Affiliation:
1Transdisciplinary Research for Drug Discovery, Sociedad Mexicana de Epigenética y Medicina Regenerativa A. C. (SMEYMER), México City 04000, México
2Centro Conjunto de Investigación en Química Sustentable (CCIQS), UAEM-UNAM, Toluca, Estado de México 50200, México
Email: ravilaa@uaemex.mx
ORCID: https://orcid.org/0000-0002-0683-073X
Explor Musculoskeletal Dis. 2025;3:1007101 DOI: https://doi.org/10.37349/emd.2025.1007101
Received: June 28, 2025 Accepted: August 05, 2025 Published: August 19, 2025
Academic Editor: Ivan Castellví Barranco, Hospital Universitari de la Santa Creu i Sant Pau, Spain
The dynamic organization of chromatin plays a critical role in regulating muscle cell differentiation. Among the molecular elements influencing chromatin architecture, long noncoding RNAs (lncRNAs) have emerged as important regulators due to their capacity to act as scaffolds, recruiters of chromatin-modifying proteins, or as transcriptional enhancers. This review aims to explore the mechanisms by which lncRNAs influence chromatin structure in the context of skeletal muscle differentiation. We classified the functional roles of lncRNAs into three main strategies: recruitment of epigenetic modifiers, assembly of transcriptional scaffolds, and regulation through enhancer-like activity. We provide specific examples of lncRNAs associated with these mechanisms and discuss their involvement in the control of myogenic gene expression. These findings highlight the complexity and specificity of lncRNA-mediated chromatin remodeling and suggest their potential as targets for therapeutic intervention in muscle-related disorders.
Adult skeletal muscle is composed of numerous multinucleated myofibrils, which are made up of hundreds to thousands of fused, postmitotic, terminally differentiated muscle cells [1]; myofibrils have the contractile machinery necessary for fundamental muscle functions such as locomotion, postural support, and breathing; also, reception, integration and transduction of metabolic signals from most of the tissues [2–4].
The growth, maintenance, and regeneration of skeletal muscle after birth rely on a specialized group of stem cells found within the muscle tissue, commonly referred to as satellite cells (SC) [1]. These cells are positioned on the surface of muscle fibers, situated between the basal lamina and the sarcolemma [5].
In muscular homeostasis, SC are in a quiescent state; after a stimulus induced by physical damage or disease, the SC are activated and re-enter the cell cycle to, on the one hand, originate myogenic precursor cells that in turn will fuse giving rise to new muscle fibers; and on the other hand to renew itself by maintaining the SC reservoir for subsequent regeneration events [6–8]. This process of muscle regeneration and differentiation is due to a complex process of transcriptional regulation modulated by the expression of the myogenic regulatory factors (MRFs). The differential expression of MRFs and their activity in the process of differentiation and muscle regeneration are controlled at multiple levels [9, 10].
At the epigenetic level, the combination of modifications in the chromatin structure exerts a dynamic and decisive role in muscle differentiation. For example, the presence of remote transcriptional regulation elements such as enhancers and promoters, the synthesis of non-coding RNAs (ncRNAs), post-translational modifications (PTMs) of regulatory proteins, as well as the combination of transcription factors and chromatin remodeling complexes, responds to different extracellular signals to execute gene expression in a given cellular context in myogenesis [11–13]. So here we highlight the role of the ncRNAs in muscle differentiation controlling the chromatin structure, which has recently generated an interest in many research groups.
Therefore, this review aims to specifically examine how long noncoding RNAs (lncRNAs) regulate chromatin structure in the context of skeletal muscle differentiation. We focus on three major functional strategies by which lncRNAs exert their influence: (1) recruitment of epigenetic regulators, (2) scaffold assembly for transcription factors or chromatin remodelers, and (3) enhancer-like activity. Unlike broader reviews on lncRNAs or general epigenetic regulation in muscle biology, this work provides a mechanistic classification of lncRNAs based on their chromatin-associated functions during myogenesis. By doing so, we aim to clarify their potential as diagnostic markers or therapeutic targets in muscle-related disorders.
In mammals, most of the transcribed genetic material does not code for proteins. Although approximately 80% of the genome undergoes transcription, only about 1% is responsible for protein production. Based on their length, ncRNAs are typically categorized into two main classes [14, 15].
Small ncRNAs are below 200 bp and include micro-RNA (miRNA), small interfering RNA (siRNA), piwi-interacting RNA (piRNA), or small nuclear RNAs (snRNA) [16]. The miRNAs are the most studied. Microarray analyses indicate that some miRNAs are enriched in muscle, such as miR-1, miR-133a, and miR-206, that are up-regulated under the influence of MRFs such as myogenic differentiation 1 (MyoD) and myogenin (MyoG) during human and murine skeletal muscle differentiation in culture cells [17, 18].
lncRNAs represent a subclass of ncRNAs defined by their length, typically exceeding 200 bp [19]. These transcripts often originate from regions closely associated with protein-coding genes, including exonic or intronic segments, and can be transcribed in either the sense or antisense direction. Some lncRNAs are also found in intergenic areas, sometimes overlapping with repetitive DNA elements. Most lncRNAs are produced by RNA polymerase II (Pol II) and may possess characteristics similar to messenger RNAs—such as a 5' cap, polyadenylated tail, and splicing sites—although some lack these features [20, 21].
The nature and secondary structures of lncRNAs allow them to interact with DNA, RNA, and proteins. And its transcriptional pattern is, in general, more developmental stage and cell type specific. lncRNAs are less studied and have a greater diversity of regulatory strategies [22, 23]. lncRNAs can act as scaffolds for regulatory proteins, recruit histone remodeling complexes, function as co-activators or co-repressors, or modulate other ncRNAs. In addition to their transcriptional roles, lncRNAs also influence chromatin architecture and three-dimensional nuclear organization by modulating the activity and localization of chromatin-associated factors [24].
The chromatin structure is dynamic and is organized by specific interactions, which have a specific role, interaction as gene to gene, promoter to enhancer or large nuclear bodies [25]. The interactions could occur across multiple chromosomes. In order to better understand the role of lncRNAs in regulating the chromatin structure, we divide lncRNAs into three strategies: lncRNAs recruiting epigenetic regulators, lncRNAs acting as scaffolds, and lncRNAs as enhancers [22] (Table 1). To facilitate understanding, Figure 1 provides a schematic summary of the three major mechanisms through which lncRNAs regulate chromatin structure in muscle differentiation.
Functional lncRNAs in skeletal muscle development that modify chromatin structure
| lncRNA | Functional role in chromatin regulation | Target genes | Biological effect | Model system | Ref. |
|---|---|---|---|---|---|
| NEAT1 | Recruits PRC2 via EZH2, promoting H3K27me3 | MyoG, Myh4, Tnni2 | Inhibits differentiation | Mouse myoblasts (C2C12) | [35–37] |
| Malat1 | Recruits Suv39h1 to MyoD, induces H3K9me3 | MyoD targets | Inhibits differentiation | Mouse myoblasts (C2C12) | [38–40] |
| SYISL | Recruits PRC2 complex to silence key myogenic genes | MyoG, Myh4, MCK4 | Inhibits differentiation | Mouse myoblasts (C2C12) | [41] |
| Dum | Recruits DNA methyltransferases to silence Dppa2 | Dppa2 | Promotes differentiation | Mouse myoblasts (C2C12) | [32] |
| Myoparr | Interacts with Ddx17 and PCAF to induce histone acetylation | MyoG | Promotes differentiation | Mouse (C2C12), human | [42, 43] |
| Linc-YY1 | Dissociates YY1/PRC2 complex, relieving repression | miR-29, MyHC, Troponin | Promotes differentiation | Mouse myoblasts (C2C12) | [44, 45] |
| Linc-RAM | Scaffolds MyoD with Baf60c/Brg1 to activate targets | MyoD targets | Promotes differentiation | Mouse (WT and KO models) | [47] |
| SRA | Forms a MyoD-p68-p72 complex to promote transcription | MyoD targets | Promotes differentiation | Mouse and human cells | [49, 50] |
| Irm | Scaffolds MyoD and MEF2D, activating downstream targets | MyoD/MEF2D targets | Promotes differentiation | Mouse (in vitro/in vivo) | [51] |
| Myolinc | Interacts with TDP-43 to modulate Filip1 expression | Filip1, Acta1, MyoD | Promotes differentiation | Human and mouse | [52] |
| CEeRNA | Enhancer RNA acting in cis to activate MyoD promoter | MyoD | Promotes differentiation | Mouse | [59] |
| DRReRNA | Enhancer RNA acting in trans to activate MyoG | MyoG | Promotes differentiation | Mouse | [60] |
lncRNA: long nonconding RNA; NEAT1: nuclear paraspeckle assembly transcript 1; PRC2: polycomb repressive complex 2; EZH2: enhancer of zeste homolog 2; H3K27me3: trimethylation of histone H3 at lysine 27; MyoG: myogenin; C2C12: mouse myoblast cell line; Malat1: metastasis-associated lung adenocarcinoma transcript 1; MyoD: myogenic differentiation 1; H3K9me3: trimethylation of histone H3 at lysine 9; Dppa2: developmental pluripotency-associated 2; Dum: Dppa2 upstream binding muscle lncRNA; Linc-RAM: long intergenic non-coding RNA activator of myogenesis; SRA: steroid receptor RNA activator; CEeRNA: core enhancer RNA; DRReRNA: distal regulatory region enhancer RNA
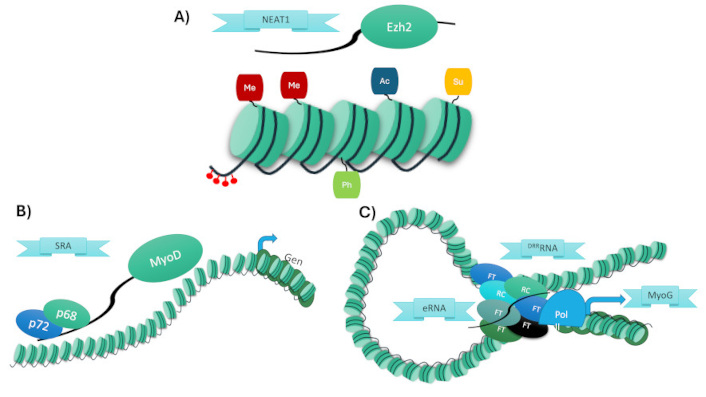
Schematic summary of the three major mechanisms by which lncRNAs regulate chromatin during muscle differentiation. (A) Recruitment of epigenetic regulators, (B) formation of scaffolds for transcriptional complexes, (C) enhancer-like function via eRNA-mediated chromatin looping. NEAT1: nuclear paraspeckle assembly transcript 1; EZH2: enhancer of zeste homolog 2; SRA: steroid receptor RNA activator; MyoD: myogenic differentiation 1; eRNA: enhancer RNA; DRReRNA: distal regulatory region enhancer RNA; MyoG: myogenin
There are two principal ways to regulate the epigenetics of specific loci: the first is controlling the DNA methylation and the second is controlling the posttranslational histone modifications, both directly control the recruitment of chromatin remodelers that modify the chromatin structure.
The DNA methylation induces, in general, a transcriptional gene silencing; and it could be for lncRNA mediation, being a conserved process where lncRNA repressive chromatin modifications to specific regions of the genome [26, 27]. One of the fundamental processes in the epigenetic regulation of muscle cell differentiation is the establishment and preservation of DNA methylation patterns, which influence gene expression [28, 29]. This process is carried out by enzymes of the DNA methyltransferase (Dnmt) family, which are typically divided into two groups: de novo methyltransferases (Dnmt3a and Dnmt3b) and maintenance methyltransferases (Dnmt1) [30, 31].
This is the case of a lncRNA called developmental pluripotency-associated 2 (Dppa2) upstream binding muscle lncRNA (Dum). Dum was identified in skeletal myoblast cells; its expression is dynamically regulated by MyoD during myogenesis in vitro and in vivo. Dum promotes muscle differentiation and regeneration during the early stages of myogenesis by silencing the upstream gene Dppa2. Mechanistically, Dum binds near the Dppa2 promoter region and recruits Dnmt1, Dnmt3a, and Dnmt3b to induce DNA methylation and repress transcription [32].
Histone modifications also play a critical role in regulating chromatin structure during myogenesis. These PTMs, including methylation, acetylation, phosphorylation, and ubiquitination, primarily occur on the N-terminal tails of histones, which are accessible and unstructured [33, 34]. These PTMs influence chromatin accessibility and help define transcriptionally active or repressive chromatin domains.
For example, nuclear paraspeckle assembly transcript 1 (NEAT1) is a lncRNA that promotes myoblast proliferation while inhibiting differentiation. NEAT1 exerts its effects by interacting with enhancer of zeste homolog 2 (EZH2), a core subunit of the polycomb repressive complex 2 (PRC2), which catalyzes trimethylation of histone H3 at lysine 27 (H3K27me3), leading to repression of myogenic genes such as MyoG, Myh4, and Tnni2 [35–37]. Beyond its regulatory role in differentiation, NEAT1 has been implicated in muscle atrophy conditions. Its overexpression in atrophic models correlates with impaired expression of myogenic genes, suggesting it may contribute to pathological inhibition of muscle regeneration.
Another lncRNA, metastasis-associated lung adenocarcinoma transcript 1 (Malat1), regulates myogenesis by recruiting Suv39h1, a histone methyltransferase that deposits H3K9me3 marks at the MyoD locus. This modification represses transcription and modulates myogenic differentiation. Knockdown of Malat1 accelerates myogenic gene expression and muscle fiber formation [38–41].
In contrast to these repressive mechanisms, some lncRNAs promote muscle differentiation through transcriptional activation. Myoparr, a promoter-associated lncRNA, interacts with the RNA Ddx17 and the histone acetyltransferase PCAF to enhance transcription of muscle-specific genes such as MyoG. This is mediated by histone acetylation, which promotes an open chromatin configuration favorable to gene expression [42, 43].
Lastly, Linc-YY1 regulates gene expression by dissociating repressive chromatin complexes. It binds to YY1 and promotes the disassembly of the YY1/PRC2 complex at the promoters of target genes like miR-29, miR-1, MyHC, and Troponin. This relieves transcriptional repression and facilitates myogenic differentiation and regeneration [44, 45].
In addition to recruiting epigenetic regulators, lncRNAs can also influence gene expression by modulating the activity of sequence-specific transcription factors. In this case, the lncRNAs act as scaffolds to proteins that modulate gene expression [46].
For example, long intergenic non-coding-RNA (linc-RNA) activator of myogenesis (Linc-RAM) is a lncRNA localized in an intergenic region and has been determined to is differentially expressed in muscle differentiation, increasing its transcription. The absence of the expression of Linc-RAM induces an abnormal SC differentiation, which generates poor and abnormal muscle regeneration after damage in knockout mice. Linc-RAM physically interacts with MyoD acting as a scaffold to form the complex MyoD-Baf60c-Brg1, to regulate the expression of multiple MyoD gene targets [47, 48].
Similarly, steroid receptor RNA activator (SRA), a lncRNA, is regulated during muscle genesis. SRA is a scaffold to form a complex SRA-MyoD-p68-p72, where p68 and p72 are RNA helicases and together with MyoD transcription factor induce the transcription of specific MyoD target genes, inducting myogenesis [49, 50].
A similar strategy is employed by lncRNA intergenic regulator of myogenesis (Irm). Irm is differentially expressed on the upside in muscle differentiation and muscle regeneration by inducing myoblast differentiation. Irm acts as a scaffold inducing the complex MyoD/MEF2D formation. The complex of these two transcriptional generates a transcriptional regulatory framework that induces the transcription of muscle-specific genes and regulates muscle differentiation through mutual reinforcements [51].
On the other hand, Myolinc, a lncRNA overexpressed in muscle, induces the progression of muscle differentiation. Myolinc interacts with TDP-43 and directly regulates Filip1. This specific complex can bind to DNA or RNA to regulate the Acta1 and MyoD targets [52].
DNA enhancers are elements that increase transcriptional output of protein-coding; the localization of enhancers is not specific; enhancers could be at a large distance of the target gene and in different genomic orientations [53]. Also, there are specific characteristics that distinct an enhancer, these characteristics could be: the presence of p300 acetyltransferase, the characteristic signature of H3K4me1 and H3K4me3 histone marks; the acetylation of histones, and the high sensitivity to nucleases [54, 55]. Some studies are associated with the presence of Pol II on enhancer regions with the probability of the transcription of enhancers, which induces the formation of enhancers of RNA called enhancer RNAs (eRNAs). We can catalogue the eRNAs by the localization of the enhancer to respect the target gene, subdivide the eRNAs in cis-eRNAs and trans-eRNAs [56, 57]. The great majority of eRNAs regulate enhancer/promoter communication by directly recruiting chromatin modifiers, remodelers, and the transcriptional machinery, altering the chromatin structure and inducing an increment transcription rate [58].
Two main eRNAs have been identified as key regulators of myogenesis by orchestrating chromatin remodeling and shaping the hierarchy of the myogenic gene regulatory network. These eRNAs are transcribed from enhancer regions located near the myogenic master regulator MyoD. The core eRNA (CEeRNA) modulates the transcription of the nearby MyoD gene [59], while the distal regulatory region eRNA (DRReRNA) functions in trans to enhance transcription at the MyoG locus [60]. At their respective target sites, both eRNAs contribute to increased chromatin accessibility and facilitate the recruitment of RNA Pol II.
In the case of DRReRNA has been demonstrated that it induces a tridimensional enhancer-promoter interaction mediated by DNA, RNA, and protein components, by the recruitment of cohesin-CTCF complex [60].
The available evidence suggests that lncRNAs act at different stages of the myogenic program, with some functioning as early differentiation gatekeepers (e.g., Dum, Linc-RAM, NEAT1) and others modulating later stages of fusion and fiber maturation (e.g., Myoparr, Myolinc, CEeRNA). This points toward a possible regulatory hierarchy among lncRNAs, in which chromatin silencing or remodeling is first established and then refined by transcriptional activation complexes. However, this model is still speculative and awaits systematic temporal validation.
Furthermore, several lncRNAs exhibit context-dependent or apparently contradictory functions. For example, Malat1 has been described as a suppressor of myogenic differentiation in vitro by recruiting Suv39h1 to silence MyoD targets, yet it is also broadly expressed in differentiated tissues and may support homeostatic functions. These dual roles may be explained by differences in expression levels, post-transcriptional modifications, or interactions with distinct protein complexes depending on cellular context. Understanding these nuances will be essential to decipher the logic of lncRNA-mediated regulation in muscle biology.
Despite the growing number of lncRNAs identified in muscle differentiation, several fundamental questions remain unresolved. For instance, do lncRNAs act redundantly, with overlapping functions, or do they operate in a hierarchical and sequential manner? How is their expression temporally coordinated during different stages of myogenesis, and what upstream signals control their activation? Moreover, it remains unclear how lncRNAs integrate into known signaling pathways and whether they participate in feedback or feedforward regulatory loops. Addressing these questions will be essential for constructing a more comprehensive model of lncRNA-mediated epigenetic regulation in muscle biology.
The role of lncRNAs in regulating muscle differentiation through the modulation of chromatin structure is becoming increasingly evident. Although only a limited number of lncRNAs have been fully characterized in this context, transcriptomic and epigenomic studies have revealed a wealth of candidates potentially involved in myogenesis.
Future research should aim to functionally validate these emerging lncRNAs, particularly in vivo models of muscle injury and regeneration. Additionally, dissecting their interactions with chromatin remodelers and transcription factors in disease states—such as muscular dystrophies, cachexia, or sarcopenia—may uncover new layers of epigenetic regulation contributing to pathology.
From a translational perspective, lncRNAs represent attractive targets for diagnostic and therapeutic development. Their tissue specificity, dynamic regulation, and ability to modulate key differentiation pathways suggest they could be used as biomarkers of muscle health or as tools for modulating gene expression in regenerative therapies.
Advancing our understanding of the lncRNA-chromatin interface will open new avenues for manipulating muscle cell fate and function, with implications not only for basic biology but also for clinical applications in neuromuscular medicine.
CEeRNA: core enhancer RNA
Dnmt: DNA methyltransferase
Dppa2: developmental pluripotency-associated 2
DRReRNA: distal regulatory region enhancer RNA
Dum: developmental pluripotency-associated 2 (Dppa2) upstream binding muscle lncRNA
eRNAs: enhancer RNAs
Irm: intergenic regulator of myogenesis
Linc-RAM: long intergenic non-coding RNA activator of myogenesis
lncRNAs: long noncoding RNAs
Malat1: metastasis-associated lung adenocarcinoma transcript 1
miRNA: micro-RNA
MRFs: myogenic regulatory factors
MyoD: myogenic differentiation 1
MyoG: myogenin
ncRNAs: non-coding RNAs
NEAT1: nuclear paraspeckle assembly transcript 1
Pol II: polymerase II
PRC2: polycomb repressive complex 2
PTMs: post-translational modifications
SC: satellite cells
snRNA: small nuclear RNAs
SRA: steroid receptor RNA activator
RDAA: Conceptualization, Investigation, Writing—original draft, Writing—review & editing, Visualization.
The author declares that there are no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
RDAA was financially supported through “Investigadoras e Investigadores COMECYT 2024” program CAT2024-0077. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
