Affiliation:
1Department of Pharmacology, Seacom Pharmacy College, Howrah 711302, West Bengal, India
Email: souravpal2525@gmail.com
ORCID: https://orcid.org/0009-0006-1532-4069
Affiliation:
2Department of Pharmaceutical Sciences, Faculty of Sciences and Engineering, Dibrugarh University, Dibrugarh 786004, Assam, India
ORCID: https://orcid.org/0000-0002-9769-3514
Explor Neuroprot Ther. 2025;5:1004111 DOI: https://doi.org/10.37349/ent.2025.1004111
Received: March 29, 2025 Accepted: June 12, 2025 Published: July 14, 2025
Academic Editor: Agnese Secondo, “Federico II” University of Naples, Italy
MicroRNAs (miRNAs) are small, non-coding RNA molecules that play a pivotal role in post-transcriptional gene regulation, influencing various biological processes such as cell division, proliferation, and apoptosis. Recent research has illuminated the significant involvement of miRNAs in neurological disorders, which encompass a wide range of conditions affecting both the central and peripheral nervous systems. These disorders, including neurodegenerative diseases like Alzheimer’s and Parkinson’s, as well as psychiatric conditions such as depression and schizophrenia, impose a substantial burden on global health. Dysregulated miRNAs contribute to disease pathogenesis by modulating neuronal differentiation and related signaling cascades. This review explores the biogenesis of miRNAs and their dysregulation in neurological disorders, highlighting specific miRNAs that serve as potential biomarkers and therapeutic targets. For instance, decreased levels of miR-125b-5p and miR-26b-5p in cerebrospinal fluid have been associated with Alzheimer’s disease progression. In Parkinson’s disease, distinct profiles of dysregulated miRNAs have been identified, including miR-7-5p and miR-153-3p, which target α-synuclein. Furthermore, studies have demonstrated the potential of miRNA-based therapies to modulate disease processes and improve clinical outcomes. This review critically evaluates current therapeutic strategies for miRNA delivery in neurological disorders, focusing on advanced platforms such as nanocarriers, exosomes, viral vectors, and ligand-mediated systems designed to overcome the blood-brain barrier. We also explore the future of miRNA research in the context of precision medicine, highlighting the importance of targeted delivery, safety optimization, and integration of patient-specific molecular profiles. A comprehensive understanding of miRNA-regulated networks will be essential for developing innovative diagnostics and personalized treatments for neurodegenerative and neuroinflammatory diseases.
A class of tiny, non-coding RNA molecules known as microRNAs (miRNAs) is essential for post-transcriptional gene control. By designating messenger RNAs (mRNAs) for translational repression or destruction, they play a crucial role in the complex orchestration of gene expression. Our understanding of how genes are regulated and how they influence various biological processes, such as cell division, proliferation, apoptosis, and immunological responses, has been significantly enhanced by the discovery of miRNAs [1]. Recent studies have shown that miRNAs play a significant role in the development of neurological illnesses, which include a wide spectrum of ailments that impact both the central and peripheral nervous systems [2]. A large portion of the burden on world health is caused by neurological illnesses, which affect millions of people and their families. These conditions include mental conditions, including depression, schizophrenia, and autism spectrum disorders (ASDs), as well as neurodegenerative illnesses like Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS). miRNAs play a variety of roles in neurological illnesses. They play a crucial role in controlling the differentiation and maturation of different neuronal cell types during embryogenesis and throughout life [3]. Additionally, miRNAs impact synaptic plasticity, a key brain mechanism controlling learning and memory functions. miRNA dysregulation is a significant contributor to neuronal malfunction, neuroinflammation, and eventually neurodegeneration in the pathogenesis of neurological diseases [4]. In AD, decreased cerebrospinal fluid (CSF) levels of miR-125b-5p and miR-26b-5p in patients, as compared to controls, underscore their diagnostic potential. Dysregulated miR-9-3p and miR-7-5p correlated with disease progression, further implicating miRNAs in AD pathogenesis [5]. Research in PD revealed distinct miRNA profiles, with 17 miRNAs dysregulated in patients and miR-7-5p and miR-153-3p targeting α-synuclein, a key pathological protein [6]. In ALS, altered expression of miRNAs associated with TDP-43, including miR-132-5p and miR-574-5p, was linked to disease mechanisms, while elevated CSF miR-27b-3p and miR-146a-5p differentiated ALS from multiple sclerosis (MS) [7]. Cerebral ischemia research demonstrated dynamic miRNA changes post-injury, with early upregulation of miR-200 family members suggesting neuroprotective roles [8]. Similarly, epilepsy models identified increased miR-132-3p levels following seizures, regulating synaptic connectivity and pathological plasticity [9]. In major depressive disorder (MDD), altered miRNA expression in the dorsolateral prefrontal cortex highlighted their role in synaptic function, with specific miRNAs such as miR-19b-3p linked to depressive symptoms [10]. Additionally, studies on homeostatic synaptic plasticity and long-term potentiation (LTP) revealed miRNAs like miR-26a-5p and miR-384-5p as crucial regulators, enhancing ribosomal S6 kinase 3 expression and sustaining LTP [11]. The overexpression of miR-431-5p in AD mouse models enhanced synaptic plasticity and memory, underscoring its therapeutic potential.
This review aims to provide a comprehensive overview of the significance of miRNAs in neurological disorders, emphasizing their crucial role in regulating gene expression within the nervous system. We will look at the specific miRNAs linked to various neurological disorders, their targets downstream, and the underlying molecular mechanisms that underlie their role in disease pathogenesis. We will also go over the potential of miRNAs as therapeutic targets for the creation of miRNA-based therapeutics, as well as diagnostic biomarkers for the early diagnosis of disease. miRNAs and neurological illnesses interact in a complex way, and understanding this interaction holds enormous potential for expanding our understanding of disease mechanisms and creating new opportunities for therapeutic approaches. It is becoming more and more clear that miRNAs represent a promising frontier in the effort to solve the puzzles of neurological illnesses and create ground-breaking approaches for their diagnosis and treatment as the area of miRNA research continues to advance.
Due to their distinct regulatory roles in gene expression and their potential as highly focused and specific therapeutic agents, miRNA-based therapies show tremendous promise in the treatment of neurological illnesses. Table 1 outlines several significant factors that highlight the importance of miRNA-based treatments in neurological disorders.
Key factors highlighting the significance of miRNA-based treatments in neurological disorders
| Significance of miRNA | Clinical outcomes | Conclusion | References |
|---|---|---|---|
| Accurate targeting | miR-379-410 cluster regulates neurogenesis by targeting multiple binding sites in the N-cadherin 3′-UTR. | This additive effect underscores how miRNAs can cooperate to exert stronger repressive actions on their targets, which is crucial during brain development. miRNAs can simultaneously target multiple mRNAs, offering a multi-targeting approach for cancer therapies. | [12] |
| Modulation of complex pathways | Reduced β-amyloid deposition, improved cognitive function, and modulated neuroinflammation using miR-124-3p. | miRNAs can modify multiple molecular pathways, providing a more comprehensive and effective intervention in AD. | [13, 14] |
| Illness modification | Downregulation of CELF2 improved motor function, reduced neurodegeneration, and sustained benefits over time. | miRNA-based treatments can alter the underlying disease process, providing long-term benefits. | [15] |
| Blood-brain barrier (BBB) penetrance | Reduced infarct size by 40%, enhanced neuroprotective signaling, and improved motor function after BBB penetration. | miRNAs can cross the BBB, providing effective therapeutics for central nervous system (CNS) conditions, such as ischemic stroke. | [16] |
| Biomarker possibilities | Downregulation of miR-125b-5p and miR-26b-5p was correlated with the severity of cognitive impairment. | miRNAs can serve as biomarkers for early diagnosis and tracking disease progression in neurological conditions. | [17] |
| Lessened off-target effects | Significant apoptosis in GBM cells with minimal neurotoxicity by targeting the Notch pathway with miR-34a mimics. | miRNA-based therapies offer high specificity, reducing off-target effects and improving safety compared to traditional therapies. | [18] |
| Versatility in delivery | Adeno-associated virus-9 (AAV9)-delivered miR-132 was stably expressed in HD brain regions, silencing MeCP2 and enhancing synaptic plasticity and neuronal survival. | AAV vectors are effective for targeted delivery, and different platforms can be used for diverse neurological disorders. | [19] |
| Personalized medicine | Reduced amyloid plaque deposition and improved cognitive function with personalized miR-29 mimic therapy. | Personalized miRNA therapies can be tailored to individual patients, enhancing treatment efficacy. | [20] |
| Combination therapies | Enhanced efficacy of miR-107, which regulates BACE-1 expression involved in amyloid-beta production. | miRNAs combined with existing therapies can reduce amyloid plaque formation and improve cognitive function in animal models. | [21] |
| Preclinical success | Reduced neuroinflammation, improved motor and cognitive function, and enhanced recovery post-injury using miR-124. | miRNA-based therapies, like miR-124, show promising preclinical results for treating neurological injuries. | [22] |
miRNA-based treatments offer a game-changing strategy to address the intricate and difficult landscape of neurological illnesses. miRNA: microRNA; mRNAs: messenger RNAs; AD: Alzheimer’s disease; HD: Huntington’s disease; BACE-1: beta-site amyloid precursor protein cleaving enzyme 1
miRNAs are small non-coding RNA molecules that play a crucial role in regulating gene expression at the post-transcriptional level. Their synthesis involves a carefully coordinated multi-step process occurring in both the cell nucleus and cytoplasm [23]. The pathway begins with the transcription of miRNA genes by RNA polymerase II (RNA Pol II) in the nucleus. These genes can be found within introns of protein-coding genes, intergenic regions, or exons of other non-coding RNA genes [24]. The initial product of transcription is a primary miRNA (pri-miRNA) transcript, a long RNA molecule with a characteristic hairpin-shaped stem-loop structure that is vital for its processing. Within the nucleus, this pri-miRNA is cleaved by the enzyme Drosha and its cofactor DGCR8 to produce a shorter precursor miRNA [25]. This pre-miRNA retains the stem-loop structure and is subsequently exported to the cytoplasm by the exportin-5 protein, a critical step for its maturation.
Once in the cytoplasm, the pre-miRNA undergoes further processing by the enzyme Dicer, which cleaves it near the stem-loop to form a double-stranded RNA duplex. One strand of this duplex, known as the mature miRNA, is selectively loaded into the RNA-induced silencing complex (RISC) [26]. The RISC, which includes Argonaute (AGO) proteins, guides the mature miRNA to its target mRNA. By binding to complementary sequences in the 3′-UTR of the target mRNA, the miRNA mediates translational repression or mRNA degradation, effectively silencing gene expression [27]. The miRNA synthesis pathway is a tightly regulated process that ensures the proper production of functional miRNAs. Dysregulation of this pathway can result in abnormal miRNA expression, contributing to the development of various diseases, including neurological disorders. Understanding the mechanisms underlying miRNA synthesis is essential for exploring their roles in gene regulation and designing therapeutic strategies targeting miRNA modulation.
Some important factors are necessary for the appropriate processing and maturation of miRNA molecules during the complicated and tightly regulated process of miRNA biogenesis. In Figure 1, we have depicted the key factors involved in the biogenesis and function of miRNA. These important factors have been discussed below.
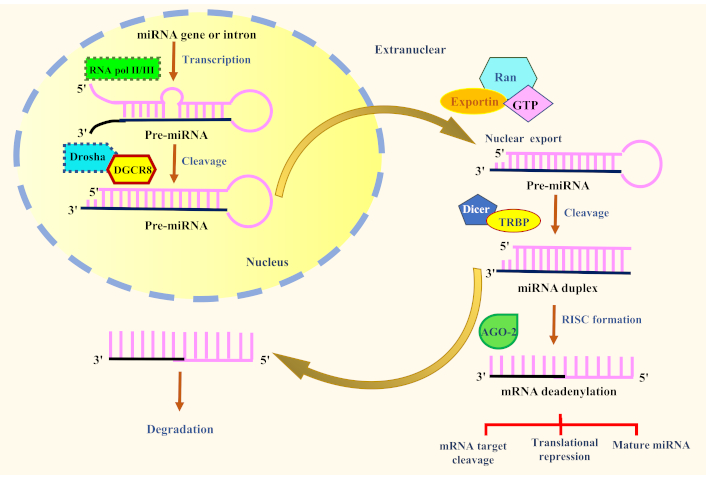
The factors involved in the biogenesis and function of miRNA. AGO-2: Argonaute 2; miRNA: microRNA; RNA pol II/III: RNA polymerase II/III; TRBP: TAR RNA-binding protein; RISC: RNA-induced silencing complex; mRNA: messenger RNA
In the cell nucleus, RNA Pol II is in charge of the transcription of miRNA genes. From miRNA gene loci, it creates the pri-miRNA transcript. As the enzyme in charge of miRNA gene transcription, RNA Pol II is essential for miRNA biogenesis [28]. miRNA genes are present throughout the genome, including in intergenic spaces, the exons of other non-coding RNA genes, and the introns of protein-coding genes [29]. The transcription of miRNA genes by RNA Pol II in the cell nucleus is the first step in the process of miRNA synthesis. The main miRNA that has been translated (pri-miRNA) is a longer RNA molecule with a stem-loop structure [30]. As a precursor to the mature miRNA, this stem-loop structure is a distinguishing quality of pri-miRNAs.
The mature miRNA is created when additional processing stages involving Drosha and Dicer enzymes are completed on the pri-miRNA. This mature miRNA can then be loaded into the RISC for gene silencing [31]. For miRNAs to develop and perform properly, RNA Pol II must correctly transcribe the miRNA genes. Alterations in miRNA expression and function can be caused by dysregulation of miRNA synthesis, including flaws in RNA Pol II transcription [28]. These diseases include cancer, neurological disorders, and cardiovascular conditions. Understanding the function of RNA Pol II in the synthesis of miRNAs can help one better understand the regulatory systems that control miRNA expression and the potential effects they may have on disease pathophysiology. Targeting RNA Pol II and other elements of the miRNA biogenesis pathway may also provide fresh treatment options for conditions characterized by aberrant miRNA expression.
Drosha is a critical enzyme in the production of miRNAs and is essential for the early processing of pri-miRNA transcripts in the cell nucleus. Drosha produces the Microprocessor complex with its cofactor DGCR8, which recognizes and cleaves the pre-miRNA to produce precursor miRNA molecules [32]. The first step in miRNA biogenesis is when RNA Pol II transcribes the miRNA genes, creating lengthy, single-stranded pri-miRNA transcripts [33]. The distinctive stem-loop structure found in these pri-miRNA transcripts, which is essential for miRNA processing, is present in these transcripts.
The junction of the pri-mRNA’s single- and double-stranded sections is recognized and bound to by the Microprocessor complex, which is made up of Drosha and DGCR8 [32]. When measuring the distance between the junction and the stem-loop structure, DGCR8 functions as a molecular ruler. Due to its exact positioning, Drosha can release the pre-miRNA hairpin by cleaving the pri-miRNA at a certain location close to the stem loop’s base [34]. Exportin-5 transports the pre-miRNA from the nucleus to the cytoplasm after Drosha cleaves it. Pre-miRNA is further processed by Dicer in the cytoplasm to create the mature miRNA duplex [33]. The RISC, which serves as the functional miRNA for target mRNA binding and gene silencing, preferentially selects and loads one strand of the miRNA duplex, known as the mature miRNA [32]. Drosha gene mutations or dysregulation of Drosha activity can result in faulty miRNA processing and changed levels of miRNA expression. These abnormalities have been linked to several diseases, including cancer, neurological disorders, and immune-related ailments. Clarifying the regulatory mechanisms of miRNA-mediated gene expression and its potential effects on disease development and therapy requires an understanding of the role of Drosha in miRNA formation [34].
Exportin-5 is a crucial protein that aids in the export of pre-miRNA from the cell nucleus to the cytoplasm, where it is involved in the biogenesis of miRNAs. It is a member of the exportin protein family, which controls how different RNA molecules leave the nucleus [35]. The pri-miRNA transcripts produced when RNA Pol II transcribes the miRNA genes go through several processing stages in the nucleus to create pre-miRNA, which is the precursor to the mature miRNA [36]. The pre-miRNA hairpin structure is released when the pri-miRNA is broken down by the enzyme Drosha and its cofactor DGCR8 in the Microprocessor complex [31]. In a complex with the GTPase Ran, exportin-5 recognizes the pre-miRNA in the nucleus and binds to it. Pre-miRNA can be exported from the nucleus to the cytoplasm via the nuclear pore complex. Pre-miRNA is further processed in the cytoplasm by the Dicer enzyme to create a brief double-stranded RNA duplex [37].
The mature miRNA, one of the duplex’s two strands, is subsequently loaded by Dicer into the RISC, which controls miRNA-guided gene silencing [38]. The mature miRNA in the RISC binds complementary regions in the 3′-UTR of particular mRNAs, which causes mRNA destruction or translational repression and controls gene expression. A crucial stage in miRNA synthesis is the ability of exportin-5 to transfer pre-miRNA from the nucleus to the cytoplasm. miRNA expression levels can change as a result of dysregulation of exportin-5 or flaws in the nuclear export of pre-miRNA, which can also contribute to some diseases, including cancer, neurological disorders, and viral infections [34]. Understanding exportin-5’s involvement in miRNA synthesis might help you better understand the regulatory systems that control the production and operation of miRNAs [36]. Targeting exportin-5 and additional miRNA biogenesis pathway constituents may present possible treatment options for conditions characterized by dysregulated miRNA expression and function.
Dicer is a key enzyme in the production of mature miRNAs, which are produced by processing pre-miRNA molecules. It belongs to the family of RNase III endonucleases and participates in the cytoplasmic stage of miRNA synthesis [39]. The pri-miRNA transcripts produced when RNA Pol II transcribes the miRNA genes are then processed in the nucleus to create pre-miRNAs, which are hairpin structures containing double-stranded RNA sections [33]. The protein exportin-5 then transports the pre-miRNAs from the nucleus to the cytoplasm. Dicer locates the pre-miRNA in the cytoplasm and binds to it [26]. In the case of canonical miRNAs, the protein TRBP (TAR RNA-binding protein) or other protein partners are used to direct the enzyme Dicer to the pre-miRNA [31]. When Dicer is attached to a pre-miRNA, it cleaves the pre-miRNA’s double-stranded RNA region, releasing a short RNA duplex. This RNA duplex has two strands; the mature miRNA strand is opted based on its thermodynamic stability and sequence properties [40]. The RISC, which includes AGO proteins, then incorporates the mature miRNA. Target mRNAs with complementary sequences in their 3′-UTR by the mature miRNA loaded into RISC. When a mature miRNA binds to its target mRNA in the RISC, the target mRNA is translated, repressed, or degraded, which controls the expression of genes [41].
Dicer is crucial for producing useful miRNAs, and dysregulation of its activity can result in incorrect miRNA processing and changed levels of miRNA expression, which can contribute to several diseases, including cancer, neurological disorders, and immune-related ailments [42]. For unraveling the regulatory mechanisms of miRNA-mediated gene expression and its possible consequences in disease development and therapy, it is crucial to comprehend the role of Dicer in miRNA production. Potential treatment strategies for conditions characterized by dysregulated miRNA expression and function include focusing on Dicer and other elements of the miRNA biogenesis pathway.
miRNAs are produced by and perform their functions thanks in large part to AGO proteins. They are a family of proteins that play important roles in the RISC, which is in charge of miRNA-mediated post-transcriptional gene silencing [39]. The mature miRNA strand, which is produced from the pre-miRNA by the enzyme Dicer, is loaded into the RISC following the miRNA biogenesis process. The fundamental elements of the RISC complex are AGO proteins [41]. A miRNA-AGO complex is created when the mature miRNA is integrated into the AGO protein. By identifying particular complementary sequences in the 3′-UTRs of the target mRNA molecules, the miRNA-AGO complex directs RISC to the target mRNA molecules [43]. There are two possible results when the miRNA binds to the target mRNA:
Translational repression: By binding to the target mRNA, the miRNA-AGO complex prevents ribosomes from converting the mRNA into protein. As a result, protein synthesis is inhibited, effectively suppressing gene expression [44].
mRNA degradation: The miRNA-AGO complex can cause the target mRNA to degrade. The target mRNA may be degraded as a result of the miRNA’s binding to the target mRNA, which may then cause the recruitment of other proteins and ribonucleases [43].
miRNAs can control the expression of many genes with complementary sequences because AGO proteins can direct RISC to particular target mRNAs. The regulation of numerous physiological processes, including development, cell differentiation, and responsiveness to environmental cues, depends on this post-transcriptional gene silencing mediated by miRNAs [45]. AGO protein dysregulation or inability to interact with miRNAs can result in aberrant miRNA-mediated gene regulation and support several diseases, including cancer, neurological disorders, and immune-related problems. To decipher the intricate regulatory networks ruled by miRNAs and create possible treatment approaches for disorders associated with dysregulated miRNA expression and function, it is imperative to comprehend the role of AGO proteins in miRNA production and gene silencing [46]. Along with other related proteins and cofactors, these significant components perform a crucial role in the precise and tightly regulated processing of miRNA molecules. Any one of these processes or elements can become dysregulated, resulting in abnormal miRNA expression and activity, which has been linked to several illnesses, including cancer [47], neurological conditions [48], and cardiovascular diseases [49]. To decipher the regulatory mechanisms of miRNA-mediated gene expression and create prospective treatment techniques based on miRNA modulation, it is imperative to comprehend the functions of these important participants in miRNA synthesis.
miRNA processing plays a vital role in orchestrating the intricate processes of neuronal development and function in the central nervous system (CNS). As small non-coding RNA molecules, miRNAs regulate gene expression at the post-transcriptional level, influencing various aspects of neural development and synaptic plasticity [48]. The tightly controlled miRNA processing ensures the precise regulation of target gene expression, enabling miRNAs to act as crucial regulators of neural circuitry and function [50]. During neuronal development, miRNA processing influences the differentiation of neural progenitor cells into mature neurons. Specific miRNAs promote or inhibit neuronal differentiation by targeting genes involved in neural fate determination and differentiation processes [51]. Additionally, miRNAs guide axon outgrowth, neuronal migration, and neurite formation, influencing the establishment of neural circuits and proper connectivity in the developing brain [50]. In mature neurons, miRNA processing continues to play a pivotal role in synaptic plasticity, a cellular mechanism underlying learning and memory [48]. miRNAs regulate the expression of synaptic proteins and receptors, modulating synaptic strength and plasticity in response to neuronal activity. They contribute to the fine-tuning of synaptic transmission, optimizing neural communication and adaptive responses to environmental stimuli.
Moreover, miRNA processing is involved in the maintenance of neuronal homeostasis. miRNAs help regulate the balance of excitatory and inhibitory inputs in neural networks, contributing to stable and functional neural circuitry [50]. They also influence neuronal survival and apoptosis, impacting the overall health and viability of neurons. Altered miRNA expression profiles can disrupt normal neural development and synaptic function, contributing to the pathogenesis of these disorders. Understanding the role of miRNA processing in neuronal development and function provides valuable insights into the complex regulatory mechanisms underlying brain development, plasticity, and neural network dynamics. Moreover, the therapeutic modulation of miRNA processing holds great potential for developing targeted interventions for neurological disorders, offering new avenues for the treatment and management of these challenging conditions.
A set of conditions known as neurodegenerative diseases is characterized by the progressive death of nerve cells in the brain or spinal cord. Normal outcomes of these illnesses include the slow deterioration of autonomic, motor, and/or cognitive abilities. AD, PD, Huntington’s disease (HD), and ALS are a few examples of neurodegenerative illnesses. Small non-coding RNA molecules called miRNAs are essential for post-transcriptional gene control. They work by attaching to mRNA molecules, which serve as the blueprints for protein synthesis [52]. This causes the mRNA to degrade or the creation of proteins to be inhibited. Alterations in miRNA expression profiles have been linked to disease progression and may help to explain the underlying causes of various disorders, according to research. The expression of genes related to neuronal survival, synaptic plasticity, inflammation, and other related processes can be influenced by miRNAs [53].
For instance, in AD, miRNA dysregulation has been seen, and some miRNAs may have a role in amyloid-beta metabolism, tau protein phosphorylation, and neuroinflammation [54]. In PD, it has been discovered that miRNAs regulate genes involved in neuroinflammation, protein handling, and mitochondrial activity in PD [55]. In AD, miR-107 has been shown to regulate amyloid-beta metabolism by targeting beta-site amyloid precursor protein cleaving enzyme 1 (BACE-1), a key enzyme in amyloid-beta production. Similarly, miR-132 dysregulation is associated with tau hyperphosphorylation, contributing to neurofibrillary tangle formation [56]. In PD, miR-155 has been implicated in modulating neuroinflammation by targeting inflammatory cytokines, while miR-34b/c downregulation affects mitochondrial function, contributing to neuronal apoptosis [57]. These findings underscore the pivotal roles of miRNAs in the molecular cascades underlying neurodegenerative disorders. In ALS, miRNAs have been linked to motor neuron degeneration and glial cell activation, two hallmarks of ALS. In ALS, miR-218 has been identified as a key regulator of motor neuron degeneration, with its downregulation contributing to impaired neuromuscular signaling [58, 59]. Additionally, miR-155 is upregulated in activated glial cells, promoting neuroinflammation and exacerbating motor neuron damage [60]. A developing field of study is figuring out how miRNA dysregulation affects neurodegenerative disorders. It may shed light on illness mechanisms, reveal possible biomarkers, and inspire the creation of potential therapeutic approaches. The complex picture of neurodegenerative illnesses, which are influenced by a combination of genetic, environmental, and epigenetic variables, is complicated, and miRNA dysregulation is just one component of it.
A stroke is a neurological condition that happens when the blood flow to a portion of the brain is cut off, causing oxygen and nutrient deprivation and damage or cell death in the affected area of the brain. For a better understanding of their involvement in illness etiology and perhaps to discover possible therapeutic targets, altered miRNA profiles have been investigated in the context of stroke and other neurological disorders. The progression and recovery from neurological illnesses like stroke have been linked to cellular processes like inflammation, oxidative stress, apoptosis, and neuroplasticity, according to research. Both the brain tissue injured by a stroke and the blood in circulation exhibit altered miRNA expression patterns, making them suitable biomarkers for diagnostic and predictive purposes [61]. Stroke and its consequences have been linked to particular miRNAs. miRNAs, including miR-21, miR-23a, miR-29a, miR-126, and miR-210, for instance, have been examined for their functions in pathways connected to stroke [62]. These miRNAs are thought to target genes that, among other things, regulate apoptosis, inflammation, and blood vessel integrity.
miRNAs can potentially affect the characteristics and operations of cellular membranes. They can control the expression of genes related to membrane transport, ion channels, and receptor signaling, all of which are crucial for intercellular communication and overall cellular function [63]. It’s crucial to remember that research in this area is continuing and that we are constantly learning more about the precise methods by which miRNA profiles alter cellular membranes in neurological illnesses like stroke. Even while miRNAs show promise as possible therapeutic targets or diagnostic markers, further research is required to completely understand their functions and prospective uses.
Small non-coding RNA molecules called miRNAs are essential for the post-transcriptional control of gene expression. They are recognized to play a role in several cellular processes, such as differentiation, proliferation, and apoptosis. Psychiatric diseases have been linked to aberrant miRNA expression in some medical situations. A category of mental health diseases known as psychiatric disorders has an impact on a person’s mood, thinking, and behavior. Depression, bipolar illness, schizophrenia, anxiety disorders, and ASDs are a few examples of psychiatric disorders [64]. These complicated illnesses are assumed to be the result of a confluence of genetic, environmental, and epigenetic variables. The expression of genes involved in neural development, synaptic plasticity, and neurotransmitter signaling, all of which are essential for maintaining healthy brain function, can be affected by miRNAs [65]. People with psychiatric diseases have abnormal miRNA expression patterns in their brains. The delicate balance of gene expression can be upset by these miRNA changes, which can also hasten the onset or progression of many illnesses [66]. Psychiatric diseases have been associated with abnormal miRNA expression. Studies on people with depression have found that some miRNAs express themselves differently. These miRNAs might target genes related to synaptic plasticity, neurotransmitter metabolism, and neurotrophic signaling. The neurobiological changes connected to depression may be influenced by altered miRNA expression [67].
Schizophrenia is a complex condition characterized by disturbances in thinking, feeling, and behavior. Schizophrenia has been linked to aberrant miRNA expression [68]. Some miRNAs may control genes involved in synaptic function, dopaminergic signaling, and neuronal migration, all of which are important in the pathophysiology of schizophrenia [69]. miRNAs have been discovered to be involved in the brain circuits and behaviors associated with anxiety disorders. The development of anxiety disorders may be influenced by the dysregulation of miRNAs that control the hypothalamic-pituitary-adrenal (HPA) axis and stress responses [70]. A neurodevelopmental illness with a significant hereditary component is known as ASD. Individuals with ASD have been found to have altered miRNA expression patterns, and these miRNAs may have an impact on the genes involved in synapse function, neural connection, and neurodevelopment [71].
Table 2 highlights key miRNA biomarkers associated with various neurological disorders, emphasizing their diagnostic and prognostic potential. miRNAs such as miR-21-5p in AD, miR-133b in PD, and miR-181c in MS demonstrate significant expression changes linked to disease progression (Figure 2). Their roles in neuronal function, inflammation, and pathology suggest their utility as biomarkers for early detection and disease monitoring.
miRNA biomarkers in neurological disorders
| Neurological disorder | Biomarker miRNA(s) | Key findings | References |
|---|---|---|---|
| AD | miR-21-5p | miR-21 was upregulated in the CSF of MCI patients who progressed to AD but not in non-AD MCI, highlighting its discriminatory potential. It was elevated in microglia, neurons, and astrocytes derived from iPSCs of PSEN1ΔE9 AD patients. Neuron-derived exosomes were enriched in miR-21, and astrocyte exosomes showed increased levels after immunostimulation. Its role in microglial activation and astrocyte reactivity was confirmed in 3D hippocampal cultures with SWE cells. | [72] |
| miR-125b-5p | miR-125b-5p was elevated 1.6-fold in AD brains and similarly high in CSF compared to healthy controls. Overexpression increased ERK1/2 activation, promoting tau phosphorylation. Studies comparing AD patients and healthy controls confirmed these findings. Induced miR-125b-5p overexpression in primary hippocampal neurons altered tau phosphatase levels, affecting Bcl-W, DUSP-6, and PP1CA, suggesting its role in AD-related tau pathology.miR-125b-5p promotes tau hyperphosphorylation in AD by reducing DUSP6/PPP1CA (~50%), elevating p-ERK1/2, tau phosphorylation, and kinase activity. Banzhaf-Strathmann (2014) [73] confirmed direct targeting of DUSP6 by miR-125b via luciferase assays, with effects abolished by binding site mutation and reversed by miR-125b inhibition. | [73, 74] | |
| miR-124 | Overexpression in AD neurons and their exosomes. Observed in mouse organotypic hippocampal slices transplanted with SH-SY5Y cells expressing the human APP695 Swedish mutation. | [72] | |
| miR-9-5p | Liu et al. (2020) [75] found that miR-9-5p is decreased in Aβ25-35-induced HT22 cells; its overexpression inhibits mitochondrial dysfunction, oxidative stress, and apoptosis by targeting GSK-3β, suggesting a protective role in AD models. | [75] | |
| PD | miR-133b | miR-133b was significantly downregulated in PD patients (p = 0.006) and linked to neurodegeneration and motor symptoms. Although its correlation with disease severity was not established, reduced levels suggest a potential role in PD. miR-133b and miR-433 were correlated (r = 0.87 and 0.85) but may not predict PD alone. A study by Zhang et al. (2019) [77] demonstrated that miR-133b directly targets the 3′-UTR of α-synuclein mRNA, leading to reduced α-synuclein levels and mitigating dopaminergic neuron injury in PD models. | [76, 77] |
| miR-124 | miR-124 was significantly upregulated in the plasma of PD patients (p < 0.001), suggesting its potential as a biomarker for early detection and disease monitoring. Its increased levels may correlate with cognitive decline and motor dysfunction, highlighting its role in PD progression. | [78] | |
| miR-34b/c | miR-34b/c was downregulated in key brain regions of PD patients, including the substantia nigra and frontal cortex, across Braak stages 1–5. This depletion was linked to mitochondrial dysfunction and oxidative stress. In SH-SY5Y dopaminergic cells, reduced miR-34b/c impaired viability and mitochondrial function, highlighting their therapeutic potential. | [79] | |
| miR-221-3p | miR-221-3p was significantly upregulated in PD patients (1.79-fold increase; p = 0.032) compared to healthy controls, suggesting its potential as a biomarker. It may help differentiate PD from other movement disorders like MSA, highlighting its diagnostic value in neurodegenerative conditions. | [80] | |
| Multiple sclerosis (MS) | miR-181c | miR levels were elevated in the CSF of MS patients compared to those with other neurologic diseases (AUC = 0.75, p < 0.0001). Higher levels were observed in secondary progressive MS (7.08 ± 0.36) than in relapsing-remitting MS (6.97 ± 0.32, p = 0.036) and primary progressive MS (6.89 ± 0.3, p = 0.046). It may indicate inflammatory activity in early MS. | [81] |
| miR-181c and miR-633 | The combination of miR-181c and miR-633 improved diagnostic accuracy for MS. A cutoff of > 6.79 for miR-181c and > 21.53 for miR-633 achieved 62% sensitivity and 89% specificity in distinguishing MS from other neurologic diseases, enhancing its potential as a diagnostic biomarker. | [81, 82] | |
| miR-145 and miR-223 | miR-145 and miR-223 were significantly upregulated in MS patients compared to healthy controls. In relapsing-remitting MS, fold changes were 2.6 and 2.7, respectively, while in secondary progressive MS, they were 1.4 and 2.2. This suggests their potential role in disease progression and as biomarkers for distinguishing MS subtypes. | [83] | |
| ALS | miR-132-5p | CSF levels of miR-132 and miR-9 were downregulated in ALS patients, particularly those with TARDBP, FUS, and C9orf72 mutations, but not in SOD1 cases. Plasma deregulation distinguished symptomatic from asymptomatic TARDBP-ALS mutation carriers. Increased peripheral expression in symptomatic subjects was observed, but not statistically significant. qPCR analysis confirmed these findings in ALS patients and healthy controls. | [84] |
| miR-155 and miR-124-3p | Cunha et al. (2018) [85] demonstrated that miR-155 is significantly upregulated in the early, pre-symptomatic phase of ALS in both SOD1G93A mouse models and patient-derived samples. In contrast, miR-124-3p, a miRNA with key roles in maintaining neuronal identity and suppressing microglial activation, was found to be upregulated predominantly during the symptomatic phase of ALS. | [85] | |
| Huntington’s disease (HD) | miR-34b | miR-34b was significantly elevated in asymptomatic HD patients, correlating with mutant huntingtin protein levels in neuronal and pluripotent cells. | [86] |
| miR-34a-5p | miR-34a-5p was deregulated in the R6/2 mouse model and human HD brain tissues, interacting with multiple HD-associated genes. Direct binding to 3′-UTRs of TAF4B, NDUFA9, HIP1, and NRF1 was confirmed via mutagenesis and protein analysis. HiTmIR identified additional targets, linking miR-34a-5p to pathways like glutamine receptor signaling and calcium ion transport. | [87] | |
| miR-10b-5p | miR-10b-5p was upregulated in PC12 Q73 cells, enhancing cell survival under apoptotic stress. Increased levels were linked to HD pathology, earlier disease onset, and CAG repeat length in postmortem brain tissue. Elevated plasma levels suggest its potential as a biomarker for predicting HD onset and severity, possibly through BDNF regulation. | [88] | |
| Epilepsy | miR-146a-5p | miR-146a-5p was significantly elevated during seizures and in drug-resistant epilepsy patients. Increased levels correlated with seizure frequency and severity, linking them to neuroinflammation and excitability. | [89] |
| miR-132 | miR-132 was significantly upregulated in epilepsy patients (p < 0.01), linking it to neuronal excitability and epilepsy pathophysiology. | [90] | |
| Disorders of consciousness | miR-150-5p | Reduced expression was observed in patients with (p = 0.003) compared to healthy controls at the time of study inclusion. Expression levels returned to normal six months post-injury in TBI patients. | [91] |
AD: Alzheimer’s disease; ALS: amyotrophic lateral sclerosis; CSF: cerebrospinal fluid; ERK1/2: extracellular signal-regulated kinase 1/2; iPSCs: induced pluripotent stem cells; PD: Parkinson’s disease; MCI: mild cognitive impairment; miRNA: microRNA; MSA: multiple system atrophy
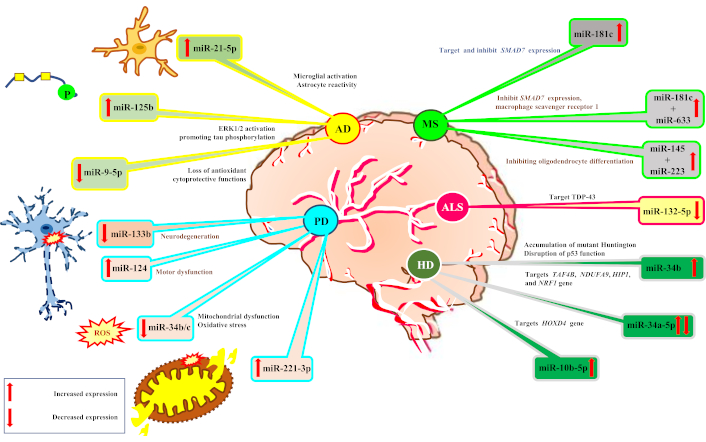
Expression of miRNA in disease progression. AD: Alzheimer’s disease; PD: Parkinson’s disease; MS: multiple sclerosis; ALS: amyotrophic lateral sclerosis; HD: Huntington’s disease
Conventional delivery methods encounter several limitations in facilitating miRNA therapeutic delivery across cellular membranes in neurological disorders. As outlined in Table 3, these challenges can substantially impede the efficacy of miRNA-based therapies.
Limitations of conventional delivery methods for miRNA therapeutics in neurological disorders
| Limitation | Description | Reference |
|---|---|---|
| BBB impedance | The BBB restricts the passage of most therapeutic agents, including miRNAs, due to its selective nature and the hydrophilic, charged characteristics of miRNAs. | [92] |
| Cellular uptake | Efficient cellular uptake of miRNAs into specific target cells in the brain is challenging due to the complex internalization mechanisms of neurons and glial cells. | [93] |
| Rapid clearance | Systemically administered miRNAs may be rapidly cleared from the bloodstream and degraded by nucleases, reducing their bioavailability and therapeutic efficacy. | [94] |
| Off-target effects | Conventional delivery methods often lack specificity, leading to off-target effects on non-neural tissues and unwanted gene regulation. | [92] |
| Stability and pharmacokinetics | miRNAs are susceptible to degradation in biological fluids, necessitating protection from enzymatic degradation to maintain therapeutic levels at the target site. | [95] |
| Immunogenicity | Repeated administration of exogenous miRNAs may elicit immune responses, potentially limiting their long-term therapeutic application. | [96] |
miRNA: microRNA; BBB: blood-brain barrier
miRNAs and other genetic material can be delivered to target cells via viral vectors, which are frequently utilized in gene therapy and molecular medicine. These vectors are altered forms of naturally occurring viruses that have been created to transport and deliver certain therapeutic payloads, like miRNAs, to host cells [97]. Viral vectors are useful tools for a variety of applications because they are highly effective at transmitting genetic material into cells. However, there are certain difficulties and things to think about when using them. Here is a summary of viral vectors used as delivery systems:
Adenoviral vectors: Adenoviruses are huge DNA-containing, non-enveloped viruses. Cells that divide and those that do not can both be effectively infected by adenoviral vectors. Because of their immunogenicity, they are frequently utilized for short-term expression [98]. Advexin and Gendicine are adenoviral vectors expressing the p53 tumor suppressor gene, showing promise in treating cancers like HNSCC and colorectal cancer [99]. Phase I/II trials demonstrated safety and anti-tumor activity, with Advexin administered intratumorally at doses up to 2.5 × 1011 viral particles, highlighting its potential as a targeted cancer therapy [100].
Adeno-associated viral (AAV) vectors: The single-stranded DNA-based AAV virus is a tiny, non-pathogenic virus. Long-lasting gene expression can be established by AAV vectors in both dividing and non-dividing cells [101]. Comparatively, they are less immunogenic than adenoviruses. A clinical trial of AAV9-SMN1 gene therapy for spinal muscular atrophy (SMA) type 1 demonstrated significant motor function improvement, milestone achievement, and a 94% two-year survival rate. Long-term follow-up confirmed sustained SMN protein expression and durable benefits [102]. The therapy was well-tolerated, with mostly mild adverse effects, marking a promising advance in SMA treatment.
Lentiviruses: Viruses having an RNA genome that, upon infection, is reverse-transcribed into DNA. To provide stable and long-lasting expression, lentiviral vectors can incorporate their genetic material into the DNA of the host cell [103]. They are especially helpful for delivering genes to dividing and non-dividing cells, such as specific kinds of stem cells. A study using lentiviral vectors to deliver the FAH gene via portal vein administration in HT1-affected pigs showed stable, long-term FAH expression, normalizing activity within weeks. Liver histology revealed no fibrosis or tumorigenicity, indicating safety and effectiveness [104].
Exosomes have shown promise as natural nanocarriers for the delivery of tailored miRNAs. Exosomes are tiny vesicles that cells release. By transporting bioactive chemicals, such as miRNAs, between cells, they aid in cell-to-cell communication. They are the subject of intensive investigation in the area of RNA-based therapies because they have various benefits as miRNA delivery mechanisms. Exosomes are naturally biocompatible because they are made of lipid bilayers, which are extremely biocompatible and well-tolerated by the body [105]. Depending on the parent cell type, exosomes may be naturally enriched with particular miRNAs. To deliver drugs precisely to particular cell types or regions, they can also be designed to show targeting ligands on their surface. Exosomes enhance the stability of miRNAs in circulation by shielding them from enzymatic activity and destruction [106]. Exosomes have a limited ability to elude immune identification, which lowers the likelihood of immunological reactions and possible negative effects. Exosome-based delivery systems offer a transformative approach for transporting therapeutic miRNAs across the blood-brain barrier (BBB) with enhanced precision and reduced immunogenicity. Recent advances in nanotechnology have enabled self-assembling nanoparticles as effective miRNA delivery vehicles for neurological disorders. These nanoparticles form stable structures via electrostatic interactions and can be functionalized with ligands like transferrin or RVG peptide to cross the BBB. Engineering strategies, including LAMP2B modification for LDLR targeting and RVG peptide functionalization for neuronal uptake, enable receptor-mediated transcytosis and cell-specific delivery. These exosomes protect miRNAs (e.g., miR-124-3p, miR-132-5p, miR-384-5p) from degradation, support scalable loading, and improve dose efficiency. By combining BBB penetration with neuronal specificity, exosome engineering holds significant clinical promise for treating neurological disorders through targeted modulation of gene expression, synaptic plasticity, and neuroinflammation, paving the way for next-generation RNA-based neurotherapeutics [107]. Exosomes have the potential to traverse biological barriers, including the BBB. This makes it easier to distribute miRNAs to hard-to-reach target areas. Exosomes can be loaded with specific miRNAs by utilizing viral vectors to express them or by directly transfecting parent cells with artificial miRNAs. The exosome synthesis then results in the miRNAs becoming enclosed within them [108].
Short amino acid sequences called CPPs can go across cell membranes and help deliver diverse payloads, such as miRNAs, into cells [109]. As prospective strategies for improving the intracellular delivery of miRNAs and other therapeutic compounds, CPPs have drawn a lot of interest. They provide a flexible and successful method for breaking down the cellular membrane barrier and facilitating effective miRNA distribution [110]. The interaction of CPPs with cell membranes and subsequent internalization via endocytosis or direct translocation constitutes the mechanism by which they promote cellular uptake. Depending on the CPP sequence, cargo, and cell type, the precise method may change. While some CPPs may interact with negatively charged elements of the cell membrane because they are positively charged, others may be taken up by receptors. CPPs offer several benefits for miRNA delivery in therapeutic applications. Firstly, CPPs enhance cellular uptake, enabling the effective transfer of miRNAs into various cell types, including non-dividing cells, which conventional delivery methods may struggle to achieve. Secondly, CPPs are versatile as they can be chemically synthesized and easily modified to carry various payloads, such as miRNAs, proteins, peptides, and nanoparticles. Additionally, CPPs improve the stability of miRNAs in the extracellular environment, protecting them from enzymatic degradation. They also allow for tissue-specific targeting by incorporating targeting ligands, ensuring the precise delivery of miRNAs to desired cells or tissues [111]. Finally, CPPs demonstrate minimal immunogenicity, reducing the likelihood of triggering immune responses.
miRNA uptake and intracellular trafficking are intricately regulated by cell-type-specific endocytic pathways, with significant implications for immune function, metabolism, and therapeutic delivery. In macrophages, actin-dependent phagocytosis enables efficient internalization of miRNA-loaded nanoparticles, a process fully inhibited by cytochalasin D. miRNAs such as miR-142-3p modulate this process by downregulating PKCα, reducing phagocytic activity and inflammatory cytokine production. Interestingly, miRNAs like miR-24 and miR-30b exert dual regulatory roles—controlling phagocytosis through transcriptional and post-translational mechanisms while also being internalized via the same pathways [112]. Pinocytosis and macropinocytosis serve as primary uptake routes for miRNA nanoparticles in non-phagocytic cells. Fluid-phase uptake dominates in hepatocytes and HEK293T cells, while receptor-mediated endocytosis via clathrin and caveolin governs uptake in epithelial cells. A recently characterized macropinocytosis-like pathway utilizes serum cationic proteins to form miRNA nanoparticles that bypass lysosomal degradation and target mitochondria via the PNPT1 transporter, enhancing mitochondrial CYB translation and increasing ATP production. Uptake mechanisms vary across cell types: macrophages rely on flotillin-1-rich domains for phagocytosis and macropinocytosis; epithelial cells depend predominantly on clathrin-mediated endocytosis; and stromal cells exhibit hybrid uptake patterns [113]. These mechanistic differences dictate miRNA bioavailability, with inefficient internalization leading to therapeutic sequestration, immune imbalance, or altered metabolic states.
Clathrin-mediated endocytosis: By producing clathrin-coated vesicles, clathrin-coated pits on the cell membrane enable the uptake of particular ligands, such as growth factors or nutrients. Some carriers may be made to take advantage of the clathrin-mediated endocytosis pathway, which enables the internalization of miRNAs into clathrin-coated vesicles [114]. Following their fusion with early endosomes, these vesicles may release miRNA into the cytoplasm. Cells can take up miRNAs, which are tiny RNA molecules that control gene expression, through endocytosis. miRNAs that are supplied exogenously can be contained in a variety of carriers, including nanoparticles, liposomes, or exosomes. Endocytic vesicles may hold onto certain miRNAs, blocking their release and subsequent action. Endocytosed miRNAs can occasionally break free from the vesicles and enter the cytoplasm, where they can then participate in the processes that silence genes. The release of miRNAs from endocytic vesicles may be accelerated by specific carriers or alterations, enhancing their bioavailability for gene regulation.
Endocytosis mediated by caveolae: Endocytosis mediated by caveolae holds significant potential for miRNA drug delivery due to the unique properties of caveolae as specialized lipid rafts in the plasma membrane. Caveolae are flask-shaped invaginations enriched in caveolin proteins, making them distinct sites for cargo uptake and intracellular trafficking. Caveolae-mediated endocytosis presents several advantages in miRNA drug delivery. First, it enables efficient cellular uptake, allowing miRNA-loaded nanocarriers or exosomes to be targeted to caveolae-rich regions of the plasma membrane, enhancing internalization [115]. Additionally, caveolae can be selectively targeted, facilitating cell-specific delivery of miRNAs, which is particularly beneficial in neurological disorders that require precise targeting within the brain. This process also offers protection from degradation, as internalization within caveolae shields miRNAs from extracellular nucleases, improving their stability and bioavailability. Furthermore, caveolae are involved in intracellular trafficking pathways, promoting the efficient delivery of miRNAs to their intracellular targets, which enhances miRNA-mediated gene regulation.
miRNAs migrate throughout cells to reach particular subcellular compartments where they carry out their regulatory tasks. This process is known as intracellular trafficking. Small RNA molecules called miRNAs are essential for post-transcriptional gene control. For miRNAs to interact with their target mRNAs and modify gene expression, proper intracellular trafficking is required. Recent research has deepened our understanding of cytoplasmic miRNA dynamics, highlighting complex regulatory mechanisms that govern their localization, stability, and function. Transport of mature miRNAs is mediated by exportin-5 for nuclear export and Importin-8, which partners with AGO-2, enabling selective nuclear re-entry [116]. RNA-binding proteins influence miRNA activity through competitive binding, chaperone-like coordination of miRNA-AGO-2 complexes, and reciprocal regulation of processing and expression. AGO-2 itself plays dual roles in gene silencing—facilitating mRNA target repression in the cytoplasm and engaging in transcriptional regulation within the nucleus. Emerging evidence also underscores spatial regulation, including miRNA sequestration in nucleoli during stress and mitochondrial localization that may modulate metabolic gene expression.
Exosomes and synthetic nanoparticles are two prominent methods for delivering miRNAs into target cells, both primarily relying on endocytosis for cellular entry. However, miRNAs must escape the endosomal compartment and reach the cytoplasm to exert their gene-regulatory functions. Endosomal escape is, therefore, a critical determinant of delivery efficiency. Once internalized, endocytic vesicles can fuse with lysosomes, exposing miRNAs to enzymatic degradation. To avoid this fate, some delivery systems are designed to promote fusion with non-degradative compartments, such as early or late endosomes and multivesicular bodies [117]. These pathways allow time for release mechanisms to activate before degradation occurs. Multiple endosomal escape strategies have been developed to enhance cytoplasmic delivery. pH-responsive polymers and ionizable lipid formulations are engineered to sense the acidic environment of endosomes and disrupt the membrane, enabling release. Some systems use enzyme-cleavable linkers activated by tumor-specific proteases, while others exploit the proton sponge effect, inducing osmotic swelling and rupture of the endosome. Carrier degradation strategies, such as the glutathione-mediated dissolution of gold nanoparticles, also contribute to controlled release within the cytoplasm. In the case of extracellular vesicle-mediated delivery, fusion of the vesicle membrane with endosomal membranes or directly with the plasma membrane enables miRNA release [118]. Modifications like CD63 surface targeting have been shown to improve fusion efficiency and boost release by up to 40% in cancer models. Additionally, alternative entry routes bypass endocytosis entirely. CPPs and other direct translocation mechanisms allow certain miRNA complexes to cross the plasma membrane and localize directly in the cytoplasm, avoiding endosomal entrapment. Once in the cytoplasm, regulatory protein interactions further modulate miRNA availability. Complexes such as TNRC6-GW182 facilitate the dissociation of miRNAs from carriers by recruiting deadenylases, while AGO-2 phosphorylation at Ser387 enhances release efficiency in neuronal systems. Molecular chaperones like HSP90 stabilize miRNA-carrier complexes during trafficking and dissociate upon release to allow regulatory activity [119]. In contrast, endogenous miRNAs synthesized by the cell follow natural processing and export pathways, reaching the cytoplasm without the need for artificial carriers. Regardless of the source, efficient cytoplasmic release remains essential for effective gene silencing. Failure to escape the endosome can lead to therapeutic resistance, prolonged oncogenic signaling, or inflammatory responses due to carrier accumulation. As a result, optimizing endosomal escape and cytoplasmic delivery remains a central goal in miRNA-based therapeutic design.
The creation of novel miRNA-based therapeutics for a variety of diseases, including cancer, cardiovascular problems, neurodegenerative diseases, and more, has been made possible by these advancements in miRNA synthesis technology (Table 4). These technologies will be essential to maximizing the potential of miRNA research and applications as the field develops.
Advances in miRNA synthesis technologies
| Advancement | Description | Reference |
|---|---|---|
| Chemical synthesis | Improvements in chemical synthesis methods have enhanced the effectiveness and precision of miRNA manufacturing, allowing for high purity and yield. | [120] |
| Enzymatic synthesis | Enzymatic techniques using RNA polymerases and ribonucleases enable large-scale production of miRNAs with site-specific labeling and modified nucleotides. | [120] |
| In vitro transcription (IVT) | IVT employs T7 or T3 RNA polymerases to produce miRNAs, allowing for the addition of modified nucleotides to enhance stability and binding affinity. | [121] |
| Chemical modifications | Advances in chemical modifications improve miRNA stability, target selectivity, and distribution, enhancing pharmacokinetics and cellular absorption. | [120] |
| DNA-encoded miRNA libraries | Synthesis of DNA-encoded miRNA libraries facilitates high-throughput screening of miRNA-mRNA interactions for broader functional studies. | [121] |
| miRNA precursor engineering | Optimizing secondary structures of miRNA precursors enhances processing efficiency and reduces off-target effects. | [25] |
| High-output synthesis | High-throughput synthesis platforms enable the rapid creation of miRNA libraries for functional genomics and drug development. | [121] |
| Genome editing tools | Advances in CRISPR-Cas9 technology allow precise modification of endogenous miRNA expression for physiological studies. | [122] |
| Profiling and next-generation sequencing (NGS) | NGS technology enables high-resolution profiling of miRNAs, facilitating detailed investigation of expression patterns and target identification. | [123] |
miRNA: microRNA; mRNA: messenger RNA
The complex and diverse character of neurological illnesses can be addressed with the use of personalized miRNA-based therapeutics. Based on each patient’s distinct genetic and molecular profile, these medicines can deliver focused and precise interventions. The following are some possibilities for tailored miRNA-based treatments for neurological disorders (Table 5).
Barriers and innovative strategies in miRNA delivery for neurological diseases
| Area of focus | Description | Opportunities | Reference |
|---|---|---|---|
| Targeting neurodegenerative disorders (NDDs) | miRNA-based therapeutics show promise in addressing AD, PD, HD, ALS, Friedreich’s ataxia, SMA, and frontotemporal dementia. These therapeutics aim to replace or inhibit dysregulated miRNAs to provide clinical benefits. | Replacing downregulated miRNAs or inhibiting upregulated miRNAs may be clinically beneficial in NDDs. Development and improvement of synthetic miRNAs, along with specific and effective delivery systems, are needed for this purpose. | [4] |
| Addressing miRNA dysregulation | miRNAs are key gene regulators, playing essential roles in biological and pathological mechanisms; their dysregulation can promote neurological deterioration and the development of NDDs. | Restoring or inhibiting miRNAs altered by disease pathology may treat neurological disorders. Manipulation of endogenous miRNAs or introduction of artificial miRNAs through oligonucleotides or viral vectors could provide effective treatment. | [59] |
| Therapeutic approaches | miRNA-based therapeutics involve using miRNA mimics, siRNAs, inhibitors, and antisense oligonucleotides for treating brain tumors and neurological diseases. Modified oligonucleotides, such as antagomirs or antimirs, can bind and disrupt endogenous miRNAs. | MRX34, a mimic of miR-34a conjugated with liposomes, has entered phase-I clinical trials for liver cancer. LNA-modified oligonucleotides targeting miR-122 delivered intravenously decreased circulating cholesterol levels with no apparent toxicity. Artificial miRNAs can be generated for the repression of specific transcripts. | [124] |
| Delivery techniques | Effective delivery systems are crucial for miRNA-based therapeutics, including viral delivery and administration of modified oligonucleotides. Systemic administration of modified oligonucleotides through intravenous injection or CSF infusion provides a relatively non-invasive and non-toxic means of harnessing miRNAs for therapeutic benefit. | Nanoparticles, exosomes, and viral vectors are being explored for targeted delivery of miRNA therapeutics to the brain. Chemical modifications, such as the fusion of cholesterol, enhance cellular uptake, stability, and integration into the RISC. | [105] |
| Specific miRNA targets | Certain miRNAs, like miR-135, have emerged as therapeutic targets for neurological diseases. Dysregulation of miRNAs, such as miR-30a-5p, may be involved in neurological disorders. | Drugs, including melatonin, can enhance the expression of miR-135 to treat various disease conditions. Morin can inhibit the expression of miR-135 to alleviate different abnormalities. miR-135 plays a role as an endogenous antidepressant in depression, epilepsy, and memory deficits. miR-30a-5p has been shown to inhibit BDNF in the prefrontal cortex, impacting antidepressant effects. | [125] |
| Impact on brain health and disease | miRNAs play major roles in brain tumorigenesis, neurodegenerative diseases, and neurodevelopmental disorders. | miR-135 can act as both an oncogene and a tumor suppressor, depending on the tissue-specific context. miRNAs are dysregulated in stroke and traumatic insults to the CNS. | [125] |
AD: Alzheimer’s disease; PD: Parkinson’s disease; HD: Huntington’s disease; ALS: amyotrophic lateral sclerosis; SMA: spinal muscular atrophy; CSF: cerebrospinal fluid; RISC: RNA-induced silencing complex; CNS: central nervous system; miRNA: microRNA
miRNA therapeutics show promise for treating a wide range of diseases, yet several challenges limit their clinical translation. Off-target effects arise from each miRNA’s ability to regulate numerous genes, leading to unintended and unpredictable consequences. Immune-related adverse events, including cytokine storms and chronic inflammation, have halted trials such as MRX34 and RG-101 [126]. Delivery barriers—such as degradation in circulation, poor endosomal escape, and toxicity of chemical carriers—further reduce efficacy. Additionally, synthetic miRNAs may disrupt gene expression networks, posing genomic and epigenomic risks. Tumor resistance mechanisms and variable pharmacokinetics complicate treatment durability and dosing. Long-term safety remains uncertain, with concerns over persistent alterations in gene regulation. To overcome these hurdles, researchers are advancing targeted delivery systems, immune-evasive modifications, and real-time biodistribution tracking [127]. These innovations aim to enhance safety, specificity, and therapeutic outcomes, making miRNA-based treatments a viable option for future clinical applications.
Recent innovations in miRNA-based technologies are redefining therapeutic possibilities in neurodegenerative disorders. Advanced gene regulation tools, such as CRISPR-dCas9 systems, allow for highly precise modulation of miRNA or disease gene expression [128]. Using CRISPR interference (CRISPRi) and epigenetic editing with dCas9-fusion proteins (e.g., DNMT3A), researchers have demonstrated effective downregulation of pathological targets like SNCA, leading to reduced α-synuclein accumulation and oxidative stress in patient-derived neurons [129]. Parallel strategies employing shRNA-miRNA constructs enable multiplexed targeting of disease pathways through stable and sustained gene silencing [130]. These molecular tools are complemented by breakthroughs in precision medicine, where differential miRNA expression is being integrated with genetic profiling. For instance, APOE4 carriers in AD show altered miR-146a and miR-155 levels that modulate neuroinflammation and disease progression [131]. Similarly, distinct miRNA signatures are observed in PD patients with SNCA or LRRK2 mutations, offering insights into risk stratification and therapy selection [132]. Together, these approaches underscore a paradigm shift toward individualized miRNA-based therapeutics, where gene-editing platforms are guided by patient-specific molecular signatures. The convergence of CRISPR-dCas9, shRNA-miRNA systems, and genotype-informed miRNA profiling offers a roadmap for precision medicine in neurology, promising not only more targeted interventions but also improved clinical outcomes in complex neurodegenerative diseases.
miRNAs represent a transformative frontier in the diagnosis and treatment of neurological disorders. Their ability to fine-tune gene expression, coupled with disease-specific expression profiles, positions them as both potent biomarkers and therapeutic agents for complex conditions such as AD, PD, ALS, and epilepsy. However, despite their potential, clinical translation is hindered by significant delivery challenges, particularly the need to navigate the BBB and achieve targeted, stable, and safe miRNA transport. Recent advances in delivery systems, including ligand-functionalized nanocarriers, engineered exosomes, and stimuli-responsive vehicles, are steadily overcoming these barriers. In particular, exosome-based approaches leveraging surface proteins such as LAMP2B and RVG peptides demonstrate promise in achieving receptor-mediated transcytosis and neuronal targeting with low immunogenicity. Nevertheless, issues of immunogenicity, scalable manufacturing, and regulatory clarity continue to pose substantial obstacles. Ensuring reproducibility, safety, and efficacy across diverse patient populations remains a pressing concern. Looking forward, the integration of miRNA-based tools into personalized medicine paradigms could revolutionize neurological care. By tailoring interventions based on an individual’s molecular profile and incorporating miRNA panels into diagnostic workflows, clinicians may soon be able to predict disease progression, monitor therapeutic responses, and adapt treatments dynamically. In sum, while challenges persist, the convergence of molecular neuroscience, nanotechnology, and clinical innovation is accelerating the path toward miRNA-based therapies. With sustained research, technological refinement, and interdisciplinary collaboration, miRNAs hold the potential to redefine therapeutic strategies and offer new hope for treating currently intractable neurological diseases.
AAV: adeno-associated viral
AD: Alzheimer’s disease
AGO: Argonaute
ALS: amyotrophic lateral sclerosis
ASDs: autism spectrum disorders
BBB: blood-brain barrier
CNS: central nervous system
CPPs: cell-penetrating peptides
CSF: cerebrospinal fluid
HD: Huntington’s disease
LTP: long-term potentiation
miRNAs: microRNAs
mRNAs: messenger RNAs
MS: multiple sclerosis
PD: Parkinson’s disease
pri-miRNA: primary microRNA
RISC: RNA-induced silencing complex
RNA Pol II: RNA polymerase II
SMA: spinal muscular atrophy
SP: Conceptualization, Supervision, Validation, Writing—original draft. SM: Visualization, Writing—review & editing.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 4351
Download: 64
Times Cited: 0
