Affiliation:
1Section of Gastroenterology, Hepatology and Nutrition, University of Chicago Medicine, Chicago, IL 60637, USA
Affiliation:
1Section of Gastroenterology, Hepatology and Nutrition, University of Chicago Medicine, Chicago, IL 60637, USA
2Inflammatory Bowel Disease Center, Department of Gastroenterology and Liver Diseases, Tel Aviv Medical Center, Tel Aviv 6423906, Israel
3Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv 6997801, Israel
Email: Nathaniel.cohen@uchospitals.edu
ORCID: https://orcid.org/0000-0001-9252-9208
Explor Med. 2022;3:219–233 DOI: https://doi.org/10.37349/emed.2022.00087
Received: January 31, 2022 Accepted: April 24, 2022 Published: May 31, 2022
Academic Editor: Lee M. Wetzler, Boston University School of Medicine, USA
The human body contains trillions of microbes which generally live in symbiosis with the host. The interaction of the gut microbiome with elements of the host immune system has far-reaching effects in the development of normal gut and systemic immune responses. Disturbances to this intricate relationship may be responsible for a multitude of gastrointestinal and systemic immune mediated diseases. This review describes the development of the gut microbiome and its interaction with host immune cells in both health and disease states.
The human microbiome is constructed of a multitude of organisms including bacteria, viruses, and fungi, which inhabit all surfaces of our bodies. Microbiomes exist on the nose, mouth, lungs, skin, stomach, sexual organs and of course the gut. The organisms comprising these microbiomes exist, largely, in symbiosis with the human host, and research over the past decade has highlighted the significant role they play in human health and disease.
The gut microbiome contains a rich and diverse microbial community consisting of over 100 trillion microorganisms [1]. In fact, the gut microbiome is considered one of the most densely populated microbial habitats and encodes for over 3 million genes as opposed to the 23,000 human genes [2, 3]. This translates to the production of thousands of metabolites which are critical to the symbiotic relationship between humans and the microbes and contribute significantly to maintaining homeostasis. The use of new technologies such as rapid throughput 16S rRNA gene sequencing and advanced analytic techniques has enabled both the identification and quantification of individual components of the gut microbiome and the dissection of their relationship to health and a multitude of human diseases [4, 5].
Indeed, these advances have revealed significant associations between perturbations in the gut microbiome and gastrointestinal diseases such as irritable bowel syndrome [6], inflammatory bowel disease (IBD) [7–9], celiac disease [10] and colorectal cancer [11] as well as a plethora of extra-intestinal diseases for example metabolic disorders such as obesity and diabetes [2, 12, 13], central nervous system diseases [14, 15] and many immune mediates diseases such as psoriasis [16], systemic lupus erythematosus [17] and rheumatoid arthritis (RA) [18] to name but a few.
The common feature of many of these diseases is that they are mediated by dysregulation of the human immune system. Recent research has shown the gut microbiome to be an important role-player in the development and modulation of host immune responses [19]. The aim of this review is to discuss the early development of the gut microbiome and how its interaction with the human immune system plays a role in both health and disease. As this topic is so vast, this review will detail some of the most important interactions and mechanisms.
Initial exposure to microbes begins in utero [20, 21] and expands dramatically following birth. There are multiple factors which influence and shape the gut microbiome in neonates and infants, and these are predominantly related to maternal-offspring exchanges of microbiota. The initial mass colonization event is childbirth. During delivery, the newborn is exposed to the vaginal, fecal and skin microbiota of the mother. As such, mode of delivery, either vaginal delivery or cesarean section, has a significant impact on the initial microbiome. Babies born by cesarean section have no vaginal microbes such as Lactobacillus, Prevotella and Sneathia spp. [22] and are instead colonized by skin bacteria such as Staphyloccocus, Corynebacterium and Propionibacterium spp. [22], in addition, there is delayed colonization by Bacteroides and Bifidobacterium spp. [23, 24] and higher levels of intestinal Clostridioides difficile (C. difficile) [25]. It is of interest that differences in composition of gut microbiome depending on mode of delivery have been seen up to 7 years after birth [26]. It is still unclear the long-term effects of mode of delivery and subsequent differences in gut microbiome have on immune development. In one study [27], the gut microbiota and T-helper (Th) cell immune responses of infants born vaginally were compared with those born by cesarean section and found that cesarean section born infants had lower microbial diversity with significantly lower abundance of the Bacteroidetes phylum and also had significantly lower type 1 T-helper (Th1) cell associated chemokines resulting in reduced Th1 cells responses which persisted during the first 2 years of life [27]. This is of interest as members of the Bacteroidetes phylum, including Bacteroides fragilis (B. fragilis), have been shown through interactions with the capsular polysaccharide, to induce the production of interleukin 10 (IL-10) and other cytokines, alter the Th cell balance and promote immunotolerance [28–30]. This alteration in Th cell balance and function may, in part, explain the association noted between cesarean section born infants and the later development of various immune mediated diseases including allergic conditions and type 1 diabetes (T1D) [31–33].
In addition to mode of delivery, factors such as antibiotic exposure and formula feeding also play an important role in shaping the gut microbiome. Formula feeding has been associated with increased prevalence of C. difficile [34], B. fragilis and Escherichia coli [34, 35] and decreased bifidobacterial [36]. It has been shown that even formula feeding in small quantities can influence the structure of the gut microbiome [37]. Antibiotic exposure has also been shown to interfere with the normal development of the gut microbiota with intrapartum antibiotics resulting in decreased bacterial diversity and lower abundance of lactobacilli and bifidobacterial [38, 39].
Until the age of 3, the gut microbiome shows high variability after which it reaches a more adult like composition [40, 41]. This coincides with an immature immune system highlighted by poorly regulated immune responses as seen in diseases such as necrotizing enterocolitis [42] as well as increased susceptibility to various infections [43]. It is therefore thought that early interaction with a structurally altered microbiome may lead to increased susceptibility to disease later in life such as asthma and IBD [44, 45].
Studies, primarily on germ free (GF) mice, have elucidated the role of this early host microbiome on the maturation of the immune system in greater detail. GF mice by definition have no microbiome, and compared with regular mice, show several immunological defects including a reduction in lymphoid cell numbers and function [46]. GF mice have fewer Th1 cells which promote cell-mediated immune responses and phagocyte-dependent inflammation to target intracellular pathogens [47, 48]. Interestingly, colonization of GF mice by a variety of microbes can restore Th1 responses. For example, Listeria monocytogenes promotes Th1 development through macrophage production of IL 12 which is a T-cell stimulating factor [49]. In addition to defective Th1 responses, GF mice also have a reduction in Th17 cells. Although generally pro-inflammatory, these cells mediate defense against extracellular pathogens and autoimmune disease [47, 48].
It must be stated that the role of the gut microbiome is not limited to the intestinal tract. The spleen and mesenteric lymph nodes of GF mice have absent lymphocyte zones [28], indicating a far-reaching and systemic influence of the gut microbiome on the host and providing insight into the possible future pathologic development of numerous diseases. Further, it appears that certain abnormalities in the immune system caused by a defective early interaction, may not be repaired by a later introduction of a normal microbiome. In GF mice, the ability to restore some of the cellular defects that occur is restricted to a short time interval in early life beyond which intestinal immune development cannot be fully achieved in an adult [50], which indicates that there may be a “window of opportunity” to influence the gut microbiome and its immune-related influences.
The immune system comprises a complex network of innate and adaptive components which have an extraordinary capacity to adapt and respond to a multitude of challenges. This essentially allows the host to maintain homeostasis and sustain or restore tissue function despite constant microbial and environmental exposures [51]. Indeed, a large proportion of the immune system’s constitutive function is aimed at controlling this interaction. One strategy the host uses to maintain this healthy interaction is through the development of structural and immunological components which form a physical barrier and minimize the contact between the microbes and the immune cells.
This barrier includes mucus, epithelial cells, secretory immunoglobulin A (IgA), numerous antimicrobial peptides and immune cells [52] and limits contact and translocation as well as impacts commensal gene expression and prevents bacterial adhesion. The components and important interactions that comprise this structural barrier are detailed in Figure 1. The gut epithelium consists of a single layer of various epithelial cells with specialized functions. These cells have a multifaceted role in maintaining the physical barrier. Certain cells secrete antimicrobial peptides such as alpha-defensins, phospholipases, and lysozyme C which are important in warding off pathogens. The rapid turnover of these epithelial cells limits pathogen adherence and gut colonization [53] and the mucous producing goblet cells lubricate and protect the epithelial lining as well as provide a milieu for antigen presentation [54]. Another important aspect of this physical barrier is the tight junction complexes which form a dynamic seal-preventing infiltration of pathogens while allowing for the exchange of water, ions and nutrients between the gut lumen and the blood stream or lymphatics [55].
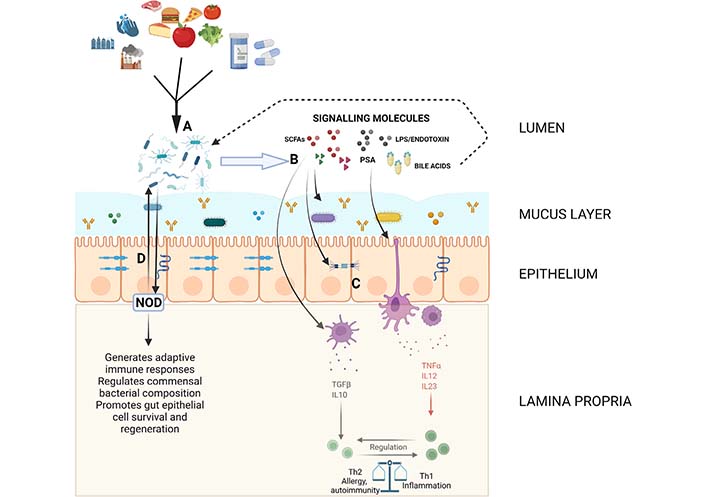
Diagram showing some of the important pathways and interactions in maintaining gut homeostasis. The mucous layer containing soluble IgA, antimicrobial peptides and some commensal organisms as well as the epithelial layer with proteins forming various intercellular junctions composes the structural barrier controlling the interaction between luminal contents and immune components. (A) Environmental and dietary factors influence the composition of the gut microbiome; (B and C) the microbiome produces or alters a variety of signaling molecules including SCFAs, LPS/endotoxin, PSA and bile acids which regulate intestinal barrier function and modulate innate immune responses which eventually lead to downstream shifts in adaptive immune function; (D) bacterial sensing molecules such as NOD molecules interact with and regulate commensal bacteria. Created with BioRender.com. LPS: lipopolysaccharide; NOD: nucleotide-binding oligomerization domain; PSA: polysaccharide A; SCFAs: short chain fatty acids; TGFβ: transforming growth factor β; TNFα: tumor necrosis factor α
The importance of this barrier, in maintaining homeostasis and the interaction with commensal and pathogenic microbes, is most notable in certain disease states. For example, during infection with Vibrio cholera, bacterial toxin stimulates the adenylate cyclase enzyme in the intestinal brush border, increasing the production of cyclic adenosine monophosphate (AMP) which in turn promotes the secretion of chloride ions and fluid accumulation in the gut lumen and subsequent severe diarrhea [56]. In addition, disruptions in this physical barrier, resulting in increased gut permeability and altered bacterial sensing have been associated with the pathogenesis of Crohn’s disease (CD) and could even be used as a biomarker for the risk of CD development [57].
The gut microbiota is intrinsically important to the function of the host’s innate immunity, both locally and systemically, by influencing the development and function of antigen presenting cells (APCs), neutrophils and other innate cell types. APCs have co-evolved with microbiota and this shapes their ability to protect the body against infection while maintaining immune tolerance to normal gut microbiota. An example of this is that dendritic cells (DCs) from Peyer’s patches produce high levels of IL-10 compared with the same cells in the spleen [58]. It is well established that IL-10 has an important role in attenuating inflammatory responses and maintaining homeostasis through T-cell regulation [59]. In addition, intestinal macrophages do not produce pro-inflammatory cytokines in response to microbial stimuli such as toll-like receptor (TLR) ligands [60] further highlighting the important role of immune tolerance development over time (Figure 1).
The gut microbiota also influences APCs outside of the gut milieu. It has been shown that microbe-derived adenosine triphosphate (ATP), via stimulation of DCs leads to the differentiation of Th17 cells [61]. These cells have a role in both promoting and attenuating inflammation and the immediate microenvironment, including the interaction with an array of various cytokines, the gut microbiome and metabolites such as SCFAs, regulating the balance between Th17 cells and regulatory T cells (Tregs) [62–64]. The microbiota also has a systemic influence over the regulation of neutrophils. It has been shown that GF rats are actually neutropenic and the neutrophils that are present have impaired phagocytic and superoxide anion and nitric oxide generation [65, 66]. These features are critical to the antimicrobial function of neutrophils. Further, it has been shown that recognition of gut microbial peptidoglycan enhanced the killing activity of bone marrow neutrophils [66]. Furthermore, the gut microbiota has also been shown to be involved in the development of other innate cell types such as natural killer (NK) cells and mast cells [67–69]. Despite these data, it is still unclear the exact influence the gut microbiome has in terms of regulating, i.e. promoting or suppressing, neutrophilic function. Future studies investigating this are required.
The gut microbiota plays a significant role in the development of the major subtypes of CD4+ T cells: Th1, Th2, Th17 and Tregs. GF mice have been shown to have a Th1/Th2 imbalance with a bias towards Th2 responses which is interesting considering the connection of gut dysbiosis to numerous atopic conditions such as asthma and eczema [44, 50, 70]. As mentioned above, specific bacterial species such as B. fragilis can induce the systemic development of a Th1 response through its PSA molecule [71]. Thus, it is of interest whether manipulating the gut microbiota can have an influence on Th1/Th2 balance and alter disease activity. A recent study investigated the use of fecal microbial transplantation in patients with active atopic dermatitis, a disease hallmarked by a shift to Th2 response, and found that most patients had significant improvement in disease severity scores with reduction in corticosteroid dependence [72]. This study, corroborated previous evidence from mice models which showed restoration of gut microbial diversity and Th1/Th2 immunologic balance, modulated Tregs, reduced levels of IgE, masts cells, basophils and eosinophils and resulted in suppression of atopic-dermatitis induced allergic responses [73].
As can be seen from the above, Tregs, are important in modulating inflammatory responses by suppressing other cell types and as such help prevent autoimmune disease [74]. Recent data show that the gut microbiota influences Tregs development. Clostridia can promote their induction [75], B. fragilis can signal Tregs to suppress pro-inflammatory Th17 responses [76] and colonic Tregs have a unique collection of T cell receptors that recognize colonic bacterial contents [77]. The gut microbiota has also been shown to be important for the presence and function of intestinal CD8+ T cells including their ability to modulate other peripheral immune cells such as marginal zone B cells, plasmacytoid DCs and NK cells [78–81]. Taken together, it can be deduced that disturbed interaction between gut microbiota and T cells can lead a more pro-inflammatory milieu within the gastrointestinal tract and beyond.
In addition to T cells, the gut microbiota, also influences B cell maturation and immunoglobulin production. Gut associated B cells are mostly found in the Peyer’s patches and are mostly IgA secreting plasma cells [47]. In GF mice, a reduced number of plasma cells and decreased level of IgA have been observed [82]. As mentioned above, the spleens of GF mice also contain fewer and smaller germinal centers where B cell differentiation and affinity maturation occur [83]. Accordingly, serum natural IgG levels are severely reduced in GF animals [84]. Furthermore, microbial exposures have also been shown to induce characteristic immunoglobulin heavy chain repertoires in B cells and systemic exposures to microbes induce diversified IgG production [85]. While the production of a diverse immunoglobulin profile to the gut microbiome is intuitive, the mechanism behind the apparent selectivity of immunoglobulins to tolerate beneficial commensal over potentially harmful bacteria, is unclear. Interestingly, IgE, the allergy associated Ig isotype has been found to be increased both in the gastrointestinal tract and systemically which is consistent with the Th2 response predisposition in GF animals [86]. Although it is clear the gut microbiota plays a role in B cell development and function, the exact interplay between the microbes and immunoglobulin diversity remains unknown and should be the subject of future studies.
One of the most important aspects of gut homeostasis remains the ability to maintain a boundary between gut luminal contents and immune cells within the submucosal space and beyond. Integral to this, is the maintenance of a set luminal pH, mucosal mucous layer and tight junctions between mucosal cells. SCFAs, namely butyrate, acetate, formate amongst others, play a vital role in maintaining this barrier. They are produced by the gut bacterial fermentation process of non-digestible carbohydrates and amino acids that have escaped digestion and absorption in the small intestine [87]. These SCFAs are then absorbed by the colonocytes and used as fuel for the colonic mucosal epithelial cells or enter the portal blood stream [88].
There are specific bacteria that are responsible for SCFA production and these have been relatively well characterized, in particular, those involved in the generation of propionate, butyrate and lactate. The production of butyrate, for example, is largely dominated by the breakdown of resistant starch by organisms such as Ruminococcus bromii, Faecalibacterium prausnitzii (F. prausnitzii), Eubacterium rectale, and Eubacterium hallii [89, 90]. Dysbiosis, resulting in alterations to the relative proportions of these and other critical bacteria reducing SCFA production, has a direct effect on gut integrity. Mechanistically, butyrate reduces mucosal level oxidative stress by consuming local oxygen and thus stabilizes hypoxia-inducible factor which is also important for barrier protection [91]. Furthermore, a decrease in butyrate which enhances intestinal barrier function through increasing expression of proteins such as claudin-1, zonula occludens-1 (ZO-1) and occludins [92], in turn increases LPS translocation promoting pro-inflammatory cascades in immune cells that lead to the further activation of downstream signaling pathways such as a nuclear factor kappa B (NF-κB) and mitogen-activated protein kinase, and cytokine production such as TNFα and IL-6 [93–95]. This resulting inflammatory state has been linked to both gastrointestinal [96] and systemic conditions including dermatologic, cardiovascular, renal and neurological diseases [16, 97–100]. More studies are needed at this time, particularly in vivo human studies, to fully elucidate the role of butyrate and other SCFAs on colonic and immune function.
Another important factor in maintaining homeostasis is the pattern recognition receptors (PRRs) which sense microbial signals and are involved in defense against pathogens, regulating the composition of the commensal microbes and maintaining physical barrier integrity [101]. TLRs, a type of PRR, are highly diverse and have a significant role in immune system regulation. For example, PSA which is produced by B. fragilis, is recognized by the TLR1/TLR2 complex and results in the downstream expression of anti-inflammatory genes [102]. Other PRRs which play an important role in intestinal homeostasis are NOD1 and 2. NOD1 serves as an innate sensor which assists in the generation of adaptive lymphoid tissues and NOD2 is a bacterial sensor which prevents intestinal inflammation through restricting growth of certain commensal bacteria and promoting gut epithelial cell survival and regeneration (Figure 1) [103–105]. Indeed, the importance in NOD2 in intestinal homeostasis is highlighted by its role in IBD pathogenesis when mutated [106]. Other NOD like receptors (NLRs) such as NOD-, leucine-rich repeat (LRR)- and pyrin domain-containing 6 (NLRP6) form inflammasomes which are important in regulating goblet cell mucus secretion and also regulate intestinal antiviral immunity [107, 108].
An important interaction between bile acids and the gut microbiome occurs resulting in both positive and negative effects on commensal bacteria. Bile acids, independent of the gut microbiome, can effect gut mucosal integrity as a result of direct damage to cells and alterations in oxidative stresses occurring in the immediate cellular microenvironment [109, 110]. Furthermore, certain bacteria, predominantly gram negatives, are more resistant to the actions of bile acids [111]. This implies that specific bile acid composition can either support or hamper the growth of certain bacterial colonies. For example, primary bile acids have been shown to restrict the growth of gram negative bacteria in the small intestine [112]. In addition, primary bile salts have also been shown to promote the germination of bacterial spores which helps in restoration of a diverse microbial population after a particular insult [113]. While generally beneficial this interaction can also lead to the germination of pathogens such as C. difficile. This complex interaction has implicated bile salts in a wide ranging spectrum of diseases including IBD [114], primary sclerosing cholangitis [115], metabolic syndrome [116], colorectal cancer [117] amongst others.
It is apparent from myriad interactions and influences of the gut microbiome on all aspects of the hosts immune system that perturbations to this symbiotic relationship are linked to the development of autoimmune and immune-mediated diseases. There is an ever-growing body of evidence showing how gut microbial dysbiosis is associated with both intestinal and extra-intestinal diseases. One of the more obvious immune-mediated diseases thought to develop from this disturbed interaction is IBD. Many studies have shown altered microbiota composition in IBD patients characterized as a reduction of Firmicutes and Bacteroides species and an overgrowth of proteobacteria [118, 119]. Further evidence of a possible causative role for certain bacteria in the pathogenesis of IBD comes from a study which showed that reductions of F. prausnitzii, increased the risk of post-operative recurrence in CD patients [9]. Further, in a mouse colitis model oral administration of F. prausnitzii led to a significant reduction in the severity of colitis and also tended to correct the associated dysbiosis [9]. It appears that these effects are induced via the secretion of metabolites which block NF-κβ activation and IL-8 production [9]. More evidence comes from studies which have shown that B. fragilis, Bacteroides thetaiotaomicron (B. thetaiotaomicron) and SCFAs can ameliorate colitis through diverse mechanisms including the production of the anti-inflammatory IL-10 and enhancing the nuclear export of peroxisome proliferator-activated receptor-ү (PPAR-ү) [29, 120–123]. Also as described above, Clostridium plays a role in the upregulation and anti-inflammatory effects of Tregs which coincided with a reduction of dextran sodium sulfate (DSS)-induced colitis in mice [75].
Beyond the gastrointestinal tract, changes in gut microbiota have been shown to be associated with many systemic autoimmune diseases. An example of this is in RA where in a spontaneous arthritis mouse model, arthritis was reduced in GF mice and introduction of a single gut microbiota species was able to trigger joint inflammation [120]. An interesting example of how gut microbiota may be protective from disease is in T1D where in a non-obese diabetic mouse model, GF mice have been found to have significantly higher rates of diabetes [121]. This is consistent with the finding that T1D is more prevalent in countries with strict hygiene practices [122]. The gut microbiota has also been shown to have distinct patterns in patients with psoriasis, with significant increases in the Firmicutes and Actinobacteria phyla compared to matched controls and increased activity in lipopolysaccharide metabolic pathways [16].
The colon harbors the highest concentration of bacteria in the gastrointestinal tract. This, together with studies showing a 12-fold increase in the risk of colorectal cancer compared with cancer in the small bowel [123], point to the important role the microbiome plays in cancer development. Certain bacteria, including Streptococcus gallolyticus and Fusobacterium nucleatum, have been directly implicated in colorectal carcinogenesis [124, 125]. These bacteria induce inflammation and alter host immune responses through various cellular adhesion molecules and suppress the function of local immune cells including macrophages and Tregs. Fusobacterium nucleatum in particular disrupts tight junctions and activates beta-catenin which promotes the transcription of multiple oncogenes [126, 127]. Taken together, these changes, provide a microenvironment which promotes carcinogenesis [125]. While certain bacteria are implicated in carcinogenesis, other bacteria appear to have an important role in anti-tumor activity. Both Bifidobacterium longum and Bifidobacterium breve have been shown to increase the function of DCs which subsequently recruit cytotoxic T-cells to a tumor microenvironment and suppress tumor growth [128]. Furthermore, bacterial metabolites such as the SCFAs, butyrate and propionate, have anti-cancer effects by inducing programmed cell death [129]. While complex, these interactions of various components of the microbiome with tumorgenesis, could potentially allow for the development of biomarkers for cancer diagnosis, predictors of treatment response and also development of adverse events on therapy.
Taken together, this review highlights the vast influence the gut microbiota has on all components of the human immune system. This influence is not limited to local gastrointestinal effects but plays a role in the development and function of systemic immune elements as well. The manipulation of the gut microbiome in the treatment of numerous diseases is the subject of intense investigation and gives hope that through the manipulation of the gut microbiome at various stages it may be possible to shape the development of the immune system—prenatally, intrapartum, during infancy and childhood and even in adulthood—and enable the prevention of certain diseases or help treat those which have already developed.
APCs: antigen presenting cells
B. fragilis: Bacteroides fragilis
C. difficile: Clostridioides difficile
CD: Crohn’s disease
DCs: dendritic cells
F. prausnitzii: Faecalibacterium prausnitzii
GF: germ free
IBD: inflammatory bowel disease
IgA: immunoglobulin A
IL-10: interleukin-10
NOD: nucleotide-binding oligomerization domain
PRRs: pattern recognition receptors
PSA: polysaccharide A
SCFAs: short chain fatty acids
T1D: type 1 diabetes
Th: T-helper
Th1: type 1 T-helper
TLR: toll-like receptor
Tregs: regulatory T cells
TC wrote major parts of the manuscript. NAC conceptualized this review, wrote and critically reviewed the manuscript. Both authors contributed to manuscript revision, read and approved the submitted version.
The authors have no conflicts of interest to declare.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2022.
Copyright: © The Author(s) 2022. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
