Affiliation:
1Departamento de Farmacología, Instituto de Investigación Biomédica de Málaga y Plataforma en Nanomedicina (IBIMA Plataforma Bionand), 29590 Málaga, Spain
†
ORCID: https://orcid.org/0000-0001-5861-9250
Affiliation:
1Departamento de Farmacología, Instituto de Investigación Biomédica de Málaga y Plataforma en Nanomedicina (IBIMA Plataforma Bionand), 29590 Málaga, Spain
2Centro de Investigación Biomédica en Red Enfermedades Hepáticas y Digestivas (CIBERehd), 28029 Madrid, Spain
†
ORCID: https://orcid.org/0000-0002-9068-0518
Affiliation:
1Departamento de Farmacología, Instituto de Investigación Biomédica de Málaga y Plataforma en Nanomedicina (IBIMA Plataforma Bionand), 29590 Málaga, Spain
2Centro de Investigación Biomédica en Red Enfermedades Hepáticas y Digestivas (CIBERehd), 28029 Madrid, Spain
ORCID: https://orcid.org/0000-0002-3421-1792
Affiliation:
1Departamento de Farmacología, Instituto de Investigación Biomédica de Málaga y Plataforma en Nanomedicina (IBIMA Plataforma Bionand), 29590 Málaga, Spain
ORCID: https://orcid.org/0000-0001-8295-6708
Affiliation:
1Departamento de Farmacología, Instituto de Investigación Biomédica de Málaga y Plataforma en Nanomedicina (IBIMA Plataforma Bionand), 29590 Málaga, Spain
ORCID: https://orcid.org/0000-0003-2706-5202
Affiliation:
1Departamento de Farmacología, Instituto de Investigación Biomédica de Málaga y Plataforma en Nanomedicina (IBIMA Plataforma Bionand), 29590 Málaga, Spain
2Centro de Investigación Biomédica en Red Enfermedades Hepáticas y Digestivas (CIBERehd), 28029 Madrid, Spain
3Service of Clinical Pharmacology, University Hospital Virgen de la Victoria, IBIMA Plataforma Bionand, Universidad de Málaga, 29010 Málaga, Spain
Email: lucena@uma.es
ORCID: https://orcid.org/0000-0001-9586-4896
Affiliation:
1Departamento de Farmacología, Instituto de Investigación Biomédica de Málaga y Plataforma en Nanomedicina (IBIMA Plataforma Bionand), 29590 Málaga, Spain
ORCID: https://orcid.org/0000-0003-4728-201X
Explor Dig Dis. 2023;2:56–76 DOI: https://doi.org/10.37349/edd.2023.00018
Received: December 16, 2022 Accepted: February 14, 2023 Published: April 26, 2023
Academic Editor: Wen-Xing Ding, University of Kansas Medical Center, USA
The article belongs to the special issue Drug-induced Liver Injury: From Bench to Clinical Application
The late event onset of a fraction of idiosyncratic drug-induced liver injury (DILI) cases and the link observed by genome-wide association studies (GWASs) of certain human leucocyte antigen (HLA) alleles with DILI due to specific drugs support the crucial role of the immune system (both innate and adaptive) in the pathogenesis of DILI. Recent advances in both flow and mass cytometry have allowed the profiling of all major immune cell types in a given sample. Therefore, determining the lymphocyte populations in samples from patients with DILI would facilitate the development of specific biomarkers for DILI diagnosis and prognosis. To date, a few studies have explored the immune landscape in DILI. In a recent study of leukocyte immunophenotyping using flow cytometry from the Spanish DILI Registry, an important role of adaptive immune response in DILI is suggested. DILI patients had significantly higher levels of T helper 1 (Th1) cells and activated helper and cytotoxic T cells than healthy controls. Furthermore, the increased expression of negative immune checkpoints and ligands in DILI patients could reflect a restoration of the immune homeostasis. Differences in the profile of cytokines in DILI patients from the Drug-Induced Liver Injury Network (DILIN) also suggest an involvement of both innate and adaptive immune systems in DILI development and prognosis. Moreover, several studies based on immunophenotyping of liver infiltrates showed a distinctive pattern of cellular infiltrates in patients with immune checkpoint inhibitors (ICIs)-DILI, with lower levels of plasma cells, CD20+ B cells and CD4+ T cells than in autoimmune hepatitis (AIH) patients. These pioneering studies highlight the importance of immunophenotyping for the mechanistic understanding of DILI. In this review, available data on immunophenotyping in DILI are gathered, and the potential clinical applications of cutting-edge, novel immunophenotyping techniques are discussed.
Drug-induced liver injury (DILI) is a hepatic condition that occurs at a relatively low frequency when an adverse reaction is triggered by exposure to certain drugs, herbs, or xenobiotics. DILI can be categorized into three types based on a combination of features: mechanism, latency, causative drugs, dosage administered, predictability, clinicopathological phenotypes, and rate of occurrence. Therefore, when hepatotoxicity is predictable, direct, and dose-dependent, DILI is classified as intrinsic. If the injury has little or no intrinsic toxicity and is caused by agents that induce hepatotoxicity only on very rare occasions, DILI is considered idiosyncratic (hereinafter, DILI) [1]. Patients who experience a DILI episode are individuals with intrinsic susceptibility; thus, it is believed that a mixture of genetic and environmental factors is responsible for the development of hepatic damage [2, 3]. The third type is indirect DILI, which was recently proposed to englobe cases induced by drugs that aggravate the underlying liver conditions. Agents that can promote immune-mediated hepatotoxicity by modifying the immune system may also elicit indirect DILI. However, this last group remains controversial [4, 5].
The incidence of DILI is difficult to estimate, since its diagnosis is uncertain, and is normally established after ruling out other causative conditions of liver injury, such as autoimmune or viral hepatitis. Therefore, there are currently no homogeneous diagnostic criteria. Another issue that makes diagnosis challenging is the lack of laboratory tests that allow the identification of the causative agent when patients have been taking a variety of drugs, herbal supplements, or other xenobiotics, as well as when they have a pre-existing liver condition [6]. For these reasons, to date, most clinical data have been obtained from retrospective database studies.
In terms of severity, DILI has a wide range of potential outcomes. Most patients make a full recovery after discontinuation of the culprit drug, but the progression of DILI can be dramatic [7]. In fact, up to 10% of patients with drug-induced jaundice fulfilling “Hy’s Law” criteria will progress to acute liver failure (ALF) [8], and some of these patients will require liver transplantation or ultimately die. Therefore, DILI reactions remain a major public concern, accounting for a significant proportion of all ALF cases [9]. In addition, DILI poses an economic challenge and is a leading cause of drug withdrawals [10].
At present, the most widely accepted idea regarding the pathogenesis underlying DILI is that a combination of different factors, whether individual, environmental, or drug-related, may increase the susceptibility to an episode. This idea is known as the “multiple determinant hypothesis” [11]. The molecular background that supports this hypothesis is that when the agent enters the organism, it can be metabolized by hepatic enzymes that can transform them into chemically reactive metabolites (CRMs) which in turn may interfere with the proper functioning of hepatocytes. CRMs can also induce cholestatic injury, which can lead to hepatocyte death by altering bile acids (BAs) metabolism and canalicular transport [12]. Additionally, CRMs can form adducts with individual proteins, generating compounds that the body recognizes as antigens. Eventually, damaged hepatocytes undergo different processes of cell death, such as apoptosis and necrosis [13]. Furthermore, intestinal permeability can be affected by changes in the gut microbiota owing to the release of pathogen-associated molecular patterns (PAMPs), which presumably trigger an inflammatory response that indirectly may lead to liver injury [14].
Moreover, immune system activation and liver immune tolerance imbalance are thought to be crucial in DILI development [15]. Since these processes are highly difficult to reproduce by using in vitro or animal models, patients’ samples are needed to study the immune profiling of the disease.
Immunophenotyping assays constitute a set of techniques based on antigen expression which is used for the identification of immune cell subsets. There is a dichotomous classification made to categorize the main immune cell populations, which consists of lymphoid-derived cells and myeloid-derived cells. Lymphoid-derived cells include B cells, T cells, natural killer (NK) cells, and invariant natural killer T (iNK-T) cells, whereas myeloid-derived cells include macrophages, neutrophils, monocytes, and dendritic cells [16, 17] (Table 1).
Human immunophenotyping standard panels. This table lists the combination of markers frequently used to analyze different immune cells populations
| Parental populations | Parental markers | Subpopulation | Subpopulation markers | Ref. |
|---|---|---|---|---|
| CD4 T cells | CD3+/CD20–/CD8−/CD4+ | Naive | CCR7+/CD45RA+ | [18, 19] |
| Central memory | CCR7+/CD45RA− | |||
| Terminal effector | CCR7−/CD45RA+ | |||
| Effector memory | CCR7−/CD45RA− | |||
| Treg | CD127low/CD25Hi/CCR4+ | |||
| Th1 | CD25low/−/CXCR5−/CD45RA−/CXCR3+/CCR6− | [19] | ||
| Th2 | CD45RA–/CCR4+/CCR6–/CXCR3+ | [20] | ||
| Th1Th17 | CD45RA–/CCR4–/CCR6+/CXCR3+ | |||
| Th17 | CD25low/−/CXCR5−/CD45RA−/CXCR3−/CCR6+ | [19] | ||
| Th22 | CD194+/CD196+/CCR10+ | [21] | ||
| CD8 T cells | CD3+/CD20–/CD8+/CD4– | Naive | CCR7+/CD45RA+ | [18, 19, 22] |
| Central memory | CCR7+/CD45RA− | |||
| Terminal effector | CCR7−/CD45RA+ | |||
| Effector memory | CCR7−/CD45RA− | |||
| Unconventional T cells | CD3+/CD20– | NKT cells | TCRVα24JαQ+ | [19] |
| MAIT cells | CD161+/TCRVα7.2+ | |||
| γδ T cells | TCRVαβ−/TCRVγδ+ | |||
| NK cells | CD3−/CD56+ | - | - | [19] |
| Granulocytes | CD45+/CD3–/CD19– | Neutrophils | CD16+ | [23] |
| Basophils | CD123+/HLA-DR– | |||
| Eosinophils | CD16– | |||
| B cells | CD3–/CD20+ | Transitional | CD10+/CD27− | [19] |
| Naive | CD27−/CD10− | |||
| Memory | CD27+/CD10− | |||
| Monocytes | CD45+/CD3–/CD19–/CD123–/HLA-DR+/CD11c+ | Classical | CD14+CD16– | [23] |
| Intermediate | CD14+CD16+ | |||
| Non-classical | CD14–CD16+ | |||
| Dendritic cells | CD45+/CD3–/CD19– | Myeloid | CD123–CD11c+/CD16–CD14–/HLA-DR++ | [23] |
| Plasmacytoid | CD123+/HLA-DR+ |
CCR7: C-C chemokine receptor type 7; CXCR5: C-X-C motif chemokine receptor 5; HLA: human leucocyte antigen; MAIT: mucosal-associated invariant T; Th1: T helper 1; Treg: regulatory T cell; -: none
Bone marrow and spleen are the main tissues of choice for studying immune cell populations in immunophenotyping procedures using rodent models and human peripheral blood [24]. However, when doing children’s diagnosis, it is suggested to use bone marrow samples for the first test [25].
Cytometric methods, such as flow cytometry, are the most frequently used for identifying human immune subsets [26]. Specific antibodies are coupled to fluorescent compounds such that it is possible to measure the expression of a particular protein within a cell population by binding the fluorophore-coupled antibody to the antigen. This enables the identification of each immune cell subtype based on a set of intracellular and/or extracellular markers [27].
In addition to classical multicolour flow cytometry, different cytometric methods have been developed for immunophenotyping, each of which has been designed for different purposes including hyperspectral flow cytometry, imaging flow cytometry (IFC), mass cytometry, and imaging mass cytometry (IMC) (Table 2) [24, 28].
Advantages and drawbacks of cytometry techniques. This table lists the most representative advantages and disadvantages of the different immunophenotyping procedures, as well as some references that collect applications of these techniques in DILI research
| Immunophenotyping techniques | Description and applications | Advantages | Drawbacks | Applications in DILI research |
|---|---|---|---|---|
| IFC | IFC platform combines features of flow cytometry and fluorescent microscopy with data-processing algorithms, allowing for the evaluation of morphological and fluorescent data and analysing protein expression in single cells in heterogeneous cell populations |
|
| [29] |
| Multicolor flow cytometry | Technology that uses multiple fluorescent markers to identify and characterize cellular subpopulations of interest with the possibility of isolating pure, viable populations by cell sorting |
|
| [30–33] |
| Mass cytometry | Mass cytometry utilizes elemental mass spectrometry to detect metal-conjugated antibodies bound intracellularly or extracellularly to antigens of interest on single cells, allowing the simultaneous analysis of a great number of cellular features |
|
| [33–35] |
| Hyperspectral flow cytometry | Single-cell analysis technique that combines ultrafast optical spectroscopy and flow cytometry. This technology uses diffraction gratings or prism-based monochromators to disperse fluorescence signals from multiple labels onto linear detector arrays |
|
| No data |
Immunophenotyping is commonly used to diagnose specific types of cancers, such as different kinds of lymphoma and leukemia [36]. Cellular biomarkers provide essential information regarding the lineage and differentiation of hematological malignancies to properly characterize the complexity and diversity of blood cancers. When diagnostics are performed, panels with a large number of markers are recommended for the detection of immunophenotypic features relevant to subsequent investigations of minimal residual diseases [37].
Recently, immunophenotyping techniques have been applied to the research and diagnosis of different diseases in which the immune system is involved, such as DILI.
In this article, we review data on immunophenotyping in DILI and discuss the potential avenues that open this approach to better understand the molecular mechanisms underlying DILI pathogenesis and to facilitate and make a more accurate and efficient diagnosis by finding immuno-fingerprints.
Idiosyncratic DILI is thought to be immune-mediated. This is mainly due to evidence emerging from genetic studies, including genome-wide association studies (GWASs), which showed different associations of DILI with HLA haplotypes [38] and polymorphisms in other genes, such as a variant in the interleukin-10 (IL-10) allele [39], a variant in protein tyrosine phosphatase non-receptor type 22 (PTPN22) [40], or the expression quantitative trait loci (eQTL) snip rs1363907, which produces a reduced liver expression of the endoplasmic reticulum aminopeptidase 2 (ERAP2) [41]. Additionally, the histological findings and clinical characteristics of the disease highlight the importance of the immune system in DILI [42].
Although innate immunity also plays an important role in DILI (especially in the early phases of these immune responses, i.e., between drug administration and the start of the adaptive response [43]), adaptive immunity is the best-characterized response that explains the mechanisms and severity of these idiosyncratic reactions, particularly the cellular arm of this immunity, although humoral immunity is also involved [15].
The formation of CRMs can cause direct damage in hepatocytes for instance, through disruption of BAs homeostasis, production of reactive oxygen species (ROS), and induction of organelle stress (such as mitochondrial and endoplasmic reticulum stress [44]) that leads to the release—mainly by extracellular vesicles—of damage-associated molecular patterns (DAMPs) [45]. These molecules, such as the high mobility group box 1 (HMGB1) protein or DNA [46], are recognized by pattern recognition receptors (PRRs) expressed on the surface of some immune cells [including Kupffer cells (KCs) or dendritic cells], causing proinflammatory effects [for example, the production of tumor necrosis factor (TNF)-α, IL-1, vasoactive amines, and ROS] [47].
In addition to these inflammatory processes, the mechanism of injury may also involve the formation of a complex by covalent binding between the parent drug itself—or its CRMs—and host proteins. This drug-protein adduct may act as a neoantigen and can be taken up by antigen-presenting cells (APCs) and presented in lymph nodes by major histocompatibility complex (MHC) class I and II to CD4+ and CD8+ naive T cells, respectively, to prime them [15, 42]. For priming to be successful, three activation signals must occur: the first is the one described above, the second is produced by the contact of naive T cells with co-stimulatory molecules (CD40, CD80, and CD86) expressed on the surface of APCs, and the third is given by the environment of cytokines that allow T cell proliferation and determine the subtype of CD4+ helper T cells produced [42]. These effector T cells then migrate to the liver and exert their function when recognizing the neoantigen presented on MHC-I (expressed in all nucleated cells, e.g., hepatocytes) for CD8+ T cell cytotoxic effects or MHC-II (expressed in APCs) in the case of CD4+ T cells, which activate other immune cells and orchestrate diverse responses via the production of different sets of cytokines [15].
Moreover, presumably, increased permeability due to microbiota changes would induce a leakage of PAMPs through the intestinal barrier into the bloodstream, potentiating the immune reaction, even leading to cell death [48]. This would induce the release of additional specific cell-death-type DAMPs that may exacerbate the response and promote a chronic immune process in the liver [49]. This theory, known as the hapten hypothesis [50], does not explain why only a small number of patients develop DILI induced by drugs known to form haptens. Furthermore, flucloxacillin, a well-known hepatotoxicant, can activate the adaptive immune system by both hapten-dependent and hapten-independent mechanisms [51], raising the possibility of additional immunological pathways involved in the pathogenesis of DILI.
Therefore, two additional hypotheses have been proposed: (i) the pharmacological interaction (p-i) concept states that the drug may interact non-covalently with HLA and T cell receptor (TCR) molecules, leading to their activation [52]. In recent years, several drugs such as sulfamethoxazole [53] and carbamazepine [54] have been described to trigger the immune system through p-i-based stimulation. Finally, (ii) the altered peptide repertoire model states that the non-covalent binding of certain drugs with their respective HLA molecules may alter its conformation (including the antigen-binding cleft) and the repertoire of self-peptides that can present [55]. However, the T cell response to the altered peptide repertoire model has only been observed for abacavir and HLA-B*57:01 [56], and may not be generalizable to other drug hypersensitivities.
Because the liver is constantly exposed to multiple exogenous antigens that come from the gastrointestinal tract via the portal vein, it is known to be an immune-privileged organ [57]. Immune tolerance is a necessary adaptation to protect liver cells from constant damage of the immune system [58] and is given by different elements that maintain liver integrity and homeostasis, such as regulatory immune cells. For instance, Tregs can suppress the activation, proliferation, and effector functions of T cells and block the release of pro-inflammatory cytokines [59]. Besides, myeloid-derived suppressor cells (MDSCs) may inhibit, in addition to the adaptive immune response, the innate immune response through the alteration of the cytokine production pattern of macrophages [60]. Other immune tolerance mechanisms involve the existence of tolerogenic APCs in the liver (such as KCs), which may not be effective enough to activate T cell responses [57]. These processes prevent progression from mild to severe injury and lead to spontaneous resolution despite continued drug intake (i.e., clinical adaptation) [58].
Therefore, disruption or impairment of immune tolerance, known as “defective adaptation” [58, 61], may explain why some patients develop a clinically relevant liver injury, whereas others do not [62]. This range of possible host responses could partially explain why the intake of a drug can provoke severe injury in some patients but not in others, although other factors may be implicated [43].
The causes of disruption of immune tolerance are not completely understood, but it is known that APCs can switch from tolerogenic to immunogenic phenotypes during an infectious process or as a response to neoantigen contact [57]. Other key events that may fail and provoke a loss of tolerance include antigen presentation control, the state of anergy, clonal deletion through apoptosis of antigen-specific T cells, and immune deviation (increase in the ratio Th2/Th1 [43, 58]). Finally, low levels of the endotoxin lipopolysaccharide (LPS) are important for maintaining the pattern of cytokines such as IL-10 and TNF-α secreted into the microenvironment by KCs [63], provoking the expansion of Tregs and hampering T cell activation, contributing to the immune tolerance status. Hence, changes in gut microbiota may affect intestinal permeability and LPS exposure in the liver, thus contributing to the breakdown of tolerance [58].
The study of the hepatic immune system has revealed the cellular landscape of the healthy liver [64, 65] and has also shed light on the mechanisms involved in the onset and progression of different hepatic diseases. Recently, Zhao et al. [66] observed a distinct intrahepatic B cell landscape in samples from patients with acute-to-chronic liver failure (ACLF) patients compared to healthy controls, highlighting the importance of B cell mediation in ACLF therapy. Liver immunophenotyping of hepatocellular carcinoma has also identified specific populations associated with poor prognosis [67]. Furthermore, novel immune-mediated mechanisms involved in liver cirrhosis have been proposed by characterizing specific subpopulations of macrophages in the cirrhotic liver [68], providing a new rationale for the discovery of therapeutic targets. Similarly, unraveling the immune microenvironment of the liver in DILI may be crucial to identifying potential treatments.
Immunophenotyping has been applied in studies of liver pathologies that share an inflammatory background. Communication between resident liver immune cells, hepatocytes, and infiltrating immune cells may affect disease progression. Characterizing liver infiltrates in the liver tissue of patients with DILI may enable a better understanding of the mechanisms underlying the disease [69]. Infiltration of CD3+CD8+ cytotoxic lymphocytes was observed in the liver tissue of a patient with floxacillin-induced DILI [70]. In another study, differences in leucocyte infiltrate in liver tissue were observed in DILI compared with idiopathic autoimmune hepatitis (AIH) and viral hepatitis. The DILI cases, as well as the other groups, showed high levels of CD8+ cytotoxic T cells and macrophages. In contrast, DILI cases presented lower numbers of B cells than other groups. In addition, DILI has been reported a lower level of NK cells than viral hepatitis, which may be explained by the importance of these cells in fighting infections [30]. However, despite these promising results, this type of study is limited because liver biopsies are not usually performed to diagnose DILI. Instead, immunophenotyping of peripheral blood could reflect inflammatory events occurring in the liver and may be an alternative to biopsy-based studies.
Following this approach, several studies have identified peripheral drug-specific T cells in peripheral blood mononuclear cells (PBMCs) from DILI patients. Monshi et al. [71] observed that PBMCs from patients with flucloxacillin-DILI were activated in vitro after flucloxacillin exposure, and detected flucloxacillin-responsive CD8+ and CD4+ clones. Using a similar strategy, Kim et al. [72] isolated both amoxicillin- and clavulanic acid-responsive CD8+ and CD4+ clones from patients with amoxicillin-clavulanate DILI, showing that both drugs contribute to the T cell response that develops in patients. Immune-based mechanisms for DILI due to antituberculosis (TB) drugs [73], tolvaptan [74], and atabecestat [75] have also been inferred following the characterization of patient PBMCs in vitro.
Recently, the first study on peripheral blood broad immunophenotyping of patients with DILI has been published by our group. In this study, we performed a broad characterization of 17 different leukocyte populations and the expression of eight immune checkpoints. This study enabled not only the comparison between DILI and controls without liver injury, but also the comparison of DILI with additional liver aetiologies, including viral hepatitis, metabolic-associated fatty liver disease (MAFLD), and AIH. Interestingly, DILI and viral hepatitis patients showed an increased proportion of activated Th cells (CD4+/HLA-DR+) and activated cytotoxic T cells (CD8+/HLA-DR+) compared to MAFLD patients and healthy controls (Figure 1). Since HLA-DR is considered a late-activating marker, its detection at elevated levels in the acute phase of injury reinforces the importance of adaptive immune response in DILI. Regarding immune checkpoints, the detected increase in the immune receptor inducible co-stimulator (ICOS) on Th cells is in line with the activation observed in both CD4+ and CD8+ T cells. This receptor is expressed on naive-T cells at low levels but increases when the cells are activated. Increased intracellular cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) expression was also observed in CD4+ T cells at an early stage of DILI compared to MAFLD patients, which is likely an effect of peripheral tolerance regulation. Although only viral hepatitis patients presented significantly higher programmed cell death-1 (PD-1) levels than healthy controls, increased levels of the programmed cell death-ligand 1 (PD-L1) in all liver damage groups compared to healthy controls were observed. Moreover, interestingly, DILI patients showed lower levels of PD-L1-expressing monocytes compared to AIH patients. PD-1 and PD-L1 binding is involved in the inhibition of the immune response while maintaining peripheral immune tolerance. The increased expression of inhibitory immune checkpoint proteins in DILI (Figure 1) supports the role of immune tolerance and reduced cell damage, in line with recuperation [21].
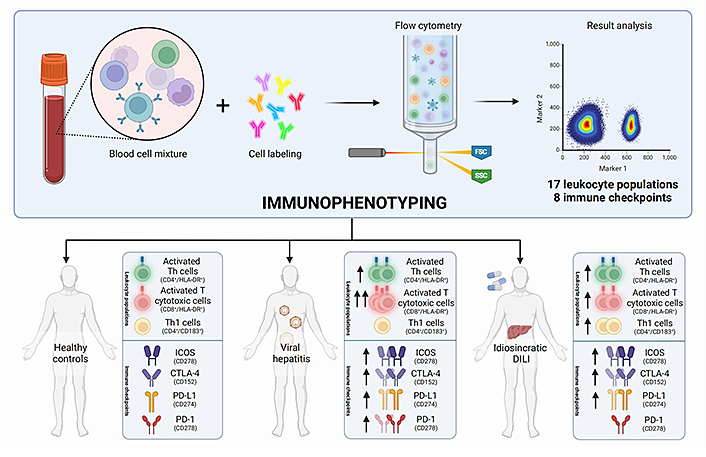
Immunophenotyping comparison between DILI patients, healthy controls, and patients with acute liver damage due to different aetiologies, including viral hepatitis and AIH. FSC: forward scatter; SSC: side scatter
No “immune fingerprint” for DILI was observed in this study. However, both viral hepatitis and DILI onset of liver injury were associated with an increase in the activation of helper and cytotoxic T cells and upregulation of CTLA-4 and PD-1 expression. These observations can shed light on shared unknown hepatotoxicity mechanisms that can be useful for understanding transaminitis commonly associated with adeno-associated virus (AAV)-based gene therapies [76]. Recently, valoctocogene roxaparvovec gene therapy (Roctavian) for haemophilia A [77] and etranacogene dezaparvovec gene therapy (Hemgenix) for haemophilia B [78] have been approved by the European Medicines Agency (EMA) and the Food and Drug Administration (FDA), respectively. Both therapies are based on AAV which contains the factor VIII or IX gene (to treat haemophilia A or B). Once administered to a patient as a one-off infusion, they carry the respective factor gene into the hepatocytes, enabling them to produce the missing factor.
The most common adverse reactions associated with Roctavian and Hemgenix include liver enzyme elevations, which occur in the majority of treated patients, usually at later time points [77, 78]. The precise pathophysiological basis for transaminitis remains unknown, in part because it has not been possible to recapitulate this toxicity in animal models [79, 80], which is similar to that observed in DILI pathophysiology [81]. Moreover, the pathogenesis of liver toxicity may differ between patients. Therefore, immunophenotyping of patients at the time of liver injury would be useful to unravel the pathogenetic details of liver toxicity and to stop using empiric aggressive therapies (such as corticosteroids or alternative immune regulatory agents, when hepatotoxicity is unresponsive to oral steroids) to mitigate these adverse events.
Subsequently, our group published another study [32] on extended immunophenotyping of DILI and MAFLD, focusing particularly on the iNK-T population. The highest level of iNK-T was found in PBMCs from MAFLD patients with significant liver fibrosis, suggesting a role for iNK-T in MAFLD progression. In contrast, PBMCs from patients with DILI had iNK-T levels similar to those of healthy controls. These results suggest a more relevant role for iNK-T cells in the pathogenesis of MAFLD than for DILI, which may represent a differential immunological feature between these two entities. Hence, immunophenotyping can provide information on a potential immunologic fingerprint of DILI, which may be different from that of other liver pathologies. Further lymphocyte population studies are required to identify unique patterns that can be used for diagnosis. On the other hand, research on cytokines profiles of patients can also give information about DILI mechanisms of progression.
Cytokines are small proteins responsible for intercellular communication, mainly between cells of the immune system, and they play a key role in the inflammatory process. Changes in the profile of cytokines and chemokines (cytokines involved in chemotaxis) have been suggested as possible biomarkers of liver pathologies such as DILI [82]. In the healthy liver, resident immune cells produce a small amount of these cytokines, but a significant increase in cytokine and chemokine expression is known to occur during damage or infection. Cytokines are produced by cells present in the blood and different tissues and are constantly released into the bloodstream. Their identification in the blood can provide information on the state of the immune system in a given organ.
Cytokine profiling has allowed the differentiation of cases of hepatocarcinoma, viral hepatitis (HBV and HCV infection), and both hepatocarcinoma and viral hepatitis [83]. The potential role of cytokines in disease prognosis has also been highlighted. Recently, in a study on the effect of MAFLD in 94 patients with severe coronavirus disease 2019 (COVID-19). Patients with concomitant MAFLD who progressed to critical COVID-19 showed a distinct cytokine profile characterized by higher levels of IL-6, -8, -10, and interferon (IFN)-β. Furthermore, IL-8 and IL-10 levels were found to have prognostic potential for time to recovery [84].
Studies identifying cytokines levels in DILI are scarce. A study analyzing polymorphisms in genes encoding the cytokines IL-10, IL-4, and TNF-α showed that DILI with severe outcomes presented a haplotype that produced lower levels of IL-10 [85]. It has been reported that Th1 cells may release IL-10 to prevent uncontrolled inflammation, and reduced IL-10 may subsequently increase the risk of cell damage [86]. Another study found that an increase in blood IL-22 levels in DILI cases was associated with an increase in Th22 levels. In addition, DILI cases with better prognoses presented higher levels of IL-22, suggesting that IL-22 may play a hepatoprotective role [87].
Other studies have investigated a more complete cytokine and chemokine profile for DILI. Steuerwald et al. [88] analysed 27 different analytes in patients with DILI and in healthy controls. The DILI group presented a diverse cytokine pattern, with an increased presence of cytokines associated with the innate immune system in some cases, and cytokines associated with the adaptive immune system in others. However, low levels of IL-9, IL-17, platelet-derived growth factor (PDGF)-bb, regulated on activation, normal T cell expressed and secreted (RANTES), and serum albumin were associated with an increased risk of fatal outcomes. Subsequently, the authors expanded the number of DILI cases to validate the previous results and analyzed the profile of these analytes in other liver pathologies. Interestingly, low RANTES and serum albumin levels maintained the ability to predict the worst outcomes in DILI [89].
Therefore, characterization of immune profiles provides novel information that may help us to understand the mechanistic roles of cytokines and chemokines in the pathogenesis of DILI.
As stated above, the study of the human peripheral immune system in DILI has recently led to important findings about the pathophysiology of the disease [21] and has even helped to improve the diagnosis [90]. However, many of these studies focused on blood samples or PBMCs are limited in their consideration of the hepatic microenvironment and the interaction of the local immune system with liver cells during a DILI episode. The liver harbors many resident immune cell populations that in many cases differ in composition and function from peripheral cells [91, 92]. Understanding the role of immune cell infiltration and tissue-resident cells in DILI may therefore be essential to fully comprehend disease mechanisms.
Due to the large contribution of individual background to the occurrence of DILI, working directly with patient-derived samples has so far been the gold standard in the study of the influence of the immune system on this disease. However, addressing the immune system in the liver environment represents an additional challenge as liver biopsies are not routinely performed in most cases of suspected DILI. Therefore, the availability of histological samples from patients for immunophenotyping studies is low. While the European Association of the Study of the Liver (EASL) Clinical Practice Guidelines [93, 94] stated that liver biopsy may be a reasonable procedure for considering DILI in some cases, the Roussel Uclaf Causality Assessment Method (RUCAM) [95], which is the most widespread case definition method for DILI, and its recent Revised Electronic Causality Assessment Method (RECAM) [96] do not consider liver biopsy essential for DILI diagnosis.
Despite the scarcity of samples, several histological studies have described the immunological features of DILI. A systematic evaluation of DILI hepatic histological findings in 249 patients showed that severe inflammation was more abundant in patients with hepatocellular injury than in those with cholestatic injury. Eosinophils and granulomas were found more often in those with milder injury, which is a potential sign of better prognosis, whereas neutrophils are associated with either severe or fatal injury. This information has helped to establish better clinical associations [7]. Wuillemin et al. [70] characterized the infiltration of cytotoxic CD3+ CD8+ lymphocytes into the liver using immunohistochemical staining of a liver biopsy from a patient with floxacillin-DILI and demonstrated the important role of activated drug-reacting T cells in DILI development. A comparison of resident macrophages from liver biopsies of acetaminophen (APAP)-DILI with either circulating macrophages or biopsies from other liver diseases showed that the macrophage population expanded in areas of necrosis, suggesting a regenerative role of these cells after injury [97]. Liver immunohistochemistry of macrophages from biopsies also showed differences in pigmented cells among the three clinical DILI injury types [98].
In recent years, immune checkpoint inhibitors (ICIs) have become an essential antitumor therapeutic resource for the treatment of various cancers. Unfortunately, they have also become a source of immune-mediated hepatotoxicity, the mechanisms of which remain unknown [99]. Immunostaining of clinical liver samples revealed important information about immune infiltration in ICIs-DILI, predominantly lymphocytes [100]. It has been found that anti-CTLA-4 infiltrating cells predominantly consist of CD8+ cells, while anti-PD-1/PD-L1 liver biopsies show a similar abundance of CD8+ and CD4+ T cells. Johncilla et al. [101] characterized the histological features of ipilimumab-DILI and observed an inflammatory infiltrate consisting predominantly of CD8+ T lymphocytes, admixed histiocytes, scattered plasma cells, and eosinophils.
A recent study by Gudd et al. [102] showed a reduction in total monocytes and an increase in soluble CD163 levels in the peripheral circulation of ICIs-hepatitis patients compared with healthy controls. A second step of transcriptional profiling of different immune cells demonstrated the presence of activated monocyte and enhanced effector CD8+ T cell populations in ICIs-hepatitis patients. Moreover, subsequent liver immunochemistry confirmed the presence of CD8+ T cells and CD163+CCR2+ macrophage aggregates in the livers of ICIs-hepatitis patients, indicating that, in those patients, peripheral changes observed are mirrored in the liver.
Histological studies based on immunophenotyping of liver infiltrates from biopsies have also served to identify differences between DILI and other non-DILI acute liver injuries. For example, DILI biopsies have shown lower levels of mature B cells than AIH and viral hepatitis, and lower NK cells than viral hepatitis [30]. Interestingly, histological studies comparing ICIs-DILI with other forms of liver injury showed that the number of plasma cell infiltrates was significantly lower in ICI-DILI than in AIH [103], whereas CD20+ B cells and CD4+ T cells were fewer in ICI-DILI than in AIH or DILI [104]. The histological patterns of ICIs-DILI may help differentiate this type of injury from AIH and DILI [105].
Immunophenotyping is a powerful diagnostic tool that can be used for a wide variety of diseases involving the immune system. When used as a clinical test, immunophenotyping is a non-invasive method to investigate the type and cause of specific immune diseases. Examples of the diagnostic abilities of immunophenotyping include acute myeloid leukemia [106], eosinophilic otitis media [107], common variable immunodeficiency [108], non-small cell lung cancer [109], and different myelodysplastic syndromes [110].
Moreover, immunophenotyping could help support the clinical diagnosis if the physician is unsure. For example, both AIH and DILI can present with acute liver injury and share several clinical features. However, the underlying pathophysiological mechanisms involving the immune system remain unclear. Recently, Bozward et al. [34] reported for the first time an NK cell population difference between patients with DILI and AIH. The authors performed deep immunophenotyping of PBMCs from patients and healthy controls using cytometry of time of flight (CyTOF) and observed that NK cells from AIH patients in remission showed significantly higher positivity for both CD27 and CXCR3 when compared with healthy controls. Although the DILI samples showed a similar tendency, the differences compared to the controls were not significant.
Additionally, obtaining an immunological fingerprint for DILI offers an opportunity to expand our understanding of the immunological mechanisms involved in the disease, and it could also become a non-invasive diagnostic and prognostic tool, considering the specific immune populations of DILI patients as biomarkers. Thus, the role of liver immune tolerance in the multiple possible clinical presentations of DILI is gaining importance. The mechanisms of liver immune tolerance have been reviewed in this review (Section “Liver immune tolerance”) and in many other studies [58, 111–113]. However, the relationship between liver immune tolerance and the occurrence of a DILI event remains a subject of study, as it remains unclear why only a subset of patients with risk‐associated HLA alleles experiences a DILI event. One proposed explanation is that clinical adaptation failure is necessary for the occurrence of DILI episodes. Then, when the state of immune tolerance against a drug or its metabolite as an immunogen fails in an individual with a predisposition to DILI, a clinically relevant liver injury episode occurs. Different animal models of DILI obtained by manipulating pathways of immune tolerance leading to a failure of adaptation support this hypothesis [81, 114–116].
Therefore, approaches comparing the immunophenotype of DILI patients due to the same drug but with different clinical manifestations (asymptomatic vs. those who develop overt liver injury) could provide clues about the role of liver immune tolerance in DILI pathophysiology and even discover new pathways in DILI.
Moreover, research on possible immunophenotypic alterations in DILI may also lead to the discovery of new genetic alterations involved in the disease that could be used as new targets for specific DILI treatments, as currently, the principal therapeutic strategy at admission is stopping the use of the drug implicated, following by glucocorticoids (GCs) treatment in selected patients.
Finally, stratification of DILI patients based on their immunophenotype is necessary to improve prognosis or even determine which therapies, if any, will benefit the patient. For example, possible differences in PD-1 and PD-L1 levels in patients with DILI may explain the distinct capacity of each individual to modify their immune tolerance, as temporary upregulation of PD-1 and PD-L1, which occurs during different acute infections, is thought to be a way of regulating peripheral tolerance [117, 118].
In recent years, several innovative technologies capable of extracting large amounts of immunophenotypic information have emerged. Examples include high-dimensional and multiparametric fluorescent flow cytometry, single-cell messenger RNA (mRNA) sequencing, and mass cytometry [119].
Mass cytometry or CyTOF is a powerful technique for immunophenotyping studies as it overcomes many of the limitations of traditional fluorescence-based flow cytometry. In CyTOF, immune cells are stained with antibodies bound to metal particles, which reduces interference by autofluorescence signals and signal overlap. In addition, the broader spectrum of available metal isotopes allows for the accurate quantification of more than 40 parameters in single cells [120]. This approach has recently been used to immunophenotype human blood samples from various immune-mediated liver diseases such as primary biliary cholangitis [120], viral hepatitis [121], and AIH [122]. McGuire et al. [33] combined immunohistochemistry and multiplex immunofluorescence with mass cytometry to comprehensively immunophenotype corticosteroid-resistant T cells after episodes of ICIs-DILI caused by PD-1 blockers. Similarly, CyTOF was also recently applied to better understand autoimmune liver toxicity after treatment with phosphatidylinositol 3 kinase inhibitors, showing that an increase in activated CD8 T cells with activation of Th17 T cells may be related to the toxicity of these drugs [35]. Therefore, CyTOF can be used to dissect the DILI immune landscape.
Immunohistochemistry and fluorescence imaging are traditional methods used to assess the local immune background in DILI. These approaches have been of great help, particularly in ruling out other forms of liver damage at diagnosis or providing prognostic information. However, the spatial characterization of the local immune response in DILI has so far been superficial. Most studies do not delve into immunophenotyping of the diversity of immune subpopulations, probably due to the low multiplexing capacity, lack of robustness, and alterations in tissue morphology due to the available fluorescence-based techniques. Following the principles of mass cytometry, IMC might be a tool for overcoming traditional limitations while deepening our understanding of the hepatic immune system in DILI [123, 124]. IMC has allowed the complex characterization of hepatic adaptive and innate immune subsets that correlate with hepatocellular injury or histological fibrosis using liver biopsies from patients with hepatitis B [125]. In a recent study, Csernalabics et al. [29] used IMC to analyze intrahepatic immune cell infiltration in patients with hepatitis due to the COVID-19 vaccine and found pathophysiological differences when compared with classical AIH.
Another promising way to assess the individual immune background behind DILI is single-cell methodologies, such as single-cell RNA-sequencing [126]. Recently, this technique was used to generate a map of the cellular landscape of the human liver, identifying 20 different cell populations of hepatocytes, endothelial cells, cholangiocytes, hepatic stellate cells, B cells, conventional and non-conventional T cells, NK-like cells, and various monocyte populations [65].
This approach has been already proposed as s way to better understand the roles of the immune system in different mice and human models of hepatic diseases, such as chronic liver injury [127], alcoholic liver disease (ALD), non-alcoholic steatohepatitis (NASH), DILI [128], and hepatitis B [129]. Both in vitro and in vivo models have been used to study the specific aspects of the immune system in DILI.
Recent attempts to study the immune system’s role in DILI in vitro have developed cell culture methods aimed at characterizing primary T cell responses to drugs. For example, Usui et al. [130] examined T cell priming with drugs associated with HLA risk alleles and control compounds by using cells derived from 14 HLA-typed healthy donors.
Moreover, in vitro models can be used to study the interaction between the liver and the immune system. Kato and Uetrecht [131] treated hepatocarcinoma cells with drugs associated with severe DILI. After putting in contact human monocytic cells (THP-1) with the supernatant of hepatic cells treated with the drugs of interest, macrophages showed an increased caspase-1 activity and production of IL-1β. The same results were observed by Oda et al. [132] and Ogese et al. [133] by using different immune and hepatic cell types.
Similarly, the use of in vivo models is gaining increasing interest in the study of the liver microenvironment in DILI, since their use may circumvent the difficulties associated with obtaining human samples. Buchweitz et al. [134] reproduced the human chlorpromazine hepatotoxicity in rats after a pre-treatment with LPS (a strategy called “the inflammagen model”). A few years later, following the hypothesis that immune tolerance prevents progression to liver failure, Metushi et al. [114] developed an amodiaquine-induced liver injury model using PD-1 knockout (KO) mice co-treated with an antibody against CTLA-4 in order to break the inhibition of cytotoxic T lymphocyte activation.
Very recently, mouse models combining both humanized liver and immune system are emerging in order to detect any human-specific DILI. The introduction of a complete human HLA system to the liver-humanized mice models displays a more complete overview of the prediction of DILI. For example, Song et al. [135] attempted to reproduce abacavir-induced liver injury in HLA-B*57:01 transgenic mice. The cotreatment with abacavir and a Toll-like receptor 9 (TLR9) agonist resulted in a marked increase in alanine aminotransferase (ALT), pathological changes in the liver, increased numbers of activated CD8 T cells, and tissue infiltration by immune cells in transgenic mice.
However, different studies have shown that mimicking genetic susceptibility in mice is not sufficient for reproducing the complex pathogenesis leading to DILI in humans [116, 136, 137], thus maybe the strategy needs to be directed toward the use of patient-derived samples.
Immunophenotyping has largely contributed to the mechanistic understanding of DILI. Infiltration of CD3+CD8+ cytotoxic lymphocytes has been observed in the liver tissue of patients with DILI due to various drugs; however, there are only a few studies using DILI liver tissue due to the scarcity of this type of sample. Drug-specific peripheral blood T cells have also been detected in PBMCs from patients with DILI due to various drugs, as well as increased levels of activated helper T cells and cytotoxic T cells compared to healthy controls. The current lack of specific markers to distinguish DILI from other liver diseases makes it difficult to apply immunophenotyping to the diagnostic criteria. To date, classical techniques, such as flow cytometry has failed to provide an “immune fingerprint” for DILI. However, the use of more complex approaches such as mass cytometry, or single-cell RNA-seq could extend the number of populations studied and allow the identification of specific markers in immune cells derived from DILI patients. Unfortunately, the use of novel immunophenotyping techniques in the field of DILI still faces several challenges, including a lack of consensus on the most appropriate procedures to be performed and the parameters to be analyzed. Furthermore, verification of the correlation between blood and liver immunophenotyping in patients with DILI is necessary to strengthen the feasibility of using peripheral blood samples to infer DILI mechanisms. Due to the low incidence of DILI, access to samples and data for researchers is limited to single centers or small consortia of DILI teams. Therefore, the establishment of a multidisciplinary network of clinicians, immunologists, and professionals with different scientific profiles is necessary to determine which parameters are more informative in DILI research and to establish standard techniques that would contribute to greater comparability across studies, with the ultimate goal of translating immunophenotyping to clinical practice.
AIH: autoimmune hepatitis
APCs: antigen-presenting cells
CCR7: C-C chemokine receptor type 7
COVID-19: coronavirus disease 2019
CRMs: chemically reactive metabolites
CTLA-4: cytotoxic T lymphocyte-associated antigen-4
CXCR5: C-X-C motif chemokine receptor 5
CyTOF: cytometry of time of flight
DILI: drug-induced liver injury
HLA: human leucocyte antigen
ICIs: immune checkpoint inhibitors
ICOS: immune receptor inducible co-stimulator
IFC: imaging flow cytometry
IL-10: interleukin-10
IMC: imaging mass cytometry
iNK-T: invariant natural killer T
KCs: Kupffer cells
LPS: lipopolysaccharide
MAFLD: metabolic-associated fatty liver disease
MHC: major histocompatibility complex
NK: natural killer
PBMCs: peripheral blood mononuclear cells
PD-1: programmed cell death-1
PD-L1: programmed cell death-ligand 1
Th1: T helper 1
TNF: tumor necrosis factor
Treg: regulatory T cell
ACS: Writing—original draft. DEDZS: Writing—original draft, Writing—review & editing. ASZ, GMC, and ABG: Writing—review & editing. MIL: Conceptualization, Writing—review & editing, Supervision. MVP: Conceptualization, Writing—original draft, Writing—review & editing, Supervision. All authors contributed to the manuscript revision, and read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
This work was supported by grants from the Instituto de Salud Carlos III cofounded by Fondo Europeo de Desarrollo Regional (FEDER), contract numbers: [PI21/01248], [PI19-00883], [PT 20/00127], [UMA18-FEDERJA-194], [PY18-3364]; and grants from Consejería de Salud de Andalucía cofounded by FEDER, contract number: [PEMP-0127-2020]. Marina Villanueva-Paz holds a Sara Borrell [CD21/00198] research contract from Instituto de Salud Carlos III (ISCIII) and Consejería de Salud de Andalucía. Alejandro Cueto-Sánchez holds an i-PFIS: Doctorados IIS-empresa en Ciencias y Tecnologías de la Salud [IFI18/00047] research contract from ISCIII. Daniel E. Di Zeo-Sánchez holds an i-PFIS: Doctorados IIS-empresa en Ciencias y Tecnologías de la Salud [IFI21/00034] research contract from ISCIII. Gonzalo Matilla-Cabello holds a Garantía Juvenil [SNGJ5Y6-09] research contract with Junta de Andalucía and the European Social Fund. Antonio Segovia-Zafra holds a Jaume Bosch Training Action 2022 from Centro de Investigación Biomédica en Red Enfermedades Hepáticas y Digestivas (CIBEREHD). Ana Bodoque-García holds a Garantía Juvenil [POEJ-00041-IBIMA] research contract from Junta de Andalucía and the European Social Fund. Spanish Clinical Research Network (SCReN) and CIBEREHD are funded by ISCIII. This publication is based on work from COST Action ‘CA17112—Prospective European Drug-Induced Liver Injury Network’ supported by COST (European Cooperation in Science and Technology), www.cost.eu. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 5719
Download: 96
Times Cited: 0
Gulcin Cakan-Akdogan ... Ozlen Konu
Hanghang Wu ... Francisco Javier Cubero
Qian Wei ... Jinsheng Guo
Miren García-Cortés ... Alberto García-García
