Affiliation:
1Department of Electrical and Computer Engineering & Technology, Minnesota State University, Mankato, MA 56001, USA
ORCID: https://orcid.org/0000-0003-1626-5639
Affiliation:
3Department of Biomedical Engineering, University of Bridgeport, Bridgeport, CT 06604, USA
4Department of Mechanical Engineering, University of Bridgeport, Bridgeport, CT 06604, USA
Email: ppatra@bridgeport.edu
Explor Med. 2020;1:355–358 DOI: https://doi.org/10.37349/emed.2020.00023
Received: May 14, 2020 Accepted: October 16, 2020 Published: October 30, 2020
Academic Editor: Derek M. Dykxhoorn, University of Miami Miller School of Medicine, USA
It is now the priority of every country on Earth to find efficient methods of treatment management and patient cure against coronavirus disease 2019 (COVID-19) [1] to ensure the longevity of the present and future generations. In recent studies, it has been found that previously existing cardiovascular and respiratory conditions are identified as high-risk factors contributing to COVID-19 infection. The spike protein on the COVID virus enters its host’s cells by utilizing the binding sites of angiotensin-converting-enzyme 2 (ACE2) in the respiratory tract. Furthermore, pre-existing health conditions enhance the expression of ACE2, which elevates the danger of cardiac distress and acute respiratory distress syndrome (ARDS) [2]. The primary defense mechanism of lungs towards any infection is type-2 pneumocyte cells. They are responsible for synthesizing, secreting, and recycling all components of the pulmonary surfactant complex (PSC) that regulates alveolar surface tension in the lungs. Pulmonary surfactant proteins and lipids in PSC provide protection against airway inflammation and oxidative stress; additionally, they reduce the surface tension between two surfaces. The surface tension between gaseous and water interface in the lung is decreased by the presence of a thin film of fluid known as pulmonary surfactant. The interaction of the spike protein on COVID-19 and ACE2 down-regulates the enzyme [3], which may lead to ARDS on account of the resulting unrestricted, damaging action of angiotensin II on lung tissue, triggering high blood pressure, inflammation, fibrosis and apoptosis.
Many case studies reported so far on COVID-19 infection show an upsurge in the concentration of anti-inflammatory proteins, including surfactant protein D (SP-D) [4, 5], which is generally an anti-inflammation and antimicrobial protein. Any disturbance in the pulmonary surfactant levels will change the molecular forces at the air-liquid interface, resulting in a high surface tension region caused by intermolecular forces between water molecules at the air-liquid interface. These results in an area of high retractile force at the surface created. Any disruption in the surfactant and lipid layer may be associated with alveolar collapse and respiratory failure.
An effective way to predict the binding sites of proteins and enzymes in pulmonary surfactant is by taking advantage of present-day high-performance computing (HPC) capabilities to identify the response of various pulmonary surfactant proteins and enzymes on COVID-19 spike proteins. The fundamental dynamics of COVID-19 and defender proteins, such as but not limited to SP-D, need to be understood at the atomic level in order to develop new target-specific drugs that can fight against the infection. Amongst many computational and bioinformatics tools available today, molecular dynamics (MD) simulation is of great importance.
We performed MD simulation of two pulmonary surfactant proteins: surfactant protein A (SP-A, PBD 1r13) and SP-D (PDB 3DBZ) using NAMD and using CHARMM27 force field parameters. Molecules were solvated using the TIP3P water model with 150 mM sodium and chloride ions. All the parameters of the MD simulation are set as suggested by Jeong et al [6]. Their primary role is fighting lung infection (bacterial, viral, and fungal pathogens). The goal of this simulation study is to help develop a carbon nanotube-(CNT) based protein sensor which detects the ultra-low quantity of pulmonary surfactant protein levels with a potential of lung disease implications. To compare the interactions of SP-A and SP-D respectively with CNT, the MD simulation data for root mean square deviation (RMSD) of the protein backbone, van der Waals (VDW) energy, and surface accessible surface area (SASA) for both the proteins were analyzed.
The RMSD between corresponding atoms of two protein chains is a commonly used measure of likeness between two protein structures. The smaller the RMSD between two structures, the more similar they are. As atoms of protein molecules get closer to the CNT surface, this increases the VDW energy of interaction. The VDW energy is negative (Figure 1a), indicating an attractive force between pulmonary surfactant protein molecules and the CNT. Usually, VDW’s interactions are nonbonding, and adsorption of protein on the surface of CNT is facilitated due to the presence of polar residues in the pulmonary surfactant protein molecule. SASA of protein atoms is defined as an accessible solvent molecule of the specified size, which can be brought into VDW’s contact. The surface topology of a protein is closely related to its functions. A portion of the protein surface is directly involved in the interaction with other molecules, and the solvent-protein interface is almost certainly related to the structure of the native molecule.
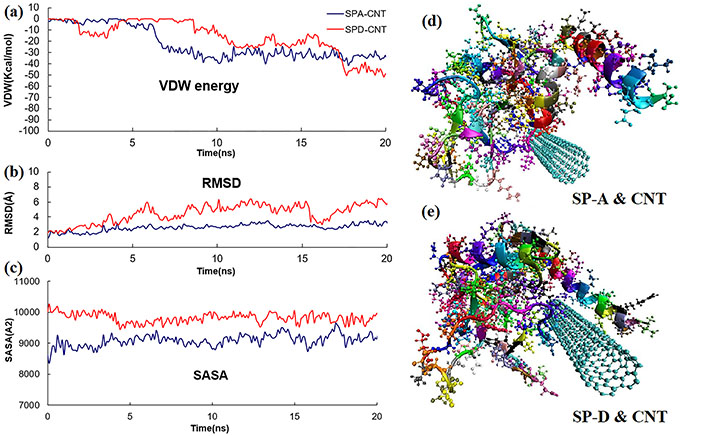
(a) VDW energy between SP-A, SP-D, and CNT; (b) RMSD of SP-A and SP-D protein backbone in the presence of CNT; (c) SASA of SP-A, SP-D in the presence of CNT; (d) molecular dynamic model for pulmonary surfactant protein SP-A and carbon nanotube after 20 ns. SP-A is illustrated in its secondary structure and atomic model; (e) molecular dynamic model for pulmonary surfactant protein SP-D and carbon nanotube after 20ns. SP-D is illustrated in its secondary structure and atomic model
The MD results show the higher reading of RMSD (Figure 1b) and SASA (Figure 1c) in the case of SP-D and CNT interactions. Figure 1d and e shown the final conformation of SP-A and SP-D with CNT after 20 ns, respectively. SP-D considers CNT to be a foreign body, and shows a higher level of protein structure changes in comparison to SP-A. This may also be the case in the presence of COVID-19 pathogens, which may be a reason for the elevated levels of SP-D. Performing similar HPC simulations with COVID-19 can give us insight into how the proteins and lipid complex inside the pulmonary surfactant are affected by this deadly pathogen. This will help physicians and other researchers to develop new drugs that can be directly targeted towards the prevention of the spread of infection or eradicate Covid-19 in the future. HPC will therefore be important in understanding the folding dynamics of COVID-19 active sites and the functional evolutions of the strains. This knowledge will eventually be able to assist virologists in developing future drug candidates.
ACE2: angiotensin-converting-enzyme 2
ARDS: acute respiratory distress syndrome
CNT: carbon nanotube
COVID-19: coronavirus disease 2019
HPC: high-performance computing
MD: molecular dynamics
PSC: pulmonary surfactant complex
RMSD: root mean square deviation
SASA: surface accessible surface area
SP-A: surfactant protein A
SP-D: surfactant protein D
VDW: van der Waals
BD and PP conceptualized and designed the study; BD and SP organized the database; BD, PP and SP performed the statistical analysis; BD wrote the first draft of the manuscript; PP and SP wrote sections of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2020.
Copyright: © The Author(s) 2020. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 3461
Download: 23
Times Cited: 0
