Affiliation:
Department of Vaccine Technology, Vaccine Institute, Hacettepe University, 06210 Ankara, Turkey
Email: sezerokay@gmail.com
ORCID: https://orcid.org/0000-0003-0355-6672
Explor Med. 2022;3:280–288 DOI: https://doi.org/10.37349/emed.2022.00092
Received: April 07, 2022 Accepted: May 18, 2022 Published: June 24, 2022
Academic Editor: Margaret M. DeAngelis, University at Buffalo, USA
Retroelements are mobile genomic components requiring an RNA intermediate which is reverse-transcribed into complementary DNA for transposition. Human genome contains a vast amount of retroelements including retrotransposons and endogenous retroviruses. These elements are categorized according to presence or absence of long terminal repeats, LTRs or non-LTRs, as well as autonomous and non-autonomous according to involvement of reverse transcriptase. The retroelements have been accumulated in mammalian genomes over all evolutionary times through vertical transmission, and many of them were inactivated through accumulation of mutations. However, the retroelements entered into genome within the last 200,000 years are mostly functional. Some of the active retroelements are associated with varying autoimmune diseases because anti-retroelement antibodies might cross-react with other proteins in the human body. For instance, autoimmunity and inflammation could be stimulated by increased expression of long interspersed element 1 (LINE-1 or L1) or decreased L1 degradation. Different regulation of L1 expression might be related to the genetic and sex-related variations or environmental factors. Activation of retroelements is also controlled by epigenetic silencing mechanisms such as histone modification. Elevated levels of L1 retroelements could trigger the production of type I interferon, a crucial innate defense mechanism in mammals against viruses, and systemic autoimmune response is induced. Loss-of-function in some deoxyribonucleases (DNases) such as three prime repair exonuclease 1 that degrades reverse-transcribed DNA is also related to autoimmune diseases. Additionally, human endogenous retroviruses also play a role in autoimmune diseases. Involvement of retroelements in autoimmune disorders is exemplified with three diseases, i.e. systemic lupus erythematosus, Aicardi–Goutières syndrome, and multiple sclerosis.
Retroelements including retrotransposons and endogenous retroviruses are mobile genetic elements. Transposition of retroelements starts with transcription by an RNA polymerase, and the RNA transcript is reverse transcribed to complementary DNA (cDNA) that integrates into a new genomic position. This process is important in proliferation and differentiation of cells such as gametes, and controlled by many factors such as non-coding RNAs, methylation machinery, transcription factors, and chromatin modification. Retroelements are classified as long terminal repeat (LTR) such as human endogenous retroviruses (HERVs) and non-LTR such as long interspersed elements (LINEs) and short interspersed elements (SINEs) according to their structures (Figure 1). Additionally, retroelements possessing reverse transcriptase for their transposition are categorized as autonomous such as HERVs and LINEs whereas non-autonomous ones such as SINEs and Alu elements (highly repetitive short repeat sequences digested by Alu endonuclease from Arthrobacter luteus) use LINE-1 (L1) machinery for their relocation [1–3]. Reverse transcription of LTR retroviral RNA includes binding of a cellular transfer RNA (tRNA) to the primer binding site in the viral RNA genome. However, retrotransposition of non-LTR LINEs occurs through a process called target-primed reverse transcription (TPRT) in which a cDNA strand is synthesized using the LINE RNA as template. Therefore, reverse transcription of LINEs takes place at the precise genomic site where retrotransposition is desired. Nevertheless, L1, only active in humans, has relaxed target site selection [4].
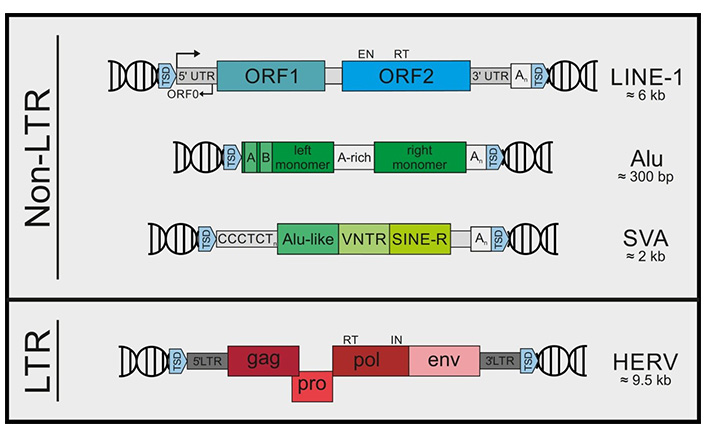
Retroelements are classified as LTR and non-LTR elements according to their structures. Non-LTR retroelements include LINE-1, Alu, and SVA (SINE-VNTR-Alu) elements. LTR retroelements mainly include HERVs. TSD: target site duplication; UTR: untranslated region; ORF: open reading frame; VNTR: variable number tandem repeat; CCCTCTn: CCCTCT repeats, C: cytosine, T: thymine; A-rich: adenine rich; An: adenine repeats; gag: the gene encoding Group Antigens, i.e. viral matrix, capsid and nucleoproteins; pro: the gene encoding protease; pol: the gene encoding reverse transcriptase; env: the gene encoding envelope. EN: endonuclease; RT: reverse transcriptase; IN: integrase
Note. Reprinted from “Recognize yourself—innate sensing of non-LTR retrotransposons,” by Lagisquet J, Zuber K, Gramberg T. Viruses. 2021;13:94 (https://doi.org/10.3390/v13010094). @ 1996–2022 MDPI. CC BY.
About half of the human genome consists of retroelements but the majority of them acquired mutations since they are not under positive selection pressures during evolutionary processes, and these altered retroelements become inactive. However, many HERVs are still found having original functions [1, 5]. Particular developmental stages, aging, inflammation and various pathologies can activate HERVs [6]. Tissue-specific expression of active retroelements also occurs. For instance, L1 has 248 functional copies in the human genome. However, the ones within an intron of an abundantly expressed gene are active [7]. Additionally, expression of these retroelements is regulated throughout the genome, especially by epigenetic silencing. The type of silencing differs according to the developmental stage and class of retroelements. For instance, LTRs are silenced in embryonic stem cells via histone modification while retroelements are silenced by DNA methylation in fully differentiated cells. Epigenetic silencing of the retroelements is strictly controlled in germ line cells to defend the genome from further expansion of endogenous retroelements [8].
A limited number of endogenous retroelements such as HERV-K are active transcriptionally and translationally. Although the majority of HERVs were integrated in the genome approximately 25 million years ago, active ones were more recently entered into the human DNA within the last 200,000 years, and they had ORFs functional for the production of viral proteins such as Gag and Pol, even forming virus-like particles (Figure 2). Some infections might induce the transcription of active retroelements in the genome, and related T and B cell responses. For instance, HIV infection might activate the transcription of many HERV proviruses through its transactivator of transcription (Tat) protein [2, 9–12].
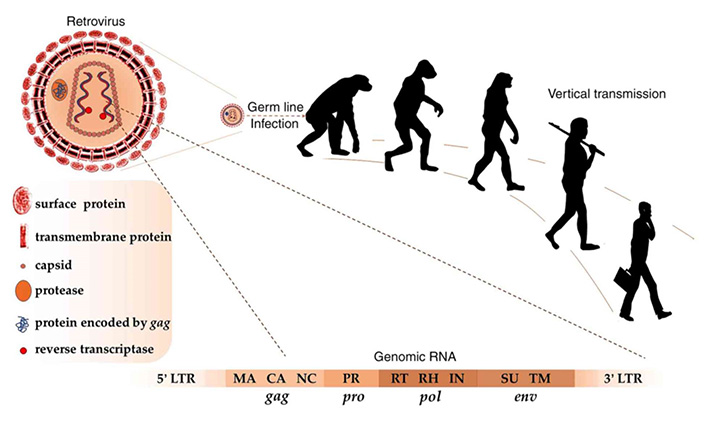
Germ line infection with retroviruses about 25 million years ago resulted in vertical transmission of HERVs in modern humans. As in retroviruses, complete HERV RNA includes the genes gag, pro, pol and env, flanked by 5’ and 3’ LTRs. Matrix (MA), capsid (CA), and nucleocapsid (NC) proteins are encoded by gag; protease (PR) is encoded by pro; reverse transcriptase (RT), Ribonuclease H (RH), and integrase (IN) are encoded by pol; surface (SU) and transmembrane (TM) proteins are encoded by env
Note. Reprinted from “Human endogenous retroviruses in cancer: expression, regulation and function,” by Gao Y, Yu XF, Chen T. Oncol Lett. 2021;21:121 (https://doi.org/10.3892/ol.2020.12382). CC BY-NC-ND.
Expression of endogenous retroelements in 32 human tissue and cell samples was investigated by transcriptome and proteogenomic analyses. All tissues investigated were shown to express retroelements at different levels. Additionally, 103 non-redundant endogenous retroelement-derived major histocompatibility complex (MHC) I-associated peptides (ereMAPs) were identified in B-lymphoblastoid cell lines (B-LCLs). Also, amino acid compositions of ereMAPS and viral MAPs were determined to have homology [13].
Antibodies produced against retroelements may cross-react with other proteins in the human body, and cause autoimmune diseases such as systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and inflammatory neurological disease. L1 retrotransposons show somatic mosaicism in many tissues, and various factors including epigenetic changes strictly control L1 activity. Increased levels of L1 expression or lowered L1 degradation might stimulate an innate immune response, and induce autoimmunity and inflammation. Different regulations of L1 expression might be due to the genetic and sex-related variations as well as environmental factors. L1-enriched nucleic acids could induce the production of type I interferon (IFN), resulting in complex series of events which cause systemic autoimmune diseases. Type I IFN is crucial for the innate defense mechanism in mammals against viruses [2, 14–16]. An autoimmune response can be induced by the retroelements in various ways such as insertional mutagenesis, sensing of retroelement RNA/cDNA, or error-prone reverse transcription of retroelement messenger RNA (mRNA), resulting in mimotopes [17].
Autoimmunity in addition to oxidative stress, fibrosis, and micro-vascular damage is observed in the systemic sclerosis (SSc). Immune responses against a viral infection are characteristics of SSc but no virus related to this disease has yet been identified. Therefore, involvement of endogenous retroelements in SSc pathogenesis cannot be excluded [18].
Alterations in the mechanisms regulating the expression or degradation of endogenous retroelements might result in various autoimmune diseases. Involvement of retroelements in the induction of autoimmunity and occurrence of related diseases was discussed on three autoimmune diseases, SLE, Aicardi–Goutières syndrome (AGS), and multiple sclerosis (MS).
SLE is a heterogenous autoimmune disease with an elusive etiology. Mucocutaneous initial symptoms and constitutional symptoms affect multiple organs such as skin, lungs, kidneys, joints, and nervous system in SLE. Genetic, hormonal, immunologic, and environmental factors are combined in the progress of this disease. Among the genomic elements, L1 retrotransposons play an important role in SLE pathogenesis. L1-expressing and virus-infected cells share the same hallmarks with specific cellular and humoral immune responses. Immunogenic ORF1p and ORF2p proteins are produced by L1 retroelements, which causes generation of type I IFN. Additionally, ORF1p and ORF2p take place in RNA-rich macromolecular structures including SLE autoantigens such as Ro60, a 60 kDa RNA-binding protein [5, 19, 20]. Expression of at least one of the L1 proteins in the organs affected in SLE was demonstrated. Additionally, IFN-β and IFN-α were detected in L1-transfected plasmacytoid dendritic cells (pDCs) or monocytes, showing the production of type I IFN [16].
Common markers of SLE include antinuclear antibody (ANA) as well as the autoantibodies targeting double-stranded DNA (dsDNA), rheumatoid factor (RF), Smith (Sm), ribosomal P, histones, ribonucleoproteins (RNPs), Ro/Sjögren’s syndrome A (SSA) and La/SSB [21]. High titers of anti-La/SSB and anti-Ro/SSA antibodies were detected in the sera of SLE patients [22]. Additionally, expression of type I IFNs and retrotransposons, such as IFN-β1 and L1, was found to be coordinated in SLE [23].
Ro60-associated RNAs were investigated in human cell lines, and it was found that Ro60 binds to an Alu-derived RNA motif. Type I IFN also affects the expression of Alu elements, and proinflammatory cytokine response is stimulated. Ro60 deficiency induces the expression of Alu RNAs and genes regulated by IFN. Expression of Alu elements was shown to be up-regulated in SLE, and related immune complexes containing anti-Ro60 also included Alu RNAs [24].
Frequently, multicomponent complexes made of nucleic acid(s) and proteins act as the autoantigens, such as spliceosomal, nucleolar, or Ro/La RNPs as well as aminoacyl-tRNA synthetase and tRNA complexes [21, 25]. RNP and chromatin particles can trigger antigen-specific immune responses because of resemblance to viral particles structurally. Therefore, SLE pathogenesis might include pseudoviral immunity driven by self nucleic acids. For instance, the mechanism that protects RNP and chromatin particles from receptor recognizing viral nucleic acids is abnormal in SLE [19].
In monogenic SLE, the most common cause is the loss-of-function in cytosolic endoplasmic reticulum (ER)-associated deoxyribonuclease III (DNase III), termed three prime repair exonuclease 1 (TREX1), which is a ubiquitous DNase for the degradation of retroelements. TREX1 is an important negative regulator of the IFN-stimulatory DNA (ISD). Digestion of single-stranded DNA (ssDNA) substrates with methylated, deaminated, or oxidized bases, as well as an abasic site at the 3’ end by TREX1 was shown. Additionally, TREX1 degrades reverse-transcribed DNA and dsDNA with methylated or deaminated bases [26–28].
Heterozygous mutations were identified in TREX1 in SLE patients [29]. TREX1-deficient mice also develop sterile inflammatory myocarditis. Anti-retroviral drugs would be expected to prevent or treat the disease; however, azidothymidine (AZT), a reverse transcription inhibitor, did not protect the TREX1-deficient mice from lethality. It was proposed that TREX1 deficiency might lead to activation of AZT-resistant reverse transcriptases, or some of the retroelements might be rescued from AZT. On the other hand, combination of the reverse transcriptase inhibitors Truvada and Viramune was shown to reduce mortality in TREX1-deficient mice [17, 28–30]. Moreover, hypomethylation of endogenous retroelements results in their activation. Therefore, the drugs such as procainamide contributing to DNA hypomethylation by blocking DNA (cytosine-5)-methyltransferase 1 (DNMT1) might be effective against autoimmune diseases [31].
Moreover, a null mutation, homozygous 2 bp-deletion c.289_290delAC (NM_004944.2) resulting in loss-of-function, in the Dnase1L3 (Dnase γ) gene causes an autosomal recessive, pediatric and rare form of SLE [32, 33]. DNA from circulating apoptotic bodies is not digested in the absence of Dnase1L3, and autoantibody responses are delayed [34].
AGS is an inflammatory encephalopathy which is heritable, and has symptoms similar to congenital brain infections of viruses, such as neurological dysfunction, skin inflammation, and psychomotor retardation. Characteristic induction of type I IFN in serum and cerebrospinal fluid in AGS is due to the alteration of sensing or metabolism of endogenous nucleic acids in response to mutations in genes encoding TREX1, RNA-specific adenosine deaminase 1 (ADAR1), sterile alpha motif (SAM) domain and histidine-aspartate (HD) domain-containing protein 1 (SAMHD1), and three subunits of the ribonuclease H2 (RNase H2). These proteins have inhibitory effect on L1 retrotransposition, and their loss-of-function because of mutations leads to accumulation of cytosolic nucleic acids. Thus, overproduction of type I IFN is triggered by ADAR1 in RNA-dependent as well as by TREX1, SAMHD1 and RNase H2 in DNA-dependent manner action [2, 7, 35].
The genes encoding these proteins in AGS have an action throughout the replication steps to inhibit the reverse transcription products. SAMHD1 degrades cellular deoxynucleoside triphosphates (dNTPs), thus depleting the nucleotide pool available for the reverse transcription. TREX1 digests DNA molecules produced after reverse transcription, and RNase H2 degrades phosphodiester backbone of RNA molecules in RNA/DNA hybrids, thus the retrotransposition is blocked [4, 7]. TREX1-deficient cells were shown to accumulate different sizes of ssDNA fragments generated during DNA replication in AGS [36].
MS is an autoimmune disease with involvement of endogenous retroelements. Analysis of the MS-associated retrovirus (MSRV) obtained from plasma of MS patients revealed a new family of HERVs, named as HERV-W, composed of gag, pol, and env genes, a primer binding site, RU5 and U3R LTRs, and a polypurine tract [37]. A total of 213 elements were discovered in HERV-W family, and grouped into three categories according to portion of LTRs and formation mechanism: 65 proviruses, 135 processed pseudogenes and 13 undefined elements [38].
Involvement of HERV-W family of retroelements in MS was shown by various studies. Serum samples obtained from 103 MS patients were analyzed by enzyme-linked immunosorbent assay (ELISA) to reveal the presence of envelope (Env) protein, and 73% of the sera were shown to contain Env antigen of HERV-W. Additionally, expression and copy number of env were found to be increased significantly in peripheral blood mononuclear cells (PBMC) of MS patients compared to healthy controls, revealed by quantitative polymerase chain reaction (qPCR). Moreover, presence of Env protein was detected via immunohistology analysis in macrophages within brain lesions of MS patients [39]. Altered expression of individual HERV-W copies in PBMC of clinically isolated syndrome (a precursor to MS) patients compared to controls was also shown by next-generation sequencing analysis [6].
Prevalence of MS is higher in women compared to men, at a ratio of 2:1 to 3:1 depending on the region. Involvement of the X chromosome in this higher ratio was investigated by mapping a 3 kb region in Xq22.3, covering the HERV-W locus, and three polymorphisms were identified as rs6622139 (T/C), rs6622140 [guanine/adenine (G/A)] and rs1290413 (G/A). Moreover, rs6622139 T/C polymorphism was found to be related to MS susceptibility and severity in women as CC structure showed lower scores than CT or TT carrying patients. Also, association between rs6622139*T genotype and higher expression of MSRV was revealed [40].
HERV-W Env interacts with Toll-like receptor 4 (TLR4) on the cells of immune system, and a pro-inflammatory and autoimmune response is activated. Therefore, blocking the interaction between Env and TLR4 using a monoclonal antibody was proposed as an effective therapeutic strategy against MS [41].
Almost half of the human genome contains retroelements started to integrate a quarter billion years ago. However, the mutations accumulated in these sequences during time inactivated majority of the retroelements. Nevertheless, the ones entered into the genome more recently preserve their function, and antibodies produced against these elements might cross-react with other proteins in the human body. This cross-reaction might induce autoimmunity and inflammation, resulting in autoimmune diseases. Mechanisms for the prevention of autoimmunity, such as degradation of nucleic acids or epigenetic silencing of the retroelement expression exist in the body. However, disruption of these mechanisms, such as increased expression of retroelements or loss-of-function in DNases may result in autoimmune disorders. Effective treatment strategies such as inhibition of reverse transcription of the retroelements might offer powerful tools against autoimmune diseases. Reverse transcriptase inhibitors are medications used in the management and treatment of HIV. However, severe adverse effects associated with their chronic use might be observed. Most retroelements are silenced by cytosine-phosphate-guanine (CpG) methylation. Therefore, many different factors affecting the methylation processes could be used for therapeutic purposes in autoimmune diseases.
AGS: Aicardi–Goutières syndrome
AZT: azidothymidine
C: cytosine
cDNA: complementary DNA
DNase III: deoxyribonuclease III
Env: envelope
HERVs: human endogenous retroviruses
IFN: interferon
L1: long interspersed element 1
LINEs: long interspersed elements
LTR: long terminal repeat
MS: multiple sclerosis
ORF: open reading frame
RNase H2: ribonuclease H2
RNPs: ribonucleoproteins
SAMHD1: sterile alpha motif domain and histidine-aspartate domain-containing protein 1
SINEs: short interspersed elements
SLE: systemic lupus erythematosus
SSc: systemic sclerosis
T: thymine
TREX1: three prime repair exonuclease 1
tRNA: transfer RNA
The author contributed solely to the work.
The author declares that he has no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2022.
Copyright: © The Author(s) 2022. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 4180
Download: 53
Times Cited: 0
