to anticancer therapy [6]. Concepts on how microorganisms and anticancer medications interact have undergone paradigm adjustments. These studies have shed light on how many systemic chemotherapies and immunotherapies work, and they have raised hopes that the microbiota may be altered to increase the effectiveness of therapy and lessen adverse effects (Figure 2).
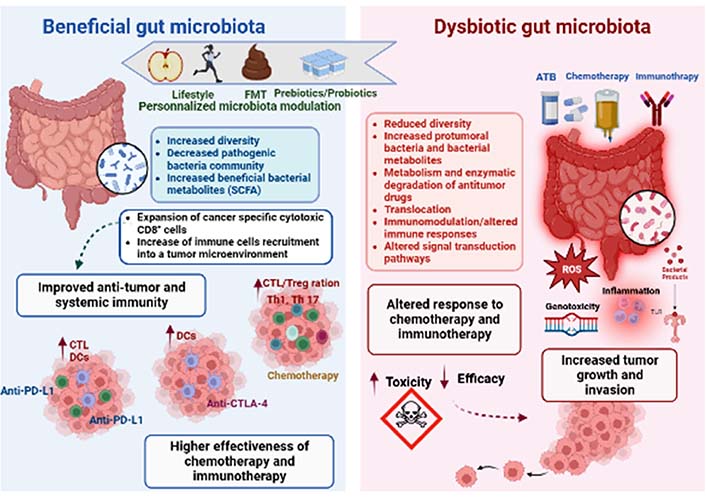
Gut microbiota impacts both anti-cancer chemotherapy and immunotherapy treatments. The gut microbiota composition and function seem to be closely related to patient responsiveness to anti-cancer therapy. The microbial composition is linked to lifestyle and could be altered by several factors, such as antibiotics (ATB), chemotherapy, or immunotherapy treatments. Thus, dysbiosis can be defined as an imbalance in number and types of microbial communities, contributing to tumor development and growth in human beings. Dysbiosis might influence the course of carcinogenesis because microbial activities appear to have an impact on genetic and epigenetic changes that result in dysplasia, clonal expansion, and malignant transformation. The dysbiotic microbiota promotes carcinogenesis in a variety of methods, including by directly activating bacterial toxins like ROS or by indirectly activating inflammatory pathways [via toll like receptor (TLR) and other pattern recognition receptors (PPR)]. Dysbiosis tends to disturb the TME leading to targeting personalized modulation of the microbiota by prebiotics/probiotics, FMT, or lifestyle (diet and exercise) intervention, which could therefore be a potential clinical strategy to enhance the anti-cancer therapeutic response. In fact, it has been proven that a favourable microbiota has the ability to enhance the efficacy of therapeutic treatments, by activating the host anti-tumor immune response. Overall, beneficial bacteria might be key determinants of improving anti-programmed cell death protein-1 (PD-1)/PD-1-ligand 1 (PD-L1), anti-cytotoxic T lymphocyte-associated antigen-4 (CTLA-4), and chemotherapy efficacy. CTL: CD8+ T cell (use only in Figure 2); Treg: regulatory T cell; Th1: T-helper 1; DCs: dendritic cells
Despite the scientific progress, we are still lacking literature on the role of the gut microbiota as an anti-cancer therapy. Little is known about microbiota-host-chemotherapy interactions in cancer. According to studies, the gut microbiota can influence how the body reacts to chemotherapy through a variety of processes, such as immunological interactions, xenometabolism, and altered community structure [4]. A study conducted by Alexander et al. [4] has demonstrated that the gut microbiota exerts, through a large suite of chemical signaling cascades, both direct and indirect effects on chemotherapy efficacy and toxicity. Although the mechanisms are not well understood, and in order to explain how bacteria affect chemotherapy treatment effectiveness in terms of translocation, immunomodulation, metabolism, enzymatic degradation, decreased diversity, and ecological variation, Alexander et al. [4] have developed a “TIMER” mechanistic framework [77].
The mechanism by which commensal or pathogenic bacteria transcend the gut barrier into the tumoral milieu, where they might increase the side effects of chemotherapy [151], was first documented in the 1960s [152]. In addition, the microbiota can modulate the chemotherapeutic response by modifying the immune response. In fact, it has been demonstrated that the continual interaction of the innate and adaptive immune systems with the gut microbiota at the mucosal surface controls inflammation and sets the immunological tone [153]. However, it has been shown that chemotherapy medications harm the mucosal epithelium, which results in bacterial translocation. This might cause a systemic infection, increasing the host’s exposure to various pathogens and priming the adaptive immune system to respond more favorably to chemotherapy [81, 82]. Immunomodulation can occur by several mechanisms. The most studied mechanisms are bacterial translocation and TH17 cell activation that enhances the action of cyclophosphamide (CTX); the activation of intraluminal myeloid cells that increases oxaliplatin action and microbiota-induced T cell activation can facilitate novel anticancer immunotherapy [154].
Numerous immunological mechanisms have been implicated in the anti-neoplastic actions of the widely used alkylating drug CTX [82, 84]. Using a mouse sarcoma model, research by Viaud et al. [155] revealed that both doxorubicin and CTX shorten intestinal villi and disrupt the intestinal barrier, causing commensal bacteria to translocate into secondary lymphoid organs. They have demonstrated that CTX can change the microbiota of the small intestine, causing bacterial species from the Firmicutes phylum, such as Roseburia, Coprococcus, and unclassified Lachnospiraaceae, as well as Lactobacilli and Enterococci, to become less prevalent [156]. Additionally, recent studies reported that a corrupted microbial barrier was more permeable to Gram-positive bacteria leading to the translocation of Lactobacillus johnsonii, Lactobacillus murinus, and Enterococcus hirae (E. hirae) from the gut into the lymphoid organs [157]. Translocated bacteria promote TH17 cell development once they reach the spleen and mesenteric lymph nodes, leading to an antitumor adaptive immune response. Recent studies have shown that vancomycin-pretreated mice had less Th17 cells in their spleens than untreated animals, and they have indicated that adoptively transferring pathogenic Th17 cells to the treated mice may restore a therapeutic response [158]. Additionally, a recent clinical study has shown that a memory Th1 immune response to E. hirae among patients with end-stage lung and ovarian cancer was proven to be a favorable predictor of progression-free survival [82, 85, 159].
Furthermore, Iida et al. [160] have investigated the impact of microbiota and myeloid cell interaction on chemotherapeutic efficacy in a murine lymphoma model. They have demonstrated that pre-treatment of mice with subcutaneous EL4 lymphoma with antibiotics could reduce both DNA damage caused by oxaliplatin and the expression of genes responsible for ROS generation by myeloid cells [161]. It leads to the conclusion that a healthy microbiota may boost the antitumor impact of oxaliplatin by prepping myeloid cells for ROS release, improving inflammatory cytokine production, and ultimately eradicating tumors [161].
The gut microbiota seems to have the potential to directly metabolize chemotherapeutic drugs and indirectly alter host-chemotherapeutic metabolism by modifying the host metabolic milieu [86, 88]. The gut microbiota seems to have the potential to directly metabolism chemotherapeutic drugs and indirectly alter host-chemotherapeutic metabolism by modifying the host metabolic milieu [86, 88]. Recent studies show that changes in the gut microbiota may be the cause of the varying clinical response to fluoropyrimidines, using the Caenorhabditis elegans (C. elegans) nematode worm as a model for symbiotic host-microbial interactions [161]. García-González et al. [162] tested the impact of either E. coli or Comamonas bacteria administration in the efficacy of camptothecin (CPT), 5-fluorouracil (5-FU), and 5-fluoro-2’-deoxyuridine (FUDR) in C. elegans. The gut microbiota seems to have the potential to directly metabolism chemotherapeutic drugs and indirectly alter host-chemotherapeutic metabolism by modifying the host metabolic milieu [86, 88]. It was found that the response to chemotherapeutic was affected by bacterial species differently. In fact, C. elegans fed by E. coli was more sensitive to the sterilizing effect of FUDR [161].
Further investigations suggested that microbial metabolism of anticancer drugs could cause severe side effects such as diarrhea, pain, and weight loss leading to dose limitation, which reduces the efficacy of anticancer treatment. Recent studies have demonstrated that irinotecan induced diarrhea in up to 30% of patients requiring dose reduction and, in many cases, the immediate termination of the drug [162]. Irinotecan (SN-38), an inhibitor of topoisomerase 1, is inactive after being subjected to hepatic glucuronidation, which results in SN-38G, which is subsequently eliminated by bile. However once in the gut lumen, SN-38G is reactivated to SN-38 by the bacterial β-glucuronidases. According to recent research, β-glucuronidase-producing bacteria including Faecalibacterium prausnitzii and Clostridium spp. contribute to the gut’s buildup of the active irinotecan metabolite (SN-38) [163].
Intestinal health and symbiosis between the host and microbiota are both maintained in large part by the composition of the bacterial community [25]. It has been reported that the microbial community is reciprocally modified by anticancer drugs. Anaerobes and streptococci were both absolute and relative microbial abundance decreasers, whereas Bacteroides were relative microbial abundance increasers, according to a research done on rats given methotrexate. Reduced villous length and diarrhea were linked to this alteration in microbial composition [164]. Additionally, a clinical study on 28 non-Hodgkin lymphoma patients who received a 5-day myeloablative chemotherapy regimen revealed profound changes in the gut microbial community’s structure and reduction of its diversity, as well as an increase in the abundance of Proteobacteria and a decrease in Firmicutes and Actinobacteria [165]. After chemotherapy, there was a decrease in taxa that have been demonstrated to suppress inflammation by altering the NF-κB pathway and by producing SCFAs. Further studies revealed a negative correlation between the quantity of Firmicutes and several metabolic pathways linked to intestinal inflammation, including cell motility, glycan metabolism, and xenobiotic degradation [165].
Cancer cells exert a variety of immunosuppression mechanisms that prevent antitumor immune responses and enhance tumor escape [166]. Among these mechanisms, cancer immuno-editing is a process tightly orchestrated by immune receptors controlling either the activation or the inhibition of immune responses, called immune checkpoints [167]. Thus, the last decade was marked by a bloom of clinical trials of immunotherapy for cancer treatment [168]. Cancer immunotherapy, based on the sensitization of the patient’s immune system, aims to boost the immune system and regulate the TME for an eventual antitumoral effect, unlike using cytotoxic treatment with chemotherapeutic agents to directly kill the tumor cells [169]. Immunotherapy has attracted attention in recent years as it is significantly advantageous to provide long-lasting anticancer effects to patients who were unresponsive to conventional therapy. In particular, the discovery and application of monoclonal antibodies against immune checkpoints have significantly advanced cancer treatment [170]. Immunological checkpoint blockade is most commonly used to inhibit the immune checkpoint regulators CTLA-4 and PD-1 or its ligand PD-L1 [171]. To date, several ICIs have received Food and Drug Administration (FDA) approval and are now widely indicated in numerous cancer types [168, 172]. In particular, metastatic melanoma and non-small-cell lung cancer are being treated with the use of PD-1 (nivolumab, pembrolizumab, and cemiplimab), PD-L1 (atezolizumab, avelumab, and durvalumab), and CTLA-4 (ipilimumab) blockers [173].
However, the patients’ responses to immunotherapy are often limited by considerable heterogeneity with resistance and immune-related adverse events [174]. Moreover, the efficacy of immune checkpoint inhibitor therapy has been shown only in 10–30% of treated patients [175]. The therapeutic outcomes depend on host and environmental factors, which have been recently connected with gut microbiota. Mounting evidence from preclinical and clinical studies highlighted the important role of gut microbiota in modulating the immune system [176], both local and systemic, the immunological response in anti-tumor activity, and immunotherapy efficacy [177].
Ipilimumab is a monoclonal antibody that works by blocking CTLA-4, mainly used for the treatment of metastatic melanoma [178]. Emergent data from preclinical studies showed that the efficacy of ipilimumab treatment is affected by gut microbiota modulation and depends particularly on distinct Bacteroides species [179]. In fact, in germ-free and antibiotics-treated mice, the response to anti-CTLA-4 treatment was compromised [180]. Besides, the efficacy of CTLA-4 blockade was restored in these in vivo models after gavage with B. fragilis, where animals responded the best to ipilimumab treatment, and the size of the tumors was negatively correlated with the outgrowth of B. fragilis [181]. It has been hypothesized that B. fragilis boosts Th1 immune response in lymph nodes that is dependent on interleukin-12 (IL-12) and encourages intratumorous DC maturation, restoring their responsiveness to CTLA-4 inhibition [182]. In other key clinical studies that strongly support the inclusion of microbial targeting in anti-tumor immunotherapy efficacy and complementing the animal models, similar observation has been reported in a good responder group of patients treated with ipilimumab [183]. The analysis of stool samples among this group showed a selective enrichment of B. fragilis [183]. In the same context, other species, like Bifidobacterium breve and Bifidobacterium longum have been correlated to the activation of immune cells in the TME during ipilimumab therapy [184].
Similarly, the clinical response to the antitumor monoclonal antibodies against-PD-1 or its ligand PD-L1 was shown to have a direct link with microbiome signature [185]. A recent animal study showed improved efficacy of the anti-PD-1/PD-L1 treatment in germ-free mice transplanted with fecal microbiota from good or poor responders and supplemented with Clostridiales and Faecalibacterium species [186]. Another report has demonstrated that the addition of Akkermansia muciniphila, Alistipes indistinctus, or E. hirae could invigorate the immunotherapy treatment in germ-free mice [187, 188]. Accordingly, a metagenomic analysis showed that the enrichment of Akkermansia muciniphila in faecal samples from non-small cell lung cancer (NSCLC) and renal cell carcinoma (RCC) patients were related to a promising clinical outcome during anti-PD-1 and anti-PD-L1 therapies [189]. Further, a high level of commensal bacteria in patients with melanoma was correlated with enhanced antitumor immune response. One suggested mechanism to explain the improvement of immunotherapy treatment is an increase in intratumoral infiltration of CD8+ T cells with enhanced tumor-specific T cell and DC-mediated immune responses [190]. On the other hand, the reduced response or the resistance to PD-1/PD-L1 immunotherapy has been associated with a high abundance of Bacteroidales [191]. Nevertheless, numerous studies pointed out the impact of antibiotics to engender dysbiosis that subsequently compromise the anti-PD-1 efficacy in cancer patients [192]. For instance, across the different cohort studies related to the anti-PD-1 or PD-L1 therapies, an overlap in the involved microbiota species has been raised. The heterogeneity of cancer types, the tumor’s microenvironment, the host genetic background and immune system, or even the sequencing approaches, may lead to biases [192]. It seems likely that a microbial responder profile reflects a combination of key species instead of a separate one.
When considered collectively, those results strongly imply that a balance between “useful” and “non-beneficial” bacteria may be able to influence the effectiveness of both immunotherapy and chemotherapeutic treatments. An anticancer medication has the potential to change the gut microbiota’s composition and the supplementation of beneficial bacterial strains may lead to better efficacy and response of the anticancer therapy.
It is conceivable to target the gut microbiota as a therapeutic strategy given its crucial involvement in cancer therapy. Recently, few studies explored microbiota modulation to improve cancer therapies response, however, the mechanisms remain less understood [193]. In the current section of the review, we concentrated on prospective methods for modifying the microbiota to increase therapeutic effectiveness.
The cornerstones of modern anti-cancer therapies, including chemotherapy, targeted therapy, immunotherapy, and radiation, can have a variety of, sometimes severe, adverse effects on patients [194–196]. In order to assess the probiotics’ overall effectiveness in reducing the risk and severity of such anti-cancer treatment-related toxicity, a number of preclinical and clinical investigations have been conducted [4]. In fact, the administration of probiotics to cancer patients, mainly Lactobacilli, is to re-populate the altered patients’ gut microbiota, thus re-establishing the composition and functionality of the commensal bacteria [197]. Probiotics are typically viewed as harmless, however, giving them to immunocompromised cancer patients runs the danger of passing on antibiotic resistance and opportunistic infections [198]. Despite this, probiotics have been proven to be effective in treating diarrhea and other gut-related side effects after anti-cancer treatment, leading to the restoration of a balanced intestinal microbiota [199]. The combination of the two probiotic species Lactobacillus johnsonii and Bifidobacterium longum was given to cancer patients for the first time in 2010. Researchers discovered that Lactobacillus johnsonii was able to adhere to the colonic mucosa, lowering the concentration of gut pathogens and modifying the local immune system [200]. In a clinical randomized control study, patients treated for a nasopharyngeal carcinoma received a combination of probiotics which resulted in an enhanced immune response and a decreased rate of oral mucositis after chemoradiotherapy. Indeed, the probiotic combination has been shown to increase the number of CD4+ T cells, CD8+ T cells, and CD3+ T cells [201].
Viaud et al. [155] found that tumor-bearing mice treated with CTX had dysbiosis, which was a breakdown of gut mucosal integrity related to chemotherapy. In mice given antibiotic treatment, they observed a decreased CTX-induced Th17 cell conversion and poorer tumor remission. But oral administration of E. hirae and Lactobacillus johnsonii to these mice restored the anti-tumor effectiveness of CTX by enhancing the T cell immune response [155]. Altogether, this study highlights that E. hirae and Lactobacillus johnsonii could be used as probiotics in combination with CTX therapy to improve treatment efficacy in cancer patients.
Good evidence in both animal and human studies supports that dietary fibers known as prebiotics could be metabolized by gut bacteria leading to an increase in the colonization and relative growth of particular bacteria and their metabolites, which may improve the anti-tumor treatment effect. The presence of certain bacteria in the human gut has been proven to be necessary for prebiotics to have any possible impact. In order to cure cancer, synbiotics, a mix of probiotics and prebiotics, appear to be helpful. According to recent research, decreased fiber intake lowered the amount of SCFA that the microbiota generated and increased the use of less-preferred substrates including amino acids and host mucins [202]. PD-1 blockade medication is currently being used to treat melanoma patients, and clinical investigations have shown that increases in bacterial diversity brought about by high-fiber diets significantly enhanced progression-free survival [203]. To firmly support the efficacy and safety of giving probiotics, prebiotics, and synbiotics during or after anti-cancer therapy, more preclinical and clinical studies may be required.
Due to its potential therapeutic value, the 4th-century Chinese idea of FMT has lately attracted attention from researchers in biology and clinical medicine [204]. FMT involves the transplantation of stool collected from a healthy donor into the intestinal tract of a patient with an altered intestinal microbiome. FMT was originally utilized to control host metabolism in obese people, outside of the setting of oncologic illnesses [205]. Indeed, the transplant of the FMT from lean donors to obese recipients has resulted in variable improvements in the insulin-sensitivity which was associated with an increased abundance of butyrate-producing intestinal microbes [206]. Interestingly, the transfer of FMT from good responders to anticancer treatment to mouse models has been reported to enhance treatment response in these animals. In fact, the study found that mice getting FMT from treatment strong responders had a larger proportion of CD8+ T cells and cells expressing CD45+, CD11b+, and lymphocyte antigen 6 complex, locus G (Ly6G+) and a lower level of myeloid cells showing CD11b+ and CD11c+ [207], suggesting a significant enrichment of adaptative and innate effector cells and a decrease of cells with suppressive activity. Further, anticancer immunity was suppressed in receivers of fecal material from poor responders because they had larger amounts of CD4+, IL-17+, Th17 cells and CD4+, forkhead box P3 (FoxP3+) Tregs in the spleen [207]. Decrease in the number of CD4+ FoxP3+ Treg cells with suppressive activity and an increase in the activity of CD8+ cytotoxic T cells. In line with this context, it has been proposed that the primary role of neutrophils in promoting antitumor immunity is to control IL-17 secretion and thus indirectly suppress tumor growth by promoting CD8+ T cell function [207].
Additionally, recipient germ free (GF) mice were given feces from three responders and three non-responders, followed by B16 implantation. Receivers of fecal material from responding patients showed increased tumor control, boosted T cell responses, and improved anti-PD-L1 treatment effectiveness in SIY melanoma cells. Two of three mouse cohorts implanted with responder feces showed slower tumor growth, whereas two mice transplanted with non-responder feces showed quicker baseline tumor growth [208]. Collectively, these results revealed that a “favorable” gut microbiome may enhance systemic and anti-tumor immune responses.
According to several clinical investigations, fecal treatment may be a potential approach in treating a variety of human ailments as FMT is used more frequently.
A novel therapeutic intervention has been recently developed known as fecal virome transplantation (FVT), where only the viral component from feces is transplanted [209, 210]. A study conducted by Draper et al. [211] showed that FVT mainly consisted of phages, and improved antibiotic-induced bacterial dysbiosis. The transplanted phages have shown to be able to colonize the gut and reshape the bacteriome similarly to a pre-antibiotic state. In FVT studies conducted in murine models [210], the gut viral composition was shown to be dominated by the order Caudovirales and the family Microviridae viruses. In another study, treatment with lytic and temperate gut phages modulated gut microbial composition. In fact, they demonstrated that lytic phages enhanced the beneficial species of gut microbiota, and temperate phages stimulated the growth of commensal in the gut [209]. More profoundly, phages are not only able to modulate the microbiome but also its associated metabolome [212]. Indeed, Hsu et al. [212] revealed that gnotobiotic mice were subjected to lytic phages after they were colonized with commensals. These findings suggested that phage-led microbiome modulation was indeed due to intense microbe–microbe (intra- and inter-microbial) interactions which resulted in remarkable changes in the metabolome.
Huge efforts are being made today to better understand the gut microbiota’s involvement in influencing how the body reacts to cancer treatments as well as the dynamic interplay between the host and gut microbiota. In the current review, we highlight the critical role of gut microbiota to modulate the course of carcinogenes and the immune response, impacting therefore the efficacy of cancer chemotherapy and immunotherapy. Dysbiosis may affect the path of carcinogenesis because microbial activities seem to have an impact on epithelial DNA damage and epigenetic modifications that result in dysplasia, clonal expansion, and malignant transformation. The dysbiotic microbiota promotes tumorigenesis in a number of ways, such as by directly activating bacterial toxins like ROS which is involved in DNA damage, CagA, and VacA that interfere with the proliferation and apoptosis pathways or by indirectly activating inflammatory pathways induced by microbe-associated molecular pattern (MAMP) activating TLR and other PPR. Interestingly, distinct beneficial bacterial species were suggested to improve the antitumor effect and to characterize good responders to cancer therapy. It has been suggested that the microbiota may exert its preventative effects by rendering carcinogens inactive and producing SCFAs, such as butyrate, and propionate. However, the exact mechanisms underlying this crosstalk remain elusive and merit a more comprehensive understanding. Besides, valuable works on the modulation of gut microbiota, based on prebiotics/probiotics/FMT, have shown promising improvement in cancer therapy response. These strategies have demonstrated promising results through a variety of mechanisms, including altering the microbiota composition, modulating the innate and adaptative immune responses, improving gut barrier function, preventing pathogen colonization, and exerting selective cytotoxicity against tumor cells. However, it should be noted that they come with drawbacks and controversies that may result in clinical problems. Although this innovative field is still limited, targeting microbiota might provide novel therapeutic strategies to treat cancers.
B. fragilis: Bacteroides fragilis
BCs: breast cancers
C. elegans: Caenorhabditis elegans
CagA: cytotoxin-associated gene A
CRC: colorectal cancer
CTLA-4: cytotoxic T lymphocyte-associated antigen-4
CTX: cyclophosphamide
E. coli: Escherichia coli
E. hirae: Enterococcus hirae
EBV: Epstein-Barr virus
F. nucleatum: Fusobacterium nucleatum
FMT: fecal microbiota transplantation
FVT: fecal virome transplantation
GI: gastrointestinal
H. pylori: Helicobacter pylori
HPV: human papillomaviruses
HSV1/2: herpes simplex virus 1/2
LMP1: latent membrane protein 1
MMR: measles, mumps, and rubella
PD-1: programmed cell death protein-1
PD-L1: programmed cell death protein-1-ligand 1
ROS: reactive oxygen species
SCFA: short-chain fatty acids
TME: tumor microenvironment
Treg: regulatory T cell
VacA: vacuolating cytotoxin gene A
SK, OZ, and ABAEG: Conceptualization, Writing—original draft. ZB, HR, AA, FBA, AM, and AC: Conceptualization, Writing—review & editing.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Mohammad A. Al-Mterin, Eyad Elkord
Maria Teresa Palano ... Lorenzo Mortara
Mohamad Omar Ashi ... Stéphanie Corgnac
Pengkun Yuan ... Bin Ma
Flora Doffe ... Pierre Savagner
Manisha Singh ... Rachana
Vanessa C. Talayero, Miguel Vicente-Manzanares
Matthew Moghaddam ... Benjamin Bonavida
Max Kam-Kwan Chan ... Patrick Ming-Kuen Tang
Ai Tsuji ... Satoru Matsuda
Sayuri Yoshikawa ... Satoru Matsuda
