Affiliation:
1Institute of Biochemistry, Food Science and Nutrition, The Hebrew University of Jerusalem, Rehovot 7610001, Israel
2Department of Bioengineering and Biotechnology, BIT Mesra, Ranchi 835215, India
Affiliation:
3French Associates Institute for Agriculture and Biotechnology of Drylands, Ben-Gurion University of the Negev, Sede-Boqer 849900, Israel
ORCID: https://orcid.org/0000-0002-8166-3572
Affiliation:
1Institute of Biochemistry, Food Science and Nutrition, The Hebrew University of Jerusalem, Rehovot 7610001, Israel
Affiliation:
1Institute of Biochemistry, Food Science and Nutrition, The Hebrew University of Jerusalem, Rehovot 7610001, Israel
Affiliation:
1Institute of Biochemistry, Food Science and Nutrition, The Hebrew University of Jerusalem, Rehovot 7610001, Israel
Affiliation:
1Institute of Biochemistry, Food Science and Nutrition, The Hebrew University of Jerusalem, Rehovot 7610001, Israel
Email: oren.tirosh@mail.huji.ac.il
ORCID: https://orcid.org/0000-0001-5407-0440
Affiliation:
1Institute of Biochemistry, Food Science and Nutrition, The Hebrew University of Jerusalem, Rehovot 7610001, Israel
Email: zvi.hayouka@mail.huji.ac.il
ORCID: https://orcid.org/0000-0003-3582-4029
Explor Dig Dis. 2022;1:118–136 DOI: https://doi.org/10.37349/edd.2022.00009
Received: May 04, 2022 Accepted: July 29, 2022 Published: October 31, 2022
Academic Editor: Natalia Nieto, University of Illinois at Chicago, USA; Jose C. Fernandez-Checa, Institute of Biomedical Research of Barcelona (IIBB), CSIC, Spain
Aim: Probiotic bacteria consumption for improving human health and for disease prevention is still controversial. There is a need to develop functional probiotic bacteria with proven efficacy for the human gastrointestinal (GI) system. The novel bacteria will lower the steady state of constant Ethanol production may lead to gut microbiota dysbiosis and liver injuries.
Methods: Herein engineered probiotic bacterium B. subtilis to enhance the secretion of human alcohol dehydrogenase-4 (ADH4) by fusion of signal peptides (SPs) was constructed. As a result, higher ADH4 secretion and Ethanol removal rates were observed in phoB SP transformant SP-64, compared to other transformants. The engineered ADH4 expressing probiotic B. subtilis was delivered as spores to evaluate various physiological, biochemical, and immuno-histochemical parameters of mice under a high-fat diet (HFD)-induced obesity and metabolic impairment.
Results: The treatment ameliorated significantly weight gain, improved glucose utilization, and prevented HFD-induced pancreatic damage. Lastly, SP-64 inoculation altered the gut microbiota, and increased the Firmicutes/Bacteroides ratio, supporting better fitness under HFD.
Conclusions: SP-64 emerged as a potential probiotic that opens a new avenue for interventions against over-nutrition-induced metabolic disorders.
A high-fat diet (HFD) severely affects cardiovascular health and is responsible for several disorders like fatty liver disease, coronary artery disease, hyperglycaemia, type-2 diabetes, etc. [1]. It also suppresses the insulin signaling pathway, promoting obesity and non-alcoholic fatty liver disease (NAFLD). NAFLD is the leading cause of chronic liver disease worldwide and represents a major socio-economic burden. Interestingly, continuous Ethanol production in the human gastrointestinal (GI) system severely affects gut microbiota and the liver. Nevertheless, continuous production of Ethanol and deleterious metabolites might lead to microbial dysbiosis and organ dysfunction [2]. However, under these circumstances, probiotic bacterial intake has been shown as a promising approach for preventing diet-induced metabolic disorders [3]. Ethanol metabolism involves its conversion to acetaldehyde, mediated by the metabolic enzymes alcohol dehydrogenase (ADH) family [4]. Besides Ethanol, ADH4 metabolizes other substrates, including retinol, hydroxysteroids, and lipid peroxidation products [5]. Therefore, enhancing ADH4 activity might improve the metabolism of alcohol in the gut and, thereby, decrease the exposure of internal organs to Ethanol.
The mammalian intestinal system is occupied by diverse microbiota, with a population of 1010–1014 that imparts several immunologic and protective functions [6]. Many diseases like inflammatory bowel disease, diabetes, obesity, heart disease, and certain types of cancers are associated with intestinal microbiota disorders [7–9]. Alteration of gut microbiota represents the sign of metabolic dysfunctions. However, these processes are not yet fully understood [10]. Nevertheless, reprogramming the gut microbiota composition make it a therapeutic intervention against metabolic and immune impairments. HFD is associated with significant gut microbiota changes with an increase in Enterobacteriaceae and bacteria populations that are positively correlated with high-density lipoproteins [11]. A recent study showed that gut microbiota dysbiosis promoted cholesterol excretion in feces, therefore clearly illustrating the role of gut microbiota in the pathogenesis of hypercholesterolemia. Enhancing the intestinal flora diversity or restoring the beneficial bacteria population like Lactobacillus spp. by probiotic supplementation ameliorated HFD-associated disorders [12, 13]. Probiotic L. rhamnosus supplementation improved lipid metabolism by enhancing Firmicutes and decreasing the Bacteroidetes population in HFD treatment [13]. The delivery of probiotic bacteria and its accumulation at the intestinal site modulate the bacterial composition and exert a beneficial effect in preventing disease progression [14, 15]. Previous studies have shown the effectiveness of engineered probiotic microorganisms in delivering bio-molecules to encounter bacterial/viral infections [16, 17] and to enhance pharmacological therapies [18]. However, most of the studies have been done in vitro, and their therapeutic evaluation using in vivo models is still lacking [19, 20]. Many of the in vivo limitations are due to their limited treatment efficacy, caused by low bioavailability and insufficient retention of probiotics in the GI tract [21]. The development of engineered bacteria using genetic and chemical approaches is an alternative to encounter many limitations [22]. Genetically engineered bacteria with higher stress tolerance have been produced to mediate the delivery of vaccines and drugs or kill the pathogens [23, 24]. To prevent probiotic bacteria from GI-tract acidity, encapsulation using synthetic materials was examined. However, many of these approaches were ineffective because of strong gastric acidic conditions, bile salts, and digestive enzymes, which cause the death of administrated bacteria. Another drawback of these coatings is that they prevent the direct contact of administrated bacteria to the intestinal mucosa, minimizing their retention and adversely affecting functionality [25]. Considering these major limitations, new strategies are urgently needed for the development of oral bacterial therapeutics.
B. subtilis is a non-pathogenic gram-positive bacterium widely known for expressing various foreign enzymes, therapeutic proteins, and metabolites with well-established functions [26]. Additionally, it is a potential substitute for antibiotics and is generally regarded as safe (GRAS) by the Food and Drug Administration (FDA) for recombinant protein production. Furthermore, ease of genetic modifications, high transformation efficiency, low nutritional requirement, and ability to produce high amounts of recombinant proteins lead to further strengthening of the host strain as per requirements. Another advantage of this bacterium is to secrete proteins directly into the used culture medium, at appropriate concentrations [27, 28]. Efforts to enhance the heterologous protein secretion in B. subtilis include the attachment of signal peptides (SPs) [29, 30], alteration of the ribosome binding site [31, 32], promoter engineering [33–36], etc. SPs play a key role in maintaining the proper transportable folded state of the precursor protein, leading it to the extracellular environment. SP-library construction and high-through screening have been reported in several secreted recombinant proteins [37, 38]. Previously, Zhang et al. [39] screened 114 Sec SP transformants for secretion of xylanase from B. pumilus using B. subtilis as an expression host.
The spore-forming member of the genus Bacillus can survive under the acidic pH of the stomach and, therefore, reach the intestine to exert its beneficial probiotic effects. Recently, probiotic Bacillus species have been used to treat metabolic disorders, including maintenance of intestinal homeostasis, exclusion of harmful bacteria, and immune modulation [40, 41].
In the present study, we aimed to enhance the secretion of ADH4 in B. subtilis driven by SPs. Our results indicated that recombinant B. subtilis with an SP was effective for in vitro Ethanol removal and effectively minimized HFD-induced metabolic disorders in an animal model. To the best of our knowledge, this is the first report showing that ADH-secreting probiotic bacteria-maintained glucose levels while executing diagnostic and therapeutical activities to alleviate diet-induced insulin resistance. Therefore, herein, we provide a potential therapeutic approach for preventing or alleviating high-fat disorders and gut dysbiosis.
B. subtilis RIK 1285 genotype and E. coli DH-5α were grown at 37°C and 200 rpm in Luria-Bertani (LB) broth (HiMedia, Israel) medium. Competent cells of E. coli DH-5α were prepared as per standard protocol. Competent cells of B. subtilis RIK 1285 were prepared as per the Takara manual (Takara Biosciences, Israel). The human ADH4 sequence was obtained from the addgene database (https://www.addgene.org/vector-database) and optimized for codon biases at GenScript. The synthesized gene sequence and primers used for cloning and sequencing are provided in Tables S1 and S2. Plasmid pBE-S (5.9 kb) and SP DNA mixture were purchased from Takara Bio Inc. Antibiotic ampicillin and kanamycin were supplied by Sigma-Aldrich (Sigma, Israel).
A 1.2 kb polymerase chain reaction (PCR) fragment of ADH4 was cloned into a pBE-S plasmid. The resulted pBE-S:ADH4 plasmid was transformed to competent B. subtilis RIK 1285 by electroporation (1.5 kv, 100 Ω, 25 μF, time constant 1.5 ms). The selected transformant RIK 1285:ADH4 (F1) was tested for its in vitro Ethanol (0.5%) removal efficiency. A short fragment of ADH4 gene of 710 bp was also cloned to use as a control and marked as RIK 1285:ADH4 (S1). Transformants were confirmed by colony PCR and further by sequencing at The Hebrew University of Jerusalem Campus [Applied Biosystems (ABI), 3730xl DNA Analyzer]. Detail about the PCR conditions and transformation procedure has been provided in the supplementary method. Changes in bacterial population were calculated by colony forming units (CFU) counting using a serial dilution method.
Wild-type RIK 1285 and ADH4 transformants were grown in 30 mL of LB medium in the presence of appropriate antibiotics. After overnight incubation at 37°C, the culture was centrifuged at 12,000 rpm for 15 min at 4°C, and proteins were extracted as described in the supplementary section, page 1. The ADH4 expression was confirmed by western blot using polyclonal antibodies. To construct the SP-library, the plasmid RIK 1285:ADH4 (F1) was randomly ligated with the SP-DNA mixture. The plasmid library was transformed into B. subtilis RIK 1285 by electroporation and plated on LB-agar medium supplemented with kanamycin (10 μg/mL). Approximately 100 clones were screened for the in vitro Ethanol removal assay and the selected efficient colony was tested for ADH activity.
A minimal basal medium (MM) containing 64 g/L Na2HPO4.7H2O, 15 g/L KH2PO4, 5 g/L NH4Cl, and 2.5 g/L NaCl with 2 mmol/L MgSO4 and 0.1 mmol/L CaCl2 was autoclaved (121°C, 15 min), cooled down at room temperature, and supplemented with 0.5% Ethanol. Approximately 1 × 107 bacterial cells were mixed in the Ethanol-supplemented minimal medium and incubated for 24 h at 37°C with shaking of 200 rpm. After the incubation period, each treatment medium was filtered with a 0.22 μm Millipore filter and analyzed on a gas chromatography (GC)-FID to measure the Ethanol concentration. A standard curve (R2 value close to 0.999) covering 0.1% to 0.5% of 95% Ethanol was used to calculate Ethanol concentrations. Detail about the condition has been provided in the supplementary method.
Overnight grown cultures (20 μL) of B. subtilis RIK 1285, RIK 1285:ADH4 (F1), and RIK 1285:ADH4 (SP-64) were inoculated into a 20 mL sterile nutrient broth medium supplemented with the appropriate antibiotics. Cultures were grown for 24 h, and the absorbance of the culture in each treatment was determined at 600 nm in an ultraviolet-visible (UV-Vis) spectrophotometer in triplicate sets using un-inoculated broth as a blank.
ADH activity was assessed in the culture supernatant following the standard protocol of nicotinamide adenine dinucleotide (NAD)+ reduction. Briefly, the supernatant (100 μL) B. subtilis RIK 1285, RIK 1285:ADH4 (F1), and RIK 1285:ADH4 (SP-64) were added to the phosphate buffer (pH 8.0), which contained NAD+ at a final concentration of 10 mmol/L. The reaction was started by adding Ethanol (0.016 mol/L), and the rate of conversion of NADH was monitored at 340 nm. The ADH activity was calculated by using the molar extinction coefficient of NADH (6.22). Protein concentration was determined by the Bradford method.
To test the stress tolerance, vegetative and spores (1 × 109) of B. subtilis RIK 1285, RIK 1285:ADH4 (F1), and RIK 1285:ADH4 (SP-64) were boiled at 80°C for 20 min. Similarly, acid tolerance was tested by growing the culture in simulated gastric fluid (SGF) of acidic pH 2.0 for 1 h. The bacterial colony was counted by serial dilution plating method. Additionally, to test the colonization ability, spores of RIK 1285, RIK 1285:ADH4 (F1), and RIK 1285:ADH4 (SP-64) were injected into 4 weeks C57BL/6J male mice (n = 3), fed on a normal diet (ND) by the oral gavage. Feces were homogenized in phosphate-buffered saline (PBS) buffer and serial dilution was cultured on an LB-agar plate with appropriate antibiotics and incubated at 37°C for 16 h. The level of colonization was calculated and expressed as the number of CFU per g of feces.
Animal experiments were done following relevant guidelines and regulations approved by the Institutional Animal Care Ethics Committee of the Hebrew University of Jerusalem. B. subtilis wild-type RIK 1285, transformants RIK 1285:ADH4 (F1), and RIK 1285:ADH4 (SP-64) were grown in Difco sporulation medium (DSM) for 48 h at 37°C and 200 rpm. Bacteria were pelleted at 10,000 rpm for 10 min at 4°C and washed in cold-sterile water followed by heat shock at 70°C for 30 min to kill the vegetative cells. Spores were washed three times with ice-cold water and stored at 4°C until being administrated to mice. Spores were counted by serial plate dilution method. On the day of administration, spores were injected into mice (C57 BL/6J) by oral gavage (1 × 109 in 200 μL PBS buffer) every third day for up to 10 weeks.
The 4–5 weeks-old male C57BL/6J mice were purchased from Harlan Laboratories (Jerusalem, Israel). C57 BL/6 J is one of the most common commercially available mouse strains widely used to assess the various diet effects and liver injury. Mice (n = 30) were randomly assigned to two groups, fed with an ND and an HFD in the presence and absence of bacterial spores (HFD is not a liver injury-related model in mice. It is more of a model of simple steatosis and metabolic impairment). Finally, a total of five groups [ND, HFD, HFD + RIK 1285, HFD + RIK 1285:ADH4 (F1), HFD + RIK 1285:ADH4 (SP-64)] were assigned. Mice were housed in sterile cages in a pathogen-free animal house at a temperature of 22°C ± 2°C and relative humidity of 50% ± 10%, under 12 h light/dark cycles. The ND and HFD composition is summarized in Table S3.
After 10 weeks on the diet, mice were fasted for 12 h, and sacrificed in random order by isoflurane overdose. Before the sacrifice, each mouse’s body weight in tested groups of ND and HFD was recorded. The collected epididymal adipose tissue and liver tissue were immediately stored at –80°C. Similarly, intestinal tissues were collected and frozen for further experimental purposes. The feces from each group (ND & HFD) were stored for microbiota analysis. Blood samples from each mouse were collected and centrifuged at 8,000 g for 10 min, at 4°C, to separate the plasma from the remaining components. The serum was used for aspartate aminotransferase (AST) and alanine aminotransferase (ALT) enzymatic assays. Additionally, total cholesterol (TC), triglycerides (TGs), and bilirubin content were recorded according to the standard protocol. Finally, the glucose content in blood was checked from the tail vein using a glucometer.
Formalin-fixed pancreases from each treatment group were cut into small (4 μm) sections and processed for immunohistochemistry. Slides were de-paraffinized in xylene and sequentially rehydrated using decreasing Ethanol concentrations down to water. Antigen retrieval required microwaving slides with 10 mmol/L citrate buffer for 30 min. Nonspecific binding was blocked by treating sections with Biocare blocking reagent (Biocare Medical, Concord, CA, USA) for 30 min at room temperature, followed by incubation with the primary antibody diluted in blocking buffer. The following primary antibodies and dilutions were used: phospho p-S6 protein (Cell Signalling Technology Inc., USA, 1:300); insulin (Dako Products, USA, 1:300); glucagon (Santa Cruz Biotechnology, USA, 1:300). The 100 uL of primary antibodies mix was loaded onto the slide, closed with parafilm, and kept overnight at 4°C. Slides were washed twice in PBS, incubated for 30 min with a secondary antibody, washed twice with PBS, stained with diaminobenzidine, and counterstained with hematoxylin. Images were captured by Nikon A1R confocal microscope and analyzed using National Institutes of Health (NIH) ImageJ software (v1.31, http://rsb.info.nih.gov/ij/index.html).
To assess the colonization, feces of each treatment were suspended to a concentration of 1 g/mL in PBS buffer and gently homogenized with a vortex for 1 min. The collected fecal suspension was heated at 80°C for 20 min to kill vegetative cells. Serial dilutions were plated on a solid LB-agar plate with the appropriate antibiotics to count the viable spore population. Plates were incubated at 37°C overnight and spores count was represented as CFU per g of feces. To check the inoculated bacterial presence in the collected feces, another set of feces from each treatment was homogenized, and the plasmid was extracted following the standard protocol (Qiagen, Germany). Finally, 50 ng of extracted plasmid was used for ADH4 amplification with a suitable primer set to confirm the presence of injected bacteria.
Fecal samples from each treatment group were collected in sterile tubes and immediately stored at –80°C. A fast DNA stool kit (Qiagen, Germany) was used to isolate DNA following the manufacturer’s protocol. DNA concentration and purity were determined with a NanoDrop™ 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, USA). Five ng per reaction was used to generate small subunit (SSU) ribosomal RNA (rRNA) amplicons of V4 variable regions using the bacterial primer set CS1_515F (5’-ACACTGACGACATGGTTCTACA_GTGCCAGCMGCCGCGGTAA) and CS2_806R (5’-TACGGTAGCAGAGACTTGGTCT_GGACTACHVGGGTWTCTAAT) [42–44]. The protocol was set as follows: denaturation at 95°C for 3 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s, and final extension at 72°C for 5 min. Library sequencing was done from both ends on an Illumina platform at the Research Resources Centre of the University of Illinois at Chicago following the standard protocols [45] (http://www.earthmicrobiome.org/emp-standard-protocols/16s/), and fastq files were generated using the illumine software package bcl2fastq.
Prior to analysis, forward and reverse primer sequences were removed from the raw fastq files with Cutadapt v.3.265 [46]. Primer-trimmed sequences were quality-filtered and merged into the error-corrected amplicon sequence variants (ASVs) using DADA2 v.1.18 [47]. The ASVs were length-filtered to keep sequences at a 251–255 bp length, to meet the expected size for the V4 variable region of the SSU rRNA gene. Chimera sequences were removed using the removeBimeraDenovo function of the DADA2 package. These high-quality sequences were annotated to the SILVA SSU rRNA reference alignment (release138) using the Ribosomal Database Project (RDP) classifier [48] as implemented in Mothur v.1.44.2 software [49] with a minimum confidence threshold of 50%. ASVs that were classified as ‘chloroplast’ and ‘mitochondria’ or did not match any taxonomic phyla level were removed from the final sequence dataset. The remaining 1,247,299 (38,978 sequences per sample ± 12,927 sequences per sample) high-quality sequences were used for downstream analysis. Alpha diversity indexes were calculated based on the number of unique ASVs (richness), Shannon’s, and Faith’s phylogenetic diversity (PD). Estimates of alpha diversity indexes were obtained, based on 9,466 randomly selected sequences per sample, and tested for significant statistical differences using a two-sided Wilcoxon test. Prior to beta-diversity analysis, the final dataset was normalized by cumulative sum scaling (CSS) [50]. An unconstrained principal coordinates analysis (PCoA) based on Bray-Curtis, weighted, and unweighted UniFrac dissimilarities were performed to quantify the major variance components of beta-diversity. The diet effect on the microbial community dissimilarity was estimated with a permutational analysis of variance (PERMANOVA) with 1,000 permutations. Statistical analysis was conducted in R v.4.0.4 using the phyloseq v1.34 and vegan v2.5-7 R packages [51, 52].
All statistically significant differences between each treatment were evaluated using variance analysis [one-way analysis of variance (ANOVA)] followed by the t-test using the significant level of P < 0.05 for all analyses. The analysis was performed with the JMP 7.0.2 and JMP Pro software suites (SAS Institute, USA).
To develop the probiotic strain featuring enhanced ADH secretion, a full-length ADH4 gene (1.2 kb) and a short fragment of 710 bp (as a control) were amplified (Figure 1A) and cloned into a pBE-S vector under a constitutive promoter. The constructed plasmid pBE-S:ADH4 was transformed into B. subtilis by electroporation and positive transformants were selected on LB-agar plates supplemented with kanamycin (10 μg/mL). The obtained transformants were screened by colony PCR and further confirmed by sequencing (Figure S1). Additionally, the selected transformant RIK 1285:ADH4 (F1) was evaluated for its in vitro Ethanol removal ability, and it showed significant (P < 0.05) Ethanol removal (60%) when compared to the wild-type strain (Figure 1B). Furthermore, CFU counting showed an increase in bacterial population, following incubation in LB-medium supplemented with Ethanol (0.5%, Figure 1C). The crude lysates of the overnight-grown wild-type culture, selected transformant RIK 1285:ADH4 (F1), and RIK 1285:ADH4 (S1) were examined for protein expression by western blotting. We observed a monomeric 21 kDa protein expression in full-length transformant RIK 1285:ADH4 (F1) (Figure 1D).
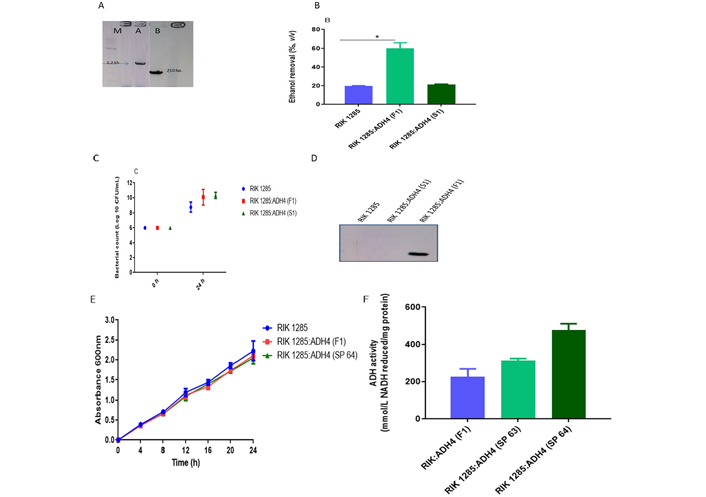
Characterization of higher ADH4 secreting engineered probiotic transformant. A. ADH4 gene amplification M; DNA ladder mix (SM0331), lanes A and B are full length (1.2 kb) and small length (710 bp) amplicon, respectively; B. to check the Ethanol removal ability, minimal media were supplemented with 0.5% Ethanol and it was inoculated with 1 × 107 CFU of wild-type RIK 1285 and engineered RIK 1285:ADH4 (F1) bacterial strain; GC-FID showed higher Ethanol removal in engineered RIK 1285:ADH4 (F1), values are mean ± standard deviation (SD), n = 3; column marked with an asterisk * represents a significant difference from control according to student’s t-test (P < 0.05); C. CFU count showed an increase in the population of tested bacterial strains (values are mean ± SD, n = 3); D. protein expression verification by western blot analysis, lane A: RIK 1285, lane B: small length cloned amplicon, lane C: full-length RIK 1285:ADH4 (F1); E. growth curve of transformants pBE-S:RIK 1285:ADH4 (SP-64) and pBE-S:RIK 1285:ADH4 (F1) the increase in turbidity was recorded at 600 nm (values are mean ± SD, n = 3); F. ADH activity in the supernatants of selected transformants. The activity was calculated with an increase in NADH absorbance at 340 nm generated after the reduction of NAD+ (values are mean ± SD, n = 3)
An SP library driven by aprE promoter was generated and further transformed into B. subtilis, yielding the recombinant strains from B. subtilis RIK 1285:ADH4 (SP-1) to RIK 1285:ADH4 (SP-100). The resulting transformants were tested for their in vitro Ethanol removal capability. The incorporation of SPs significantly enhanced the ability of the transformants to reduce Ethanol concentration. While some of the SP transformants like SP-1, SP-7, SP-13, SP-15, SP-27, SP-31, SP-33, SP-37, SP-55, SP-72, SP-74, etc. showed 35% to 40% of Ethanol removal, the SP-43, SP-46, SP-50, SP-67, and SP-70 transformants were able to consume up to 60% of added Ethanol. Finally, transformants SP-63 and SP-64 showed the highest Ethanol removal in the range of 70% to 75%, among all transformants (Figure S2). Based on these observations, the B. subtilis transformant SP-64 was selected for downstream analysis.
Growth comparison studies of selected transformant RIK 1285:SP-64 with wild-type RIK 1285 showed no significant plasmid effect on the host growth rate under the evaluated conditions (Figure 1E). The NADH reduction assay was used to measure ADH activity, having found the highest ADH enzymatic activity in SP-64 transformants (470.68 U/mg protein, Figure 1F). Time-dependent ADH4 secretion showed the ADH4 secretion in the culture supernatant by the SP-64 (Figure S3). SP sequence analysis identified the phoB SP in the selected transformant SP-64.
As B. subtilis is the spore-forming bacteria, heat and acid resistance of RIK 1285:ADH4 (F1), and RIK 1285:ADH4 (SP-64) were evaluated (Figure S4). After confirming the viability of these bacterial strains in simulated gastric conditions, we did an animal trial study to test the colonization of engineered bacterial strains in mouse model, and also the dosing interval when supplementing as spores. This was done in order to evaluate the effectiveness of the GI tract colonization following gavage supplementation. Administrated bacterial spores in feces were detected after 1 day to 3 days with a gradual decrease (Figure S5).
Based on higher ADH activity and in vitro Ethanol removal efficacy, we selected ADH4-secreting SP-64 strain for in vivo study. After a 10-week diet protocol, mice in the HFD group were shown to gain more weight than that in ND groups. Mice inoculated with RIK 1285:ADH4 (SP-64) resulted in the lowest (P < 0.05) body and liver weight gain among HFD groups (Figure 2A, 2B). The changes in the body weight throughout the 10 weeks have been provided in Figure S6. Additionally, the adipose tissue mass in the ND group was substantially lower than that in the HFD-treated group (Figure 2C). Moreover, the HFD group treated with SP-64 spores showed significantly (P < 0.05) reduced adipose tissue and caecum weight gain when compared to the other HFD treatments (Figure 2D). Finally, the fasting glucose levels of SP-64-treated mice were similar to those in the ND group and significantly lower among the tested HFD groups (Figure 2E).
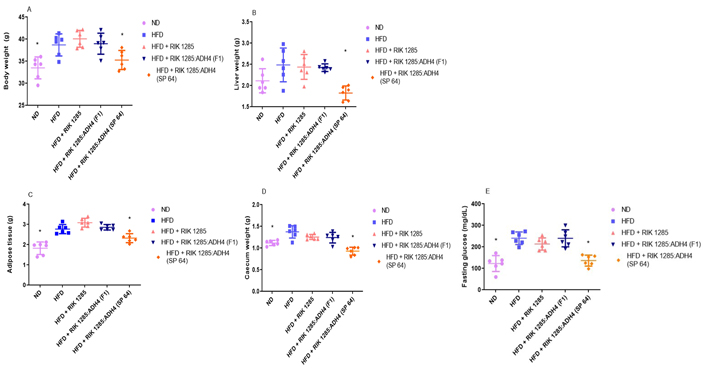
Oral administration of engineered probiotic SP-64 minimized the high-fat disorders parameters. A. Mice orally administrated with SP-64 spores showed less weight gain compared to the control; B. effect of SP-64 on liver weight; C. effect of SP-64 on total adipose tissue weight; D. effect of SP-64 on caecum weight; E. effect of SP-64 on fasting glucose level. All values are expressed as mean ± standard error of the mean (SEM, n = 6 in each treatment). *: P < 0.05
The analysis revealed a significant (P < 0.05) decrease in the cholesterol level, liver damage markers, and ALT and AST enzymatic activities in the SP-64-treated group. On the other hand, high ALT and AST levels were found in the untreated HFD group (Figure S7A, S7B). Nevertheless, all bacterial treatments [i.e. RIK 1285, RIK 1285:ADH4 (F1), and RIK 1285:ADH4 (SP-64)] were able to lower ALT and AST levels. Additionally, all bacterial treated groups showed a lower level of bilirubin, cholesterol, and TG levels in the HFD group (Figure S7C–E).
After we observed a significant reduction in the glucose blood level in the ADH4-secreting bacteria-treated mice, the pancreas beta cells integrity was analyzed and compared between the treatments. Pancreas immunohistochemistry staining showed that HFD induced higher phospho p-S6 level. S6 is an effector of mammalian target of rapamycin (mTOR) that impaired insulin signals indicating insulin resistance (Figure 3A). In the HFD-treated mouse, insulin-containing beta cells density was lower within the beta cell islet. HFD-treated mouse with ADH4-secreting probiotic bacteria-maintained islet high density of insulin expressing cells and lower level of phosphor p-S6 staining intensity even below ND treated mouse control level (Figure 3B).
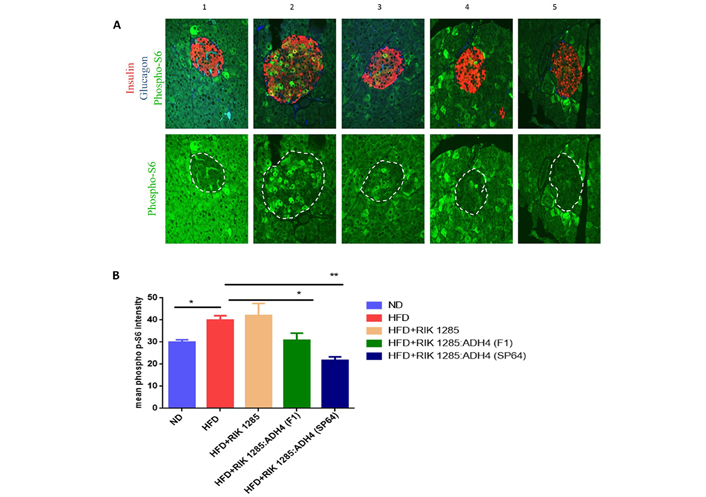
Analysis of pancreatic dysfunction. Immunohistochemistry of pancreas. A. Representative immunostaining of pancreatic cells. Phospho p-S6 (green), insulin (labeling β cells, red) and glucagon (labeling α cells, blue) in pancreatic sections of islets of mice fed with ND (group 1), HFD (group 2), HFD+RIK 1285 (group 3), HFD+RIK 1285:ADH4 (F1) (group 4), HFD+RIK 1285:ADH4 (SP-64) (group 5). All treatments were analyzed in triplicates, figure resolution 96 pixels per inch, n = 3; B. mean fluorescent intensity (MFI) of p-S6 staining in β-cell islets of mice from each group. Columns in each group represent mean ± SEM (n = 3). Unpaired student’s t-test was used to assign P values, *: P < 0.05; **: P < 0.01. Raw data are in the supplementary material
B. subtilis administrated spores’ population decreased following oral administration, and it was lowest on the 3rd day. CFU counting showed that the spores’ population decreased over time (Figure 4A). To further support the presence of the probiotic bacteria, PCR with the ADH4 gene showed the expected amplification in RIK 1285:ADH4 (F1) and RIK 1285:ADH4 (SP-64) spores-inoculated group, showing its presence in feces (Figure 4B). There was no amplification in inoculated RIK 1285 spore from the ND and HFD treated groups.
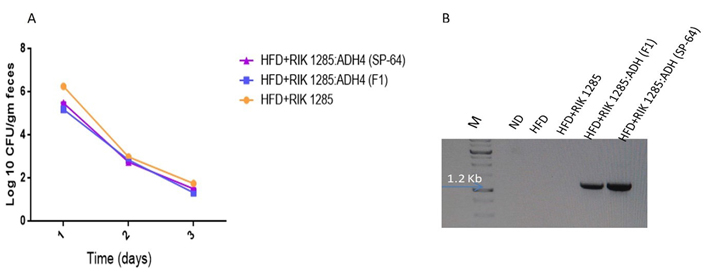
Test of colonization of administrated bacteria. A. Bacterial colonization was assessed by serial dilution of fecal homogenate. Results are average from three independent experiments (values are mean ± SD, n = 3); B. plasmid was isolated from the fecal homogenate from each treatment followed by PCR with ADH4 gene primers to monitor the presence of the inoculated bacteria. The 1.2 kb amplification which ensures the presence of our administrated bacteria in the feces was observed in RIK 1285:ADH4 (F1) and RIK 1285:ADH4 (SP-64) treated group
Gut microbiota imparts various roles in homeostasis, metabolic abnormalities, and inflammation. Therefore, we evaluated the microbiota changes following oral administration of tested bacterial spores at different time points, using SSU rRNA genes high-throughput sequencing techniques. High-throughput sequencing of SSU rRNA genes yielded a total of 1,247,299 high-quality sequences (range 9,466–67,795; median 34,936 sequences per sample), revealing profound changes in microbial community diversity and composition associated with mouse diets and bacterial treatments. Rarefaction curve-based alpha diversity analysis also indicates that mouse age and diet significantly affected microbial community species richness (Figure 5A). Overall, more diverse prokaryotic communities were observed at the initial stages when compared to later stages (Figure 5A). A variation in gut-microbiome diversity was observed, featuring a decrease in PD in the first week, followed by progress toward a phylogenetic dispersion at 6 weeks, under both ND and HFD-treated mice (Figure 5B). The microbial community dispersion (weighted UniFrac distances) was higher after 6 weeks when compared to week 1 and week 10 (Figure 5C). Fecal microbiota of the HFD groups clustered differently from ND, illustrating a dominant effect of diet. To determine the influence of diet and probiotic treatment on microbial communities, unconstrained PCoA ordination and PERMANOVA analyses based on the weighted UniFrac-distance matrices were performed. These analyses underlined the important role of mouse physiological age on the distribution of microbial communities. Accordingly, mouse age explained almost 50% of the variation in the prokaryotic community composition (R2 = 0.5, P < 0.0001, Figure 5C).
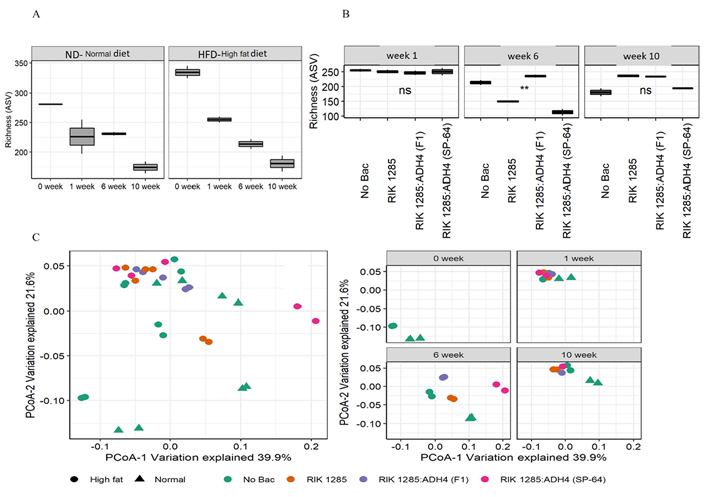
Administration of probiotic bacteria on mice microbiome. A. Alpha diversity indices were calculated based on randomly selected 9,466 sequences per sample for. The ‘Normal’ and ‘High fat’ indicate ND and HFD, respectively; B. diversity was further assessed following bacterial treatments under HFD. The asterisks ** indicate statistically significant differences between observations based on a Wilcoxon signed-rank test; C. characterization of the similarity between microbial communities across assemblages. Community similarity is visualized using principal component analysis of 32 prokaryotic communities based on SSU rRNA gene amplicon sequencing. The abundances of 1,247,299 high-quality sequences were normalized by CSS, and beta diversity indices were estimated based on weighted UniFrac distances. While the left panel summarizes the microbial community relationship of all samples of this study, the right panel shows the microbial community relationship based on the sample collection point. A PERMANOVA test on weighted UniFrac distance metrics with 1,000 permutations analyzed significant differences in beta diversity. *: P < 0.05; **: P < 0.01; ns: no significant differences
The vast majority of retrieved bacterial SSU rRNA gene sequences were affiliated with the phyla Proteobacteria, Bacteroidota, Actinobacteria, and Firmicutes, which comprised up to 100% of bacterial sequences (Figure 6A). At the phylum level, Firmicutes were enriched at week 1 and week 10 (75% on average) under HFD, whereas at 6 weeks Bacteroidota and Proteobacteria were dominant. Interestingly, SP-64 inoculation significantly increased Firmicutes and decreased Bacteroidota compared to other treatments. Under ND, they maintained a constant presence. Phylum Actinobacteriota and Verrucomicrobiota were not observed among bacterial-treated groups under HFD. Bacteria treatments significantly reduced the Bacteroidota family.
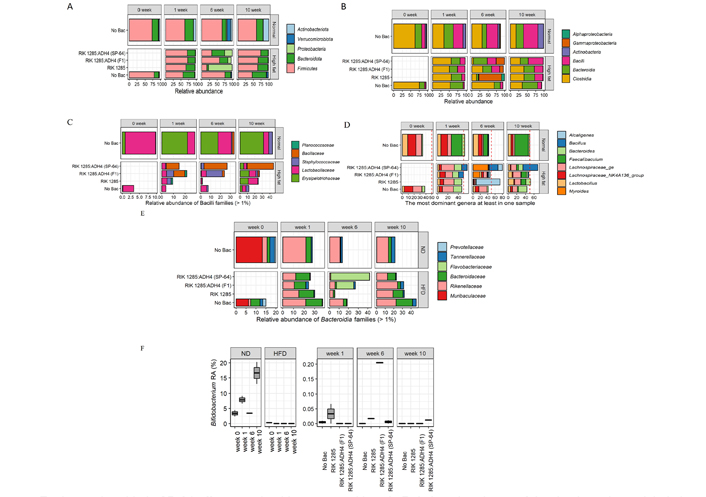
Engineered probiotic SP-64 effect on microbiota composition. A. Relative abundance of the dominant bacterial phyla. The distribution of 1,247,299 high-quality SSU rRNA gene amplicons from 32 prokaryotic communities were assigned to SILVA v138 data set; B. a substantial increase in proinflammatory taxa (α- and γ-Proteobacteria) observed after 6 weeks of an HFD; C. an HFD negatively affected the relative abundance of Bacilli families and stimulated propagation of proinflammatory members of the Staphylococcaceae family. B. subtilis treatments partially restored the overall quantities of Bacilli families; D. abundance of most dominant bacterial genera presents at least in one sample; E. HFD positively affected the relative abundance of Bacterioidia families. B. subtilis treatments partially restored the overall quantities of Bacterioidia families; F. Bifidobacterium population at different time points under ND and HFD
An HFD substantially affects the microbial taxonomic composition. A significant taxonomic shift was observed after 6 weeks with an increase in proinflammatory taxa of γ-Proteobacteria, Bacilli, and Bacteroidia (Figure 6B). Bacilli population was found to be higher after 10 weeks, among the bacteria-treated groups under HFD. HFD negatively affected the relative abundance of Bacilli families and stimulated the propagation of proinflammatory members of the Staphylococcaceae family. Nonetheless, B. subtilis treatments partially restored the overall quantities of Bacilli families (Figure 6C). While the Staphylococcaceae family increases their relative abundance, Erysipelotrichaceae was negatively affected by an HFD. These opposite responses of the Bacilli families resulted in an overall increase in the relative abundance of Bacilli due to prolonged exposure to an HFD (Figure 6C). However, the highest population of Bacillaceae was recorded in the SP-64 treated group.
A total of eight genera were observed as the most prevalent in the microbial community. The cumulative abundance of these taxa gradually increased with time. The quantity of these genera in the spore-unamended treatment increased from 34.6% ± 1.1% to 50% ± 3.9% under an ND and from 38.1% ± 1.3% to 43% ± 0.2% under an HFD (Figure 6D). A similar taxonomic succession was observed in the spores-amended treatments. Nevertheless, RIK 1285:ADH4 (F1) was associated with the highest increase in cumulative abundance from 38.1% ± 1.3% to 79.8% ± 0.4% (Figure 6D). While the dominant genera composition was stable in mice on an ND, we detected a substantial variation in mouse microbiomes under HFD. The relative abundance of the Faecalibacterium genus gradually increased from 0.8% to 31.2% ± 10.5% under an ND. When comparing the taxonomic profile of the 8 groups at the genus level, we found that with the exception of Bacillus and Lachnospiraceae, the relative abundance of other genera (> 1% relative abundance in at least one sample) decreased in the entire sample after 10 weeks. This variation pattern of the genera reveals that Bacillus was a more effective colonizer among all groups, and it was the major driver of the community composition. However, some genera showed distinct patterns regarding the variation of their relative abundance among different treatment groups. Bacteroides species were positively associated with an HFD. Under ND conditions, Bacteroides abundance remained low (3.4% ± 2.4%) after 10 weeks. On the other hand, under HFD conditions, Bacteroides contributed to almost 20% of the microbial community (Figure 6D). Supplementation of an HFD combined with Bacilli spores, suppressed the development of Bacteroides populations. Among the Bacteroides under an HFD, SP-64 inoculation decreased the population of Flavobacteriaceae and Bacteroidaceae (Figure 6E).
An HFD negatively affected the relative abundance of the probiotic genus Bifidobacterium, taxonomically affiliated with the Actinobacteria phylum. Under ND conditions, the Bifidobacterium relative abundance rose in an age-dependent manner from 3.4% ± 1.5% at week 0 to 16.6% ± 5.1% by the end of the experiment (Figure 6F). In contrast, after 1 week of feeding, an HFD resulted in a decline of the Bifidobacterium to undetectable levels. Nevertheless, feeding mice with Bacilli spores resulted in the recovery of the Bifidobacterium abundances. Furthermore, after 6 weeks on an HFD supplemented with Bacilli spores, Bifidobacterium was detected in all spores-amended treatments, but absent in spores-unamended treatments (Figure 6F). These results suggest that the improvement in HFD- induced metabolic disorders by SP-64 supplementation was associated with significant gut microbiota composition changes.
B. subtilis does not produce endotoxins and secrete industrial-relevant recombinant proteins in the surrounding media. Furthermore, this microorganism has been shown to be well suited for heterologous protein expression, as it can grow in low-nutrient media, be easily modified genetically, and allow for quick protein recovery [53]. Previous studies have shown that homologous protein production and its expression level were higher when compared to heterologous proteins [54, 55]. However, the attachment of SPs enhances the level of heterologous proteins by guiding them to the extracellular environment. Different proteins have different recognition mechanisms for SPs. Therefore, the effectiveness of SPs w.r.t. to the enhanced protein secretion must be evaluated with the protein of interest [37]. Here, we utilized an approach using a combination of biochemistry, microbiomes, and animal study to dissect how the designed SP-64 is able to protect the host under HFD-induced obesity and metabolic abnormalities. In the present study, we assessed the SPs ability to mediate ADH4 secretion from B. subtilis cells. B. subtilis RIK 1285 is a protease-deficient strain that promotes protein stability [56]. The nucleotide sequence of ADH4 was codon optimized to enhance expression level in prokaryotes like B. subtilis. In the optimization process, the codon usage bias was changed to increase its adaption index, GC content, ribosome binding sites as well as stability of mRNA. The optimized ADH4 gene was transformed into B. subtilis. Gel-electrophoresis and western blotting confirmed heterologous protein expression in prokaryotic hosts like B. subtilis. A functional assay for Ethanol metabolization further confirmed the activity of ADH4 protein.
The SP library was propagated in E. coli and transformed into B. subtilis. All the B. subtilis SP clones were propagated on a selective marker plate for its functional in vitro Ethanol removal assay. Following an evaluation, we observed that most SP clones were able to remove 30% to 40% of Ethanol. Outstandingly, propagated clones SP-63 and SP-64 showed a considerably higher level of Ethanol removal. It is evident from the functional assay that ADH activity varies, depending on SPs nature to target enzyme secretion in B. subtilis. Our results align with previous observations where a library of SPs was screened for optimal enzyme secretion [57, 58]. Hemmerich et al. [38] showed that these SPs can be functional in low GC content microorganisms like Bacillus, but also in high GC content microorganisms, such as C. glutamicum. Furthermore, over-expression of protein secretion components leads to enhanced protein secretion in B. subtilis [59]. Colony PCR identified the phoB SP in a selected clone. The results herein obtained fit with the previous work developed by Zhang et al. [39], where the phoB SP leads to higher xylanase secretion in B. subtilis. A previous study has shown that the overexpression of prsA enhanced the secretion performance when compared to other overexpressed components [60]. Additionally, combinatorial overexpression of prsA and dnaK operons significantly enhanced protein expression [58]. These studies illustrated that SP performance during SP library screening might be enhanced by overexpression of factors resulting in enhanced secretion of a specific protein.
HFD often results in abnormal lipid metabolism in serum, leading to atherosclerosis, diabetes, obesity, and gut microbiota dysbiosis. However, probiotic bacteria like Bacillus colonize the intestine, improving lipid metabolism and regulating gut flora [61]. The use of probiotic bacteria such as Bacillus directly modulates the gut flora composition, complementing the native population with therapeutical species. Bacillus species are spore-forming bacteria that can survive under unfavorable environmental conditions, such as acidic pH, high temperature, and salt concentration [21, 62]. Moreover, these bacteria are important regulators of intestinal homeostasis capable of preventing pathogenic infections and remaining viable while passing through the stomach. In recent years, many experimental studies have shown the potential role of intestinal flora to counteract the effects of HFD-induced obesity and lipid metabolism. Therefore, gut microbiota of mice under different diet conditions following probiotic administration was analyzed to study the mechanisms responsible for preventing hyperlipidaemia.
An HFD often leads to fat accumulation, which can result in metabolic dysfunctions such as obesity, insulin resistance, and hepatic steatosis. Indeed, our study confirmed such a tendency, having that all mouse groups exposed to HFD gained weight when compared to those fed on ND. Nonetheless, recombinant SP-64 treatment showed the lowest weight gain and liver injury among all HFD-treated groups leading to the idea that the treatment improved the adverse effects of the diet. SP-64-mediated protection could be exerted by improving the antioxidant system, decreasing inflammation which lowers the likelihood of hepatic damage, but also regulating microbiota composition. These findings are consistent with previous data that showed lower hepatic damage following probiotic Bacillus administration [21, 63]. Based on these results, we hypothesize that oral SP-64 administration could ameliorate HFD-induced abnormalities, like TC level, bilirubin, ALT, AST, and TG content. HFD-induced hepatic steatosis and hepatic insulin resistance are closely associated phenomena. Phospho p-S6 levels could be explained by a higher mTOR activation, through changes in IR signaling or adenosine monophosphate-activated protein kinase (AMPK) activity in response to the HFD. The presence of spores in the gut was evaluated by their prevalence in the feces after administration when compared to untreated mice. Spores’ persistence suggests that they were present in the gut, where they germinated and subsequently sporulated. This finding suggests that spores germinated and that vegetative cells expressing ADH4 secretion showed a protective effect against HFD-induced disorders. Thus, the role of SP-64 in counteracting adipose tissue gain, as well as other parameters like fasting blood glucose increase, prompted us to examine whether SP-64 also attenuated gut bacterial dysbiosis. Previous studies demonstrated that B. subtilis are able to colonize the human gut [64], grass carps [65], and dogs [66]. These outcomes illustrate the safety aspect of the use of recombinant Bacillus spores.
Accumulating evidence suggests the role of gut microbiota as a new target for prevention of diet-induced metabolic abnormalities. Short chain fatty acids (SCFAs) are well known for their anti-inflammatory responses, and we found that probiotic SP-64 administration increased SCFAs-producing bacteria. The elevated level of SCFAs down-regulates inflammatory cytokines and suppresses T helper 17 (Th17) polarization [67]. Therefore, our microbiota analysis provided strong evidence that probiotic bacterium intake modulated the microbial community in mice and supports better fitness to the host. At the phylum level, we observed an increased level of Firmicutes, while the relative abundance of Bacteroidetes decreased the result of exposure to HFD when compared to ND, leading to an increased ratio of Firmicutes/Bacteroidetes, which is in line with previous reports [68, 69]. Noteworthy, probiotic SP-64 intake in HFD-fed mice attenuated the increase of Firmicutes/Bacteroidetes ratio. At the genus level, the relative abundance of Faecalibaculum decreased, while Alcaligenes, Bacillus, Lachnospiraceae, and Myroides increased in probiotic-treated groups. Alcaligenes and Lachnospiraceae can produce SCFAs, such as acetic acid and butyric acid, which can reduce intestinal pH and inhibit the growth of pathogenic bacteria [70]. Relative changes to these microbiotas could be responsible for alleviating body weight gain in HFD-fed mice. Additionally, the produced SCFAs can reach to liver through portal vein, thereby activating the Nrf-2 and AMPK pathways to inhibit lipid accumulation [71]. Moreover, SP-64 supplementation enhanced the abundance of Bifidobacterium. Bifidobacterium is well known for reducing or preventing obesity, inflammation, and other metabolic disorders. The increased population of Bifidobacterium resulted in improved gut barrier function, hepatic inflammation, and oxidation. Based on the observed results, we speculate that probiotic SP-64 administration might reduce liver lipidic accumulation and oxidative stress by regulating microbiota and metabolites.
In conclusion, we highlighted that the administration of probiotic B. subtilis SP-64 ameliorated several metabolic abnormalities induced by HFD while improving metabolic functions. Our findings showed that ADH4-secreting B. subtilis SP-64 is a potential probiotic candidate, displaying anti-hypercholesterolemic and hyperglycaemic effects. However, there are several challenges and limitations to probiotics’ development including the absence of a clear mechanistic understanding of disease progression. With the development of molecular biology tools, the use of engineered probiotic microorganisms in food and medicine needs to be established. Future studies using metabolome or metatranscriptome analysis could improve the understanding of the effects of engineered probiotics on the gut metabolism, and subsequently, on other tissues beyond the intestinal tract. We believe that our study provides valuable insight into counteracting diet-induced metabolic abnormalities which could be applied to the prevention of liver injuries.
ADH4: alcohol dehydrogenase-4
ALT: alanine aminotransferase
AST: aspartate aminotransferase
ASVs: amplicon sequence variants
CFU: colony forming units
GC: gas chromatography
GI: gastrointestinal
HFD: high-fat diet
LB: Luria-Bertani
NAD: nicotinamide adenine dinucleotide
ND: normal diet
PBS: phosphate-buffered saline
PCoA: principal coordinates analysis
PCR: polymerase chain reaction
PERMANOVA: permutational analysis of variance
rRNA: ribosomal RNA
SCFAs: short chain fatty acids
SPs: signal peptides
SSU: small subunit
TGs: triglycerides
The supplementary materials for this article is available at: https://www.explorationpub.com/uploads/article/file/10059_sup_1.pdf.
RPS: conceptualization, methodology, analysis, writing; MK: microbiome analysis and writing; MB: activity assay; NBD: immunohistochemistry; SG: Illumina sequencing; AH: immunohistochemistry; OT: supervised the project and writing; ZH: supervised the project.
The authors declare no competing financial interests.
The mice study was approved by the ethical committee of Hebrew University of Jerusalem AG-19-15587-4.
Not applicable.
Not applicable.
The Illumina-generated SSU rRNA gene amplicon sequences are available from the BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession PRJNA702247. The authors declare that all relevant data supporting the findings of this study are provided within the manuscript and its supplementary information files, or from the corresponding author upon request.
This work was supported by the Ministry of Agriculture (ARO), Israel Grant (12-05-0030) to OT and ZH and was partially funded by Tnuva Ltd., Israel. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2022.
Copyright: © The Author(s) 2022. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
