Affiliation:
1Departamento de Psicología Social y de las Organizaciones, Universidad Nacional de Educación a Distancia (UNED), 28040 Madrid, Spain
Email: a.borrego@psi.uned.es
ORCID: https://orcid.org/0000-0002-4699-3031
Affiliation:
2Departamento de Microbiología, Universidad de Málaga, 29071 Málaga, Spain
ORCID: https://orcid.org/0000-0002-2174-0652
Explor Dig Dis. 2025;4:100584 DOI: https://doi.org/10.37349/edd.2025.100584
Received: June 23, 2025 Accepted: July 22, 2025 Published: August 07, 2025
Academic Editor: Raquel Soares, University of Porto, Portugal
The article belongs to the special issue Gut Microbiota towards Personalized Medicine in Metabolic Disease
Celiac disease is an immune-mediated disorder with significant metabolic implications. Several factors have been proposed to explain the association between celiac disease in patients following a gluten-free diet and metabolic disorders, including metabolic syndrome. Growing evidence suggests a pivotal role of gut microbiome dysbiosis in the onset of celiac disease and its associated metabolic disturbances. The present narrative review examines (i) the connections between celiac disease and metabolism-related comorbidities, including metabolic syndrome and metabolic dysfunction-associated steatotic liver disease; (ii) the role of the gut microbiome in celiac disease, including the outcomes of gut microbiome dysbiosis in celiac children and adults; and (iii) the potential of microbial therapeutic strategies within the context of personalized medicine for patients with celiac disease and comorbid metabolic conditions. A synthesis of existing studies highlights several protective factors and interventions for future celiac disease prevention research. Adopting plant-based, health-promoting dietary patterns such as the Mediterranean or vegetarian diet within the first two years of life reduces celiac disease risk. These fiber- and phytochemical-rich diets support beneficial gut microbiota growth and short-chain fatty acid production, which maintain intestinal barrier integrity by enhancing mucus and tight junction proteins. Short-chain fatty acids also modulate immunity by inducing Tregs that secrete IL-10, suppressing pro-inflammatory Th1 responses and autoantibody production. Precision probiotics offer diverse therapeutic benefits in celiac disease by reducing inflammation, restoring beneficial microbes, and degrading immunogenic gliadin peptides. Postbiotics complement these effects by reinforcing barrier integrity and counteracting gliadin-induced inflammation. Thus, integrating clinical models with microbial biomarkers promises to improve celiac disease diagnosis and monitoring, enabling better risk stratification, earlier detection, and personalized management of this heterogeneous disease.
Celiac disease (CeD) is an immune-mediated disorder that leads to chronic inflammation of the small intestine in response to gluten intake. Gluten is a protein complex specific to wheat, while structurally related prolamins, such as secalin in rye, hordein in barley, and avenins in oats, may also provoke immune activation in genetically predisposed individuals [1, 2]. This susceptibility is primarily linked to the presence of HLA-DQ2 and/or HLA-DQ8 alleles. CeD is also characterized by the production of autoantibodies against tissue transglutaminase (tTG) type 2, as well as immunoglobulin A (IgA) anti-endomysium and intestinal IgM antibodies targeting gluten [3]. Gastrointestinal symptoms typically manifest upon the ingestion of gluten-containing foods [4].
The global prevalence of CeD, based on serological testing, is estimated at 1.4%, making it the most common immune-mediated disorder affecting the gastrointestinal tract [5]. Currently, the only effective treatment for CeD is strict adherence to a gluten-free diet (GFD). Nevertheless, the restrictive nature of the GFD presents multiple challenges, including financial burden, social isolation, and potential adverse health effects such as macro- and micronutrient deficiencies and long-term metabolic complications [6–8]. Previously, CeD was recognized as a condition associated with significant weight loss, micronutrient deficiencies, and low body mass index (BMI) [9]. However, recent findings have revealed that CeD can present with extraintestinal manifestations and may occur without obvious signs of malnutrition [9]. According to Verma [10], malnutrition is common among individuals with CeD, both at diagnosis and during long-term follow-up. In this context, malnutrition may result from impaired nutrient absorption due to intestinal inflammation and/or the inadequate nutritional quality of the GFD [8, 11].
Furthermore, although the link between CeD and malnutrition has been well-established, recent research indicates that a wider range of patients, including those with normal weight or overweight, are now being diagnosed with the condition [12, 13]. A systematic review reported that 14% of CeD patients present overweight and 6% are obese at diagnosis [14]. Another concern is the rising incidence of metabolic syndrome (MetS) and liver disorders among CeD patients adhering to a GFD [15–17]. Several factors have been proposed to explain the association between CeD in patients following a GFD and metabolic disorders. First, the reduction in intestinal inflammation and restoration of absorptive capacity following a GFD often lead to improved nutrient absorption, sometimes described as a compensatory hyperphagic state [18]. Second, gluten-free processed foods frequently contain high amounts of saturated fats added to enhance palatability, which contributes to increased caloric intake [19]. Third, the GFD is generally associated with higher consumption of simple carbohydrates and saturated fats and lower intake of complex carbohydrates and dietary fiber [20]. Fourth, stress-related emotional eating may partly contribute to weight gain, especially among children and adolescents [14, 21]. Moreover, studies suggest that adherence to a GFD may increase the risk of metabolic complications such as weight gain, obesity, and MetS due to these dietary imbalances [16, 22, 23], as well as nutritional deficiencies, toxicity, morbidity, mortality, and mental health problems [24].
A paucity of studies has been conducted on the evaluation of risk factors for MetS in patients diagnosed with CeD. An investigation involving Italian patients found that a high BMI at diagnosis and exposure to proton pump inhibitors (PPIs) were the only factors significantly associated with MetS development in a multivariable logistic regression model. Variables such as age at diagnosis, baseline waist circumference, sex, insulin resistance, hyperglycemia, hypertension, hypercholesterolemia, hypertriglyceridemia, and elevated liver enzymes were not linked to MetS onset [25]. The mechanisms through which PPIs influence MetS risk remain to be fully elucidated. Emerging evidence indicates a possible connection between PPIs and gut microbiome (GM) alterations, which may promote intestinal dysbiosis and impair nutrient absorption. These changes could contribute to abdominal obesity, dyslipidemia, insulin resistance, and hepatic fat accumulation [26]. The role of medications in modulating MetS risk among CeD patients requires further investigation and represents a promising area for future research.
One of the non-gluten environmental factors implicated in the development of CeD is the GM, defined as the complex community of microorganisms residing in the gastrointestinal tract that contributes to the host’s immune, metabolic, and physiological functions [27–29]. Multiple studies have shown that the GM in CeD patients undergoes significant alterations, characterized by an increase in opportunistic bacterial taxa alongside a reduction in beneficial bacteria, leading to a dysbiotic state [30].
Therefore, CeD is a systemic disorder with significant metabolic implications, and growing evidence suggests a pivotal role of GM dysbiosis in its onset and associated metabolic disturbances. The present narrative review examines (i) the connections between CeD and metabolism-related comorbidities, including MetS and metabolic dysfunction-associated steatotic liver disease (MASLD); (ii) the role of the GM in CeD, including the outcomes of GM dysbiosis in celiac children and adults; and (iii) the potential of microbial therapeutic strategies within the context of personalized medicine for patients with CeD and comorbid metabolic conditions.
MetS comprises a cluster of interrelated conditions, including abdominal obesity, dyslipidemia, insulin resistance, hypertension, and elevated fasting glucose, all of which increase the risk of cardiovascular disease (CVD) and type 2 diabetes mellitus (T2DM) [31]. Insulin resistance and inflammation associated with excess central adiposity result in impaired metabolism of glucose, lipids, and other energy substrates across multiple organ systems, potentially contributing to the development of CVD and T2DM progressively [31, 32].
In individuals without CeD who have MetS, metabolic disturbances such as hypercholesterolemia, hypertriglyceridemia, and hyperglycemia are linked to insulin resistance. These changes contribute to oxidative stress, steatosis, lipid peroxidation, and increased cytokine production, resulting in inflammation and necrosis. Whether MetS in patients with CeD arises from similar pathophysiological mechanisms remains unclear [8]. However, it is well established that CeD is associated with a heightened risk of coronary artery disease, which correlates with risk factors including dyslipidemia, male sex, hypertension, obesity, and T2DM [33]. Patients with MetS display evidence of a persistent, subclinical inflammatory state characterized by elevated levels of cytokines and other inflammatory markers indicative of endothelial dysfunction and increased cardiovascular risk. These include elevated interleukins (IL-6, IL-10, and IL-18), adiponectin, C-reactive protein, leptin, fibrinogen, and tumor necrosis factor alpha (TNF-α), collectively defining the inflammation-obesity-insulin resistance triad [32, 34]. Additional contributors to obesity have been identified, including dietary habits and patterns, as well as obesogenic hormones such as estrogens, leptin, androgens, insulin, and incretins. Moreover, cytokines, physical activity, and alterations in the GM have also been implicated. However, studies addressing these factors specifically in the context of CeD remain limited [35].
Recent research has reported an increased prevalence of MetS among patients with CeD, particularly following the initiation of a GFD. A meta-analysis by Aggarwal et al. [16] found that the pooled prevalence of MetS increased from 4.3% before GFD to 21.3% afterward. Similarly, a prospective observational study from Italy observed an increase in MetS prevalence among newly diagnosed CeD patients, from 2% at baseline to 29.5% after one year on a GFD [36]. In contrast, a study conducted in the United States reported a relatively low MetS prevalence of 3.5% in individuals with CeD, significantly lower than the 12.7% prevalence found in age-, sex-, and ethnicity-matched controls [37]. A key limitation of this study was the lack of control for GFD initiation and adherence, which could have influenced the results. Further investigations into MetS parameters have produced mixed findings regarding the relationships between high-density lipoprotein (HDL) and triglyceride levels, waist circumference, glycemic control, and blood pressure [38]. Likewise, Yerushalmy-Feler et al. [39] reported that fat percentage, rather than weight status, is associated with the risk of developing MetS components in individuals with childhood-onset CeD. A systematic review assessing the effects of the GFD on CVD risk factors in CeD patients found increases in HDL, fasting glucose, and BMI [40]. However, results related to low-density lipoprotein (LDL), triglycerides, and blood pressure were inconsistent, and the overall quality of evidence was rated as low. A recent study proposed that adherence to a GFD may contribute to MetS development in CeD patients, although the MetS rate remains lower than that observed in the general population [41].
MASLD, formerly termed non-alcoholic fatty liver disease, is a condition defined by hepatic steatosis in combination with one or more metabolic abnormalities, including overweight or obesity, T2DM, and insulin resistance [31, 42]. MASLD is a leading cause of liver disease globally, with an estimated prevalence of 25.2% [43]. However, its relationship with CeD remains under investigation. Aggarwal et al. [16] reported that MASLD can co-occur with CeD both prior to and following adherence to a GFD, finding a pooled prevalence of 18.2% in treatment-naive CeD patients and 28.2% among those on a GFD. Longitudinal studies indicate an increase in MASLD prevalence from 15.3% to 29.1% after GFD initiation, although the duration of dietary treatment varied widely, ranging from 6 months to 36 years. Other studies have shown that patients with CeD have an elevated risk of developing MASLD compared to the general population, with reported hazard and odds ratios of 2.8 and 3.21, respectively [44, 45]. In an Italian cohort matched for age, sex, and other MASLD risk factors, the odds ratio for MASLD in CeD patients was 2.9 [46]. These findings indicate that CeD may predispose individuals to MASLD independently of conventional metabolic risk factors and that MASLD frequently occurs in patients with normal or low BMI.
The pathophysiological mechanisms underlying the relationship between CeD and MASLD are complex and not yet fully understood. A key hypothesis centers on the role of the gut-liver axis, which describes the bidirectional relationship between the gastrointestinal tract and the liver via the portal vein and biliary system, facilitating the transport of nutrients, microbial products, and immune mediators directly to the liver [47, 48]. Intestinal damage caused by CeD may induce gut dysbiosis, disrupting this axis and allowing bacterial endotoxins and inflammatory mediators to translocate into the portal circulation. This process can trigger hepatic inflammation, lipid accumulation, and fibrosis via the activation of Kupffer cells [47]. In addition, systemic chronic inflammation associated with active CeD may further promote hepatic inflammation, contributing to MASLD development [49]. While inflammation typically decreases following the initiation of a GFD [18], the diet itself may involve increased consumption of fructose and saturated fats [22, 50], nutrients implicated in de novo hepatic lipogenesis [46, 51]. Moreover, gluten-free processed foods, often based on refined grains, tend to have a higher glycemic index, which can cause postprandial hyperglycemia. This condition may increase the risk of insulin resistance and hepatic fat accumulation, further contributing to MASLD development in individuals with CeD [18].
A GFD has been shown to alter the GM through a decrease in beneficial bacterial species and an increase in potentially harmful ones [30]. The link between CeD, MetS, and MASLD may be related to increased intestinal permeability (i.e., the “leaky gut” phenomenon) and the subsequent development of small intestinal bacterial overgrowth (SIBO) induced by dysbiosis [52]. The translocation of luminal microbiota or microbial products across a compromised intestinal barrier has been shown to initiate immune responses that contribute to the onset of MetS and MASLD. Proposed mechanisms involve alterations in the bile acid pool, decreased production of short-chain fatty acids (SCFAs), and reduced activation of the farnesoid X receptor in the distal small intestine by bile acids, all of which are associated with impaired intestinal barrier integrity [53].
In the context of CeD, the abundance of protective bacteria, including bifidobacteria and members of the phylum Bacillota such as the families Lactobacillaceae and Streptococcaceae, is reduced in comparison to healthy controls. Conversely, the prevalence of harmful bacteria (i.e., pathobionts) belonging to the phylum Bacteroidota, including Bacteroides and Prevotella, as well as members of the phylum Pseudomonadota such as Escherichia, Haemophilus, Serratia, and Klebsiella, is elevated [54, 55]. Consequently, the disruption of metabolic processes due to dysbiosis can elevate the risk of metabolic diseases, including MetS and MASLD [56].
The sustained inflammation or the overgrowth of bacterial pathobionts may disrupt the regulation of adhesion molecules at tight junctions. This disruption facilitates the translocation of foreign microorganisms and toxic substances, promoting the release of partially digested gliadin peptides into the lamina propria [28, 30, 57]. CeD causes structural changes in the small intestine, characterized by focal defects in the epithelial barrier, increased apoptosis, and altered expression of tight junction proteins [58]. These alterations affect barrier permeability, leading to the loss of ions and water into the gut lumen. Specifically, barrier-forming claudins (claudin-3, claudin-5, and claudin-7) are downregulated, while channel-forming claudins (claudin-2 and claudin-15) are upregulated, resulting in increased selective paracellular solute transport [59]. In addition, zonulin, which is a protein that reversibly regulates intestinal permeability by modulating tight junction molecules, has been linked to CeD [60]. Gluten peptides and certain enteric bacteria, such as Escherichia coli (E. coli), have been found to induce zonulin, suggesting its involvement in CeD pathogenesis [61]. Moreover, pro-inflammatory mediators, including TNF-α and interferon-gamma (IFN-γ), have been identified as contributors to the downregulation of barrier and tight junction proteins [59].
Furthermore, microbial dysbiosis has been demonstrated to augment the magnitude and complexity of gliadin peptides, a process attributed to the differential proteolytic activity of the GM [62, 63]. Recent studies indicate that peptidases from various microbial sources can degrade gluten and its derived peptides [62, 63]. In this context, certain gut bacteria, such as Bifidobacterium spp., Lactobacillus spp., and Rothia spp., possess the ability to degrade gluten, alter intestinal permeability, and activate the host immune response, all of which contribute to the pathogenesis of CeD. Therefore, maintaining an eubiotic GM composition may help modulate symptoms associated with gluten-related disorders (GRDs) [62].
Although evidence supports a role for the GM in the pathogenesis of CeD, there remains no clear consensus regarding the specific microbial alterations associated with the condition. Previous research has primarily focused on characterizing the GM composition in infants to identify potential predictors of CeD development [64–66]. Moreover, factors such as the timing of initial gluten exposure and other environmental influences, including premature birth, mode of delivery, type of infant feeding, antibiotic use, and early infectious exposures, have been shown to affect epigenetic regulation through modifications of the GM ecosystem. These alterations can disrupt the maturation of the intestinal barrier, gut-associated lymphoid tissue (GALT), and the balance of innate and adaptive immune responses [66–68].
Premature birth has been proposed to result in delayed gut bacterial colonization, reduced microbial diversity, decreased abundance of obligate anaerobic commensals such as Bifidobacterium and Bacteroides, and an increased prevalence of facultative and pathogenic anaerobes, including Enterobacter, Enterococcus, Escherichia, Klebsiella, Clostridioides difficile, and Staphylococcus. This microbial profile favors an inflammatory gut environment that may promote the development of CeD [69]. Similarly, the mode of delivery is a critical determinant of neonatal GM colonization [70]. Cesarean section results in neonatal microbial colonization predominantly from environmental and maternal skin sources, characterized by increased Enterococcus faecalis, and decreased Bacteroides spp. and Parabacteroides spp., changes associated with a heightened risk of CeD [71]. Within this context, Tanpowpong et al. [72] reported an adjusted hazard ratio of 1.39 for CeD in cesarean-born infants compared to those delivered vaginally.
Infant feeding type significantly influences early GM composition, playing a key role in its initial structuring. Breastfeeding has been shown to increase the prevalence of genera such as Lactobacillus, Bifidobacterium, Enterococcus, Corynebacterium, Propionibacterium, Streptococcus, and Sneathia, while reducing Bacteroides and Staphylococcus [70]. Cenit et al. [73] observed that continued breastfeeding at the time of gluten introduction correlates with increased transfer via milk of immunomodulatory factors, including IL-12p70, transforming growth factor-β1 (TGF-β1), secretory IgA (sIgA), and Bifidobacterium spp., which may delay or reduce the risk of CeD development. However, large epidemiological studies have not consistently demonstrated a protective effect of breastfeeding against CeD onset in genetically predisposed children [74].
In addition, multiple studies have linked early antibiotic exposure to an increased risk of chronic autoimmune and inflammatory bowel diseases, including CeD [64, 75]. Lindfors et al. [76] demonstrated a cumulative effect in which enterovirus infection combined with gluten exposure increased the risk of CeD development in children. Viral pathogens such as rotavirus, enterovirus, adenovirus type 12, and orthoreovirus have been identified as potential triggers of CeD by activating innate immunity via Toll-like receptor 3 (TLR3), leading to intestinal inflammation and loss of tolerance to gliadin peptides [77, 78].
Furthermore, researchers have identified that certain microbial species, metabolites, and pathways undergo alterations regarding abundance in infants at high risk of developing CeD prior to the manifestation of the disorder, thereby indicating that HLA-DQ alleles can exert an influence on early GM composition [79]. Specifically, these alterations in taxa abundance have been observed to result in an increase of members belonging to the phylum Pseudomonadota in patients diagnosed with CeD, accompanied by a concurrent decrease in members of the Bacillota and Actinomycetota phyla [80–83].
Figure 1 shows a proposed model for the pathogenesis of CeD. This model incorporates a series of factors that contribute to the development of CeD. Specifically, it considers the influence of host genetics, environmental factors, and gluten consumption. The consumption of gluten has been proven to result in an increase of pathobiont colonization, a reduction in autochthonous gut microbiota, and a disruption in GM dysbiosis. This, in turn, results in a disruption of immune homeostasis and gut integrity. Consequently, this disruption favors the onset of CeD and its clinical manifestations.
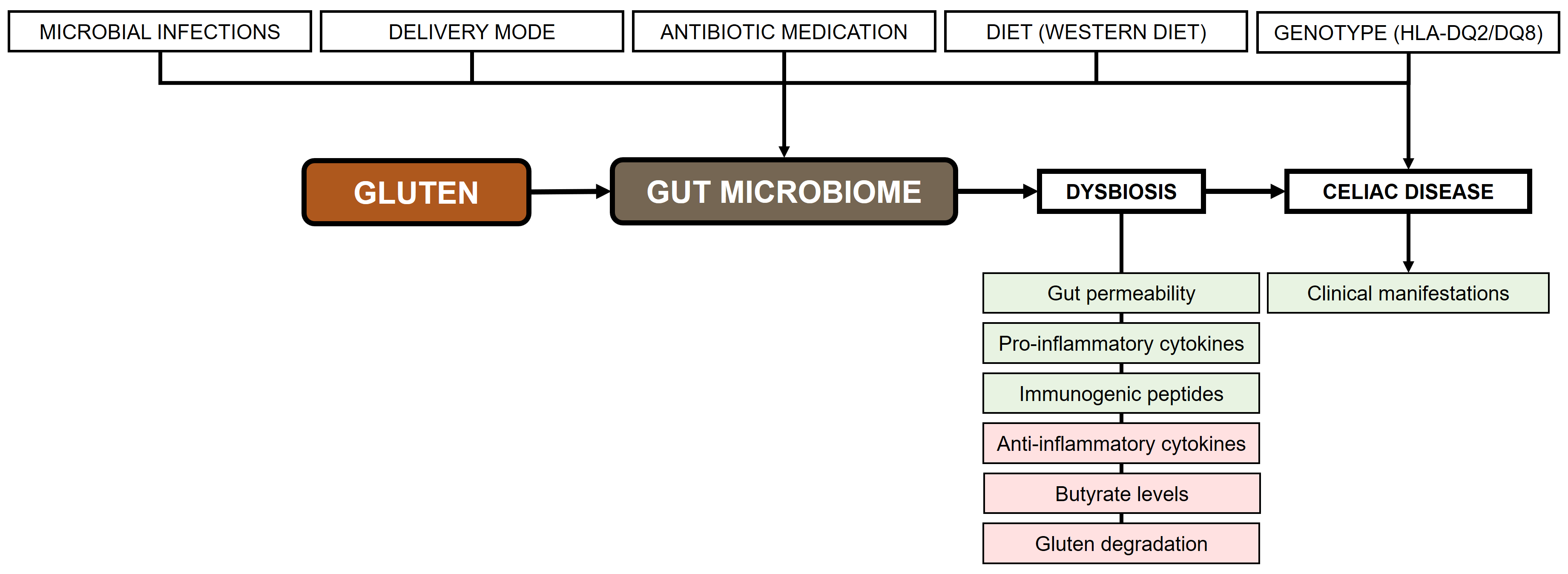
Pathogenesis of celiac disease. Rectangles in green: increase; rectangles in red: decrease. Adapted from [73], © 2015 by the authors
A substantial body of research has examined the microbial composition in patients with active CeD compared to healthy controls. To this end, samples are typically obtained from duodenal biopsies, intestinal aspirates, and stool specimens to analyze GM composition. In addition, salivary and pharyngeal microbiota have been examined, although these studies were conducted under specific research questions [84]. The methodologies employed for microbial identification in these studies are highly heterogeneous, each presenting distinct advantages and limitations. Common techniques include culture-based approaches (culturomics), quantitative polymerase chain reaction (qPCR), next-generation sequencing (NGS) methods (e.g., targeted amplicon sequencing, shotgun metagenomics, shallow metagenomics), denaturing gradient gel electrophoresis (DGGE), temperature gradient gel electrophoresis (TGGE), fluorescence in situ hybridization (FISH), flow cytometry, gas chromatography, and 16S–23S rRNA intergenic spacer region analysis [85, 86]. Beyond methodological differences, other factors influencing GM composition in CeD include disease activity status, adherence to a GFD, and patient age [87]. While stool samples are commonly employed as proxies for GM composition, notable discrepancies often exist between fecal microbiota and the actual microbial communities adhering to the intestinal mucosa. Despite its invasiveness, biopsy sampling is generally regarded as providing a more precise representation of the GM content [88].
In children diagnosed with CeD, several seminal studies have demonstrated a change in their GM, both in stool and duodenal samples. An increase in the abundance of the species belonging to the genera Bacteroides (B. fragilis), Clostridium leptum, Staphylococcus (S. epidermidis and S. haemolyticus), E. coli, Klebsiella spp., Latilactobacillus (formerly Lactobacillus) curvatus, Leuconostoc carnosum, Leuconostoc mesenteroides, Prevotella spp., Salmonella spp., and Shigella spp. has been observed in stool specimens. In addition, it has also been observed a decline in bacterial species of the genera Bifidobacterium (B. longum and B. fragilis subsp. ovatus), Clostridium histolyticum, Enterococcus (E. faecium), Faecalibacterium prausnitzii (F. prausnitzii), Lacticaseibacillus (formerly Lactobacillus) casei, and Romboutsia (formerly Clostridium) lituseburense [61, 89–95]. In contrast, the composition of the GM in duodenal biopsies has been reported to exhibit weak differences. The abundance of certain species, including Actinomyces graevenitzii, Bacteroides (B. vulgatus), Blautia (formerly Clostridium) coccoides, Clostridium spp., E. coli, Haemophilus spp., Klebsiella oxytoca, Prevotella spp., Serratia spp., and Staphylococcus (S. pasteuri), has been found to be augmented. Additionally, it has been observed decreases in the abundance of the following species: Bifidobacterium (B. catenulatum), Enterococcus faecium, Lactiplantibacillus (formerly Lactobacillus) plantarum, Papillibacter cinnamivorans, Prevotella oralis, Proteus spp., Ruminococcus bromii, Streptococcus anginosus, and Thermoclostridium (formerly Clostridium) stercorarium [81, 91, 92, 96–99]. Furthermore, de Meij et al. [100] found that the composition and diversity of the mucosa-associated duodenal microbiome were comparable between children with untreated CeD and controls. The results of the study revealed increases in the abundance of Clostridium, Lactobacillus, and Streptococcus in both groups.
In recent years, advancements in generation sequencing, including targeted amplicon, shotgun metagenomics, and shallow metagenomics sequencing, have facilitated the investigation of GM dysbiosis in children diagnosed with CeD. In this respect, Olivares et al. [101] conducted a prospective study, including 22 breastfed and vaginally delivered infants with either high genetic risk (HLA-DQ2 carriers) or low genetic risk (non-HLA-DQ2/8 carriers) of developing CeD. The fecal microbiota of infants was subjected to analysis through 16S rRNA gene pyrosequencing and real-time quantitative PCR. Their findings indicated that children with a high genetic risk had significantly higher abundance of members of the phyla Bacillota and Pseudomonadota, as well as lower proportions of members of Actinomycetota phylum compared to children with a low genetic risk. At the genus level, high-risk children exhibited a significantly lower abundance of Bifidobacterium and a higher abundance of the genera Gemella, Clostridium sensu stricto, Corynebacterium, unclassified Enterobacteriaceae, and Raoultella. Moreover, in high-risk children, a negative correlation was identified between Bifidobacterium species and several genera belonging to the phyla Pseudomonadota (Escherichia/Shigella) and Bacillota (Clostridium).
This same research group conducted another case-control study including 10 children with CeD and 10 children who did not develop the disease after a 5-year follow-up [64]. The fecal microbiota of children with CeD was assessed using a high-throughput 16S rRNA gene amplicon sequencing. The findings revealed that children who remained healthy showed a progressive increase in bacterial diversity over time, marked by a greater abundance of Bacillota families. In contrast, those who developed CeD failed to exhibit this increase in microbial diversity. In addition, children who developed CeD experienced a significant decline in sIgA levels over the study period, whereas healthy children showed elevated levels of TNF-α, which correlated with Bifidobacterium spp. Furthermore, a higher relative abundance of B. longum was detected in healthy controls, while increased proportions of Bifidobacterium breve and Enterococcus spp. were associated with a greater risk of CeD onset.
In a recent study, Leonard et al. [79] performed both cross-sectional and longitudinal analyses of the GM in a cohort of 10 children who developed CeD and a matched group of 10 unaffected controls. The cross-sectional analysis at CeD onset revealed altered abundances of several microbial species between cases and controls, including Bacteroides uniformis, Bacteroides vulgatus, Enterocloster bolteae, B. longum subsp. longum, and Streptococcus thermophilus, although no significant changes in overall microbial species abundance were observed. Conversely, the longitudinal analysis identified several microbial taxa with increased abundance prior to CeD onset, such as Dialister invisus, Parabacteroides spp., and members of the family Lachnospiraceae. On the other hand, other taxa, including Enterocloster clostridioformis (formerly Clostridium clostridioforme), F. prausnitzii, and Streptococcus thermophilus, were found to be decreased before the development of CeD.
A study conducted by El Mouzan et al. [102] aimed to determine whether a distinct GM profile is associated with CeD in children from Saudi Arabia. The study included 40 children diagnosed with CeD. Comprehensive analyses comparing the microbial composition between CeD patients and controls revealed significant differences at both fecal and mucosal levels. Fecal samples exhibited greater microbial diversity and abundance compared to mucosal samples. At the phylum level, members of Pseudomonadota were more abundant in duodenal mucosal samples, whereas Bacillota and Bacteroidota predominated in stool samples. At the species level, children with CeD showed increased abundance of Acinetobacter lwoffii, Bifidobacterium angulatum, Corynebacterium ihumii, Corynebacterium tuberculostearicum, Kocuria rhizophila, Lactobacillus acidophilus, Ralstonia pickettii, and Staphylococcus aureus. In contrast, Roseburia intestinalis was significantly enriched in non-CeD controls. A total of 169 distinct bacterial species were identified in fecal samples, exhibiting significant abundance differences between CeD and non-CeD children. Notably, Actinobaculum massiliense, Blautia hydrogenotrophica, Corynebacterium pyruviciproducens, Klebsiella michiganensis, and Prevotella sp. BV3P1 were elevated in CeD patients, while Actinomyces sp. ICM58, Alistipes inops, Anaerostipes caccae, Bacteroides pyogenes, Coprobacter fastidiosus, Enterobacter sp. MGH38, and Raoultella ornithinolytica were reduced in this group.
Salamon et al. [103] analyzed the bacterial microbiota profile by employing NGS targeting the V3–V4 regions of the 16S rRNA subunit. Biopsy samples were collected from the stomach and duodenum of children newly diagnosed with CeD (N = 40) and from a control group (N = 20). At the phylum level, Pseudomonadota was the dominant phylum in both the stomach and duodenum. No significant differences were detected in the relative abundance of most bacterial phyla between CeD and control groups or between anatomical sites, except for Campylobacterota, which was exclusively identified in the stomachs of children with CeD. In the duodenal microbiota, a positive correlation was found between the presence of the HLA-DQ8 allele and the abundance of bacteria from the genus Blautia, with statistical significance observed specifically for Blautia wexlerae.
A body of research has previously identified alterations in the GM composition in adults. Nistal et al. [104] reported a decrease of Latilactobacillus (formerly Lactobacillus) sakei and Bifidobacterium spp. in stool specimens of CeD patients. These authors reported a reduction in the abundance of Mycobacterium spp. and Methylobacterium spp. in samples from duodenal biopsies. In contrast, Wacklin et al. [105] observed an increase in members belonging to the phylum Pseudomonadota, and a decrease in the abundance of the Bacillota and Bacteroidota phyla in duodenal biopsies of GFD-treated adults diagnosed with CeD.
More recently, D’Argenio et al. [106] investigated the GM composition in duodenal biopsy samples from 20 adult patients with active CeD, 6 CeD patients adhering to a GFD, and 15 healthy controls using 16S rRNA gene sequencing. In addition, cultured and isolated bacterial species were identified via mass spectrometry. The GM profiles of active CeD patients were predominantly composed of bacteria from the phylum Pseudomonadota, whereas Bacillota and Actinomycetota were among the least abundant phyla. At the species level, Neisseria flavescens emerged as the most prevalent Neisseria species in the duodenum of patients with active CeD.
In a separate study, bacterial communities were characterized by analyzing 16S rRNA extracted from duodenal biopsies of untreated adult CeD patients and non-CeD controls using pyrosequencing [107]. Bacterial richness and diversity were found to be higher in non-CeD controls compared to untreated CeD patients. Taxonomic classification revealed that the bacteria predominantly belonged to the phyla Bacillota and Pseudomonadota. Nevertheless, no statistically significant differences were observed in the composition of bacterial communities in the upper small intestine between untreated CeD patients and non-CeD controls.
Garcia-Mazcorro et al. [108] examined the GM in Mexican individuals affected by GRDs. Using ultra-high-throughput 16S rRNA marker sequencing, the study comprehensively characterized the duodenal and fecal microbiota of patients with CeD (N = 6), non-celiac gluten sensitivity (NCGS) (N = 12), and healthy controls (N = 12). Linear discriminant analysis effect size revealed that the genus Actinobacillus and the family Ruminococcaceae were significantly enriched in the duodenal and fecal microbiota of patients with NCGS, respectively, whereas Novispirillum was more abundant in the duodenum of patients with CeD.
Bodkhe et al. [80] utilized 16S rRNA gene sequencing to investigate the microbial diversity across three distinct groups: individuals with CeD, those in a pre-disease state, and healthy adult controls. Although no statistically significant differences in overall microbial diversity were observed among the groups, specific alterations in amplicon sequence variants (ASVs) were identified between the pre-disease and disease groups. Duodenal biopsies revealed more pronounced differences in ASV profiles compared to fecal samples, indicating a greater microbial disruption at the primary site of disease manifestation. The duodenal microbiota in the pre-disease group was enriched in ASVs belonging to the genera Actinomyces, Anaerostipes, Bifidobacterium, Gemella, Granulicatella, and Parvimonas. In contrast, the CeD group showed a higher abundance of ASVs from Helicobacter and Megasphaera. Fecal microbiota analysis of CeD and pre-disease groups demonstrated a reduction in ASVs associated with Akkermansia and Dorea compared to healthy controls. Furthermore, predicted functional metagenomic analysis suggested a decreased capacity for gluten degradation in the fecal microbiota of CeD patients relative to both the pre-disease and control groups.
In a study assessing adherence to a GFD, fecal samples were collected from 46 individuals with CeD who had maintained a GFD for a minimum of two years, along with 30 samples from healthy controls [109]. Adherence to the GFD was associated with a restoration of alpha-diversity among CeD individuals. However, the beta-diversity analysis revealed a microbial composition that remained distinct from that of the control group. Specifically, the GM of CeD patients exhibited a reduced abundance of several taxa, including B. longum and multiple members of the Lachnospiraceae family, whereas the genus Bacteroides was comparatively more prevalent.
Shi et al. [110] utilized 16S rDNA sequencing and metabolomics to examine the fecal microbial composition and metabolomic profile of patients diagnosed with CeD in Northwest China. The analysis revealed a substantial divergence in GM composition between CeD patients and healthy controls. At the genus level, the CeD group exhibited increased relative abundances of Allisonella, Lactobacillus, Streptococcus, and Veillonella. In contrast, the genera Anaerostipes, Blautia, Faecalibacterium, Gemmiger, and Ruminococcus were significantly reduced in CeD patients. Using a random forest model, the authors identified four bacterial genera (Allisonella, Clostridium cluster IV, Ruminococcus, and Christensenella) and six differential metabolites as potential biomarkers, highlighting strong correlations between these microbial taxa and metabolomic alterations.
In an observational study, Francavilla et al. [111] applied small RNA and shotgun metagenomic sequencing to stool samples collected from 63 treated CeD (tCD) patients, comprising 51 individuals adhering strictly to a GFD with negative TG serology (tCD-TG−), and 12 symptomatic individuals with non-strict or short-term GFD adherence and positive TG serology (tCD-TG+), as well as from 66 healthy controls. In the tCD-TG− group, notable alterations in the GM were observed, including an increased abundance of members of the phylum Bacteroidota and the species Roseburia inulinivorans, alongside a reduction in members of the phyla Actinomycetota and Verrucomicrobiota, and in the species B. longum, Eubacterium sp. CAG274, Roseburia sp. CAG309, Ruminococcus bicirculans, and Ruminococcus callidus. In the tCD-TG+ group, microbial shifts were also evident, with a decreased abundance of members of the phylum Euryarchaeota and the species Ruminococcus bicirculans, Haemophilus parainfluenzae, Streptococcus sanguinis, Veillonella atypica, and Veillonella tobetsuensis. The presence of specific molecular patterns in stool samples has been identified as a potential diagnostic biomarker for individuals with CeD, reflecting either long-term effects of dietary treatment or ongoing intestinal inflammation due to poor adherence to the GFD.
Currently, the most effective treatment for CeD involves strict and lifelong adherence to a GFD. However, evidence indicates that GFDs may be nutritionally inadequate, often lacking essential nutrients such as protein, folate, iron, niacin, riboflavin, thiamine, vitamin B12, zinc, selenium, and dietary fiber [8, 112]. Moreover, an improperly balanced GFD has been associated with adverse metabolic outcomes, including impaired glucose and lipid metabolism, as well as heightened risk of MetS and obesity [113]. As a result, ongoing research is exploring novel therapeutic targets with the aim of developing alternative or adjunctive therapies for CeD [114].
A deeper understanding of the role of the GM in the pathogenesis of CeD is expected to facilitate the development of novel preventive strategies, particularly via the early correction of dysbiosis prior to the onset of increased intestinal permeability. A multitude of review studies have highlighted the pivotal role of the GM in gluten metabolism, modulation of immune responses, and regulation of intestinal barrier integrity [78, 115–117]. These investigations have focused on targeting active CeD via modulation of the GM, including the use of probiotics, postbiotics, and synbiotics to restore beneficial microbial communities and promote SCFA-producing commensals, as well as the utilization of naturally occurring gluten-degrading microbial enzymes. Table 1 summarizes human studies that have employed microbial-based interventions in the therapeutic management of CeD.
Microbial tools used for celiac disease treatment in human studies
| Study | Microbial tool | Main outcomes |
|---|---|---|
| Gluten-degrading bacteria | ||
| Caminero et al. [118] | Cultivable gut microbiota | Thirty-five bacterial species were involved in gluten metabolism. The main genera were Lactobacillus, Streptococcus, Staphylococcus, Clostridium, and Bifidobacterium. |
| Francavilla et al. [119] | Eighteen commercial strains of probiotic lactobacilli | Ten bacterial strains provided the peptidase repertory required to completely degrade the immunogenic gluten peptides involved in CeD. |
| Herrán et al. [62] | Bacterial species isolated from duodenal biopsies | Thirty-two bacterial species showed extracellular proteolytic activity against gluten protein. They were included within the genera Actinomyces, Bacillus, Bifidobacterium, Lactobacillus, Neisseria, Prevotella, Pseudomonas, Staphylococcus, Stenotrophomonas, Streptococcus, Veillonella, and Virgibacillus. |
| Moreno Amador et al. [120] | A bacterial strain belonging to the species Chryseobacterium taeanense isolated from the rhizosphere | The strain showed the presence of prolyl endopeptidases and the hydrolytic capacity of the gluten immunogenic peptides. Glutenase activity was detected in the extracellular medium, where gel electrophoresis and gliadin zymography identified the presence of about 50 kDa gluten-degrading enzyme. |
| Probiotics | ||
| Francavilla et al. [121] | N = 109 diagnosed patients with CeD with IBSTreatment for 6 weeksProbiotic cocktail: Lacticaseibacillus casei (L. casei), Lactiplantibacillus plantarum (L. plantarum), Bifidobacterium infantis (B. infantis) subsp. lactis, and Bifidobacterium breve (B. breve) (2 strains) | Gastrointestinal symptoms and the severity of IBS substantially decreased in the probiotic-treated group compared to the placebo. Lactic bacteria, Staphylococcus, and Bifidobacterium increased in patients receiving probiotic treatment. |
| Håkansson et al. [122] | N = 78 children with CeD autoimmunityTreatment for 24 weeksProbiotics: L. plantarum strain HEAL9 and Lactocaseibacillus paracasei (L. paracasei) strain 8700:2 | Daily oral administration of probiotics modulates the peripheral immune response in children with CeD autoimmunity. Over time, median levels of IgA-tTG decreased more markedly in the probiotic group compared to the placebo group, whereas an opposite trend was observed for IgG-tTG levels. |
| Harnett et al. [123] | N = 45 diagnosed patients with CeDTreatment for 12 weeksProbiotic cocktail: VSL#3 consists of Streptococcus thermophilus, B. breve, Bifidobacterium longum (B. longum), B. infantis, Lactobacillus acidophilus, L. plantarum, L. paracasei, and Lactobacillus delbrueckii subsp. bulgaricus | The primary outcome indicated that the probiotic formulation did not result in significant alterations in the GM composition between baseline and week 12. |
| Jenickova et al. [124] | N = 78 (40 genetically predisposed children having tTG autoantibodies and 38 healthy controls)Treatment for 24 weeksProbiotics: L. plantarum strain HEAL9 and L. paracasei strain 8700:2 | The findings indicate a modest yet significant impact of probiotic supplementation on the fecal metabolome, primarily affecting proteolytic pathways within the gut. Over the six-month intervention period, stool concentrations of 4-hydroxyphenylacetate increased in the probiotic group compared to controls, whereas levels of amino acids such as threonine, valine, leucine, isoleucine, methionine, phenylalanine, aspartate, and the intermediate fumarate were reduced. |
| Klemenak et al. [125] | N = 49 children diagnosed with CeDTreatment for 12 weeksProbiotics: B. breve strain BR03 and strain B632 | Probiotic intervention using B. breve strains demonstrated a beneficial effect by reducing the production of the pro-inflammatory cytokine TNF-α in children with CeD adhering to a GFD. |
| Lionetti et al. [126] | N = 96 children diagnosed with CeDTreatment for 12 weeksProbiotics: L. casei, L. plantarum, B. infantis subsp. lactis, and B. breve (2 strains) + GFD | Treatment with a multispecies probiotic resulted in a more rapid and pronounced increase in BMI among children newly diagnosed with CeD. |
| Olivares et al. [127] | N = 36 children diagnosed with CeDTreatment for 12 weeksProbiotic: B. longum strain CECT 7347 | Decreased peripheral CD3+ T lymphocytes and slightly reduced TNF-α concentration were obtained in the experimental group. Comparison between the groups revealed that the administration of probiotics reduced the numbers of the Bacteroides fragilis group and the content of sIgA in stools compared to the administration of a placebo. |
| Pinto-Sánchez et al. [128] | N = 24 untreated CeD patientsTreatment for 6 weeksProbiotic: B. infantis subsp. lactis strain NLS-SS | The patients treated with GFD for 1 year showed a decrease in duodenal macrophages, whereas probiotic treatment decreases Paneth cell counts and expression of α-defensin-5 in CeD patients. |
| Primec et al. [129] | N = 40 children with CeDTreatment for 12 weeksProbiotics: B. breve strain BR03 and strain B632 | Probiotic administration showed a negative relationship between Bacillota and pro-inflammatory TNF-α. In addition, probiotic effect exposed new phyla, particularly Synergistota, which negatively correlated to acetic acid and total SCFAs, indicating a potential role in microbiome restoration. |
| Quagliariello et al. [130] | N = 40 children with CeDTreatment for 12 weeksProbiotics: B. breve strain BR03 and strain B632 | The effects of the probiotics produce an increase in members of the phylum Actinomycetota and a re-establishment of the physiological Bacillota/Bacteroidota ratio. |
| Synbiotics | ||
| Tremblay et al. [131] | Commercial Synbiotic: probiotics Lactocaseibacillus helveticus strain Rosell®-52, B. infantis subsp. lactis strain Rosell®-33, and Bifidobacterium. bifidum strain Rosell®-71 with the prebiotic fructooligosaccharides | A review of twelve studies demonstrated that synbiotic administration significantly enhances the efficacy of standard diarrhea treatments, independent of the underlying etiology. In eight of these studies, synbiotic use was associated with improved immune function, as evidenced by increased levels of various immune competence and mucosal immunity markers, alongside a reduced incidence of common infections. Furthermore, probiotic supplementation was found to improve the therapeutic outcomes of iron deficiency anemia. |
| Postbiotics | ||
| Freire et al. [132] | Patient-derived organoids monolayers. Microbiota-derived bioproducts from Bacteroides fragilis, including butyrate, lactate, and polysaccharide A | Monolayers derived from CeD organoids exposed to gliadin showed increased intestinal permeability and enhanced secretion of pro-inflammatory cytokines compared to non-celiac controls. Microbiota-derived bioproducts, butyrate, lactate, and polysaccharide A, improved barrier function and reduced gliadin-induced cytokine secretion. These bioproducts can be used to modulate the epithelial response to gluten. |
BMI: body mass index; CeD: celiac disease; GFD: gluten-free diet; GM: gut microbiome; IgA: immunoglobulin A; tTG: tissue transglutaminase; IBS: irritable bowel syndrome; SCFAs: short-chain fatty acids; sIgA: secretory IgA; TNF-α: tumor necrosis factor alpha
Certain bacterial taxa have been identified for their dual roles in gluten metabolism: some degrade gluten peptides that trigger strong immune responses, while others possess the ability to detoxify these peptides. A combination of Lactobacillus and Bifidobacterium strains has been shown to hydrolyze the immunogenic gliadin 33-mer peptide generated during gluten digestion by pepsin and trypsin. In vitro studies using Caco-2 cell lines demonstrated that these bacterial strains produce low-molecular-weight peptides and inhibit the release of the pro-inflammatory cytokine IL-6, as well as the differentiation of cytotoxic T cells [133]. Another study utilizing the same cell model revealed that B. longum and B. bifidum strains reduced gliadin-induced activation of the NF-κB p65 signaling pathway, alongside decreased production of TNF-α and IL-1β. This protective effect was mediated by the degradation of gliadin peptides, resulting in diminished cytotoxicity [134]. Similarly, in a mouse model replicating CD4+ T cell-mediated enteropathy in response to gliadin, administration of B. longum strain CECT 7347 increased IL-10 levels and decreased CD4+ T cell populations, thereby mitigating gliadin’s deleterious effects [135]. In addition, co-culturing this strain with Caco-2 cells exposed to gliadin enhanced cell viability and prevented gliadin-induced alterations in key proteins, including regulator of G-protein signaling 5, actin filament-associated proteins, sorting nexin-20, and T cell receptor R chain V region CTL-L17, which are typically upregulated during pro-inflammatory responses [136].
Another potential mechanism by which bacteria may influence CeD development involves the production of aryl hydrocarbon receptor (AhR) ligands. The AhR is activated in various cell types by indole-containing ligands derived from tryptophan metabolism, many of which are produced by the GM. AhR plays a critical role in modulating host immune responses. Notably, a study demonstrated that AhR activation by bacterial tryptophan metabolites inhibits the activation of actin-regulatory proteins MyoIIA and ezrin [137]. This inhibition helps maintain the integrity of tight and adherens junctions, which are essential for preserving the structural stability of enterocytes and, consequently, the intestinal barrier. Preservation of these junctions contributes to decreased intestinal permeability, a key factor in CeD pathogenesis.
In the intestinal mucosa of patients with active CeD, AhR expression is decreased. A study reported that these patients exhibited reduced levels of AhR ligands in stool samples, and their GM displayed a diminished capacity to activate this receptor compared to non-celiac controls [138]. Using a murine model expressing the HLA-DQ8 susceptibility gene, the researchers modulated the intestinal microbiota via a tryptophan-enriched diet. This intervention enhanced the production of AhR ligands and subsequent receptor activation, which mitigated gluten-induced immunopathology. Furthermore, a study utilizing intestinal organoids co-cultured with lamina propria lymphocytes demonstrated that a metabolite produced by a strain of Limosilactobacillus reuteri stimulated lamina propria lymphocytes to secrete IL-22 through AhR activation. This process promoted the proliferation of intestinal stem cells and facilitated epithelial recovery following TNF-α-induced damage [139]. Another beneficial bacterial mechanism has been identified in a preclinical study using a DQ8 mouse model of gluten sensitivity, where the protective effect of bifidobacteria was attributed to the production of a serine protease inhibitor, known as serpin, which prevented gliadin-induced immunopathology [140]. In addition, other microbial metabolites have shown the ability to modulate both the epithelial barrier and the immune system. Freire et al. [132] demonstrated that organoids derived from CeD patients exhibited a distinct response to gliadin and an improvement in barrier function when treated with bacterial metabolites such as lactate and butyrate, as well as B. fragilis polysaccharide A. These microbial bioproducts, termed postbiotics, enhanced the expression of genes involved in mucin production, trefoil factor 1 (TFF1), and claudin-18, genes known to be downregulated in CeD organoids. Furthermore, Serena et al. [141] identified a direct correlation between the alternative splicing of FOXP3 isoforms and the beneficial bacterial metabolite butyrate. FOXP3, which is a key transcription factor regulating T cell development and function, exists in multiple splicing variants in humans, and its deficiency is a critical factor in systemic autoimmunity. This study showed that butyrate, together with IFN-γ, upregulated FOXP3 isoforms in the intestinal tissue of CeD patients.
Microbial TG (mTG) is a commonly used food additive and has been identified as a potential inducer of autoimmune and neurodegenerative diseases [142, 143]. This enzyme increases intestinal permeability, suppresses mechanical (mucus) and immunological (anti-phagocytic) enteric protective barriers, stimulates luminal bacterial growth, and enhances the uptake of gliadin peptide. mTG and gliadin molecules are co-transcytosed through the enterocytes and subsequently deposited subepithelially. Additionally, mucosal dendritic cell surface TG induces gliadin endocytosis, and enzyme-treated wheat products elicit immune reactivity in CeD patients [144, 145]. Recently, Lerner et al. [146] found that sequence similarity and cross-reactivity between mTG and various tissue antigens could underlie the link between mTG and autoimmune disorders. Furthermore, cross-reactivity and sequence homology between gluten/gliadin peptides and human epitopes may contribute to molecular mimicry, potentially triggering autoimmunity. A GFD has been shown to prevent these phenomena via various mechanisms [147].
CeD is linked to both internal genetic factors and potential external influences, such as dietary habits and antibiotic use [148]. These factors can alter the microbial composition, leading to dysbiosis, which may increase the risk of developing CeD later in life [149]. The diagnosis of CeD involves a combination of clinical, serological, and histopathological data. In children, diagnosis can be performed without biopsy, and it is based on strict criteria, including small bowel symptoms, positive HLA-DQ2/DQ8, as well as IgA and tTG levels [150]. In elderly subjects, the diagnosis still requires the presence of duodenal villous atrophy, and it is carried out through the analysis of IgA/IgG anti-tTG and anti-endomysium antibodies in a small intestinal biopsy [151].
The findings of this review underscore several important implications for managing patients diagnosed with CeD. First, the results challenge the traditional belief that CeD patients universally suffer from poor nutritional status due to malabsorption. Instead, it is common for patients with CeD to present with MASLD and MetS at diagnosis. Second, the rising prevalence of MASLD and MetS after starting a GFD requires serious attention, as the severity of these conditions may worsen with prolonged adherence to GFD. Third, both MASLD and MetS have been shown to increase the risk of CVD, stroke, T2DM, and cirrhosis [16, 152, 153]. Therefore, it is crucial to take preventive steps to avoid the onset of MASLD and MetS in CeD patients. Specifically, patients should be screened for MASLD and MetS at diagnosis using standardized tests, enabling closer monitoring for those already affected at the start of GFD. Furthermore, ongoing monitoring after beginning GFD is essential to identify and manage any delayed complications. Lastly, patients must be informed about the risks of developing metabolic complications and consistently counseled to maintain a balanced diet and engage in regular physical activity.
The only current treatment for CeD is a strict GFD, which is often difficult to maintain and costly, leading to high rates of non-adherence. Moreover, despite following a GFD, 25–50% of patients fail to show significant clinical improvement [68]. The requirement for continuous monitoring of food intake has been shown to negatively impact patients’ quality of life, highlighting an unmet need for adjunctive therapies [154]. Therefore, ongoing research aimed at discovering novel and supplementary treatments for CeD is imperative [155]. It is crucial to recognize the need for additional prospective studies with larger sample sizes and standardized definitions of MetS. Such studies are essential for thoroughly assessing the impact of a GFD on MetS development in CeD patients. Furthermore, the pathophysiological mechanisms underlying MetS in individuals with CeD remain unclear. Thus, it has to be determined whether the same processes that drive MetS in non-CeD populations also contribute to its development in CeD populations or if distinct immunologic or inflammatory pathways are involved [8].
Notably, adherence to a GFD has been shown to negatively affect microbial homeostasis in healthy individuals [156]. However, a major limitation in current research is the lack of longitudinal studies analyzing GM composition in CeD patients before and after the initiation of a GFD. The typical GFD often relies heavily on ultra-processed and refined foods that are high in fat and sugar while being low in dietary fiber, folic acid, iron, calcium, selenium, magnesium, zinc, niacin, biotin, riboflavin, pyridoxine, and vitamin D [157]. This highlights the broader shortcomings of the Western diet, which are particularly detrimental for individuals with CeD. As an alternative, CeD patients should be encouraged to adopt a Mediterranean or vegetarian dietary pattern that emphasizes the consumption of seasonal, organic vegetables and foods rich in fiber, micronutrients, and bioactive vitamins [158, 159]. In this context, incorporating pseudocereals such as quinoa, amaranth, and sorghum, as well as naturally gluten-free cereals, is recommended due to their richness in fiber, minerals, thiamine, carotenoids, flavones, tannins, proteins, and healthy fats [157]. In addition, ketogenic diets have been recognized for their efficacy in managing various metabolic conditions, as well as for their potential to modulate autoimmune diseases by reducing inflammation [160]. Building on these dietary patterns, a plant-based ketogenic diet has been proposed as a potential health-promoting approach [161]. For patients with CeD, this diet may offer promising benefits when appropriately adapted to exclude gluten and meet individual patient requirements. However, the potential benefits of ketogenic diets for CeD are primarily based on theoretical considerations, and research in this area is needed to determine their efficacy. Furthermore, nutraceutical supplementation, including the targeted use of probiotics, has emerged as a promising strategy to support both nutritional balance and gut health in CeD patients [162].
A range of probiotics has been investigated in the context of CeD. However, current evidence from clinical studies remains inconclusive regarding their efficacy. A recent meta-analysis concluded that probiotics may alleviate gastrointestinal symptoms in individuals with CeD, yet emphasized that higher-quality studies are necessary before firm recommendations can be made, given the heterogeneity among existing trials [163]. Notably, most probiotics studied thus far have been selected based on their general anti-inflammatory properties rather than their specificity to CeD pathophysiology. It is important to emphasize the specificity of probiotic strains, such as B. longum CECT 7347, which have shown efficacy in clinical applications [127, 135]. In this respect, the therapeutic potential of probiotics is strain-dependent, and selecting the appropriate strain is crucial for achieving the desired clinical outcomes. Future probiotic and microbial-based interventions should focus on strains that target pathways directly implicated in CeD. For example, the enzymatic degradation of immunogenic wheat proteins represents a promising research direction, as it could mitigate the inflammatory responses triggered by gluten and other wheat-derived peptides. In addition, the development of combination therapies using multiple strains that act synergistically, or a single probiotic engineered to affect multiple targets, holds potential. Nevertheless, these strategies must be guided by a clear mechanistic rationale to optimize both efficacy and safety. In this context, the use of genetically engineered microbes customized to modulate key immune or metabolic pathways in CeD also requires further investigation. Precision probiotics and postbiotics represent a promising avenue, but their role in CeD and related metabolic conditions remains to be fully defined, as their use is still at an early stage and more evidence is needed to clarify their clinical relevance.
The next generation of microbial therapeutics represents an emerging class of pharmaceuticals, encompassing live biotherapeutic products (LBPs) and genetically modified microorganisms engineered to express or secrete bioactive molecules relevant to the pathogenesis of CeD [164]. The therapeutic efficacy of engineered bacteria has been demonstrated in animal models of CeD. However, their translation to human use remains limited, primarily due to safety concerns associated with plasmid-based gene delivery systems [165]. Recent advances have proposed novel approaches in which genes of interest are stably integrated into the chromosomal DNA of probiotic strains, thereby minimizing the risk of horizontal gene transfer and enhancing their potential suitability for clinical application [166]. In addition to the detoxification of immunogenic gluten peptides, emerging microbial therapeutic strategies for CeD include the restoration of AhR signaling through microbial modulation of tryptophan metabolism, as well as the reestablishment of intestinal proteolytic homeostasis via the production of serine protease inhibitors [138]. These mechanisms offer promising avenues for the development of adjuvant therapies aimed at modifying disease progression and improving outcomes in patients with CeD.
A recent review by Herrera-Quintana et al. [167] outlined several emerging therapeutic strategies for the prevention and treatment of CeD. Among these, oral enzyme therapy has garnered attention for its ability to degrade immunogenic gluten peptides within the gastrointestinal tract. Agents such as latiglutenase, zamaglutenase, and AGY-010 are currently under investigation for their capacity to neutralize gluten toxicity by hydrolyzing immunodominant epitopes in the stomach prior to their interaction with the intestinal immune system [168]. Another promising therapeutic avenue involves targeting tissue TG2, an enzyme central to CeD pathogenesis. TG2-mediated deamidation enhances the binding affinity of gluten peptides to HLA-DQ2/DQ8 molecules, thereby promoting CD4+ T helper cell activation and the subsequent release of pro-inflammatory cytokines [169, 170]. Inhibition of TG2 enzymatic activity has thus emerged as a viable strategy for mitigating gluten-driven immune activation [171]. Additionally, monoclonal antibody (mAb)-based therapies targeting inflammatory mediators have shown clinical promise. IL-15, which is a key cytokine implicated in CeD, plays a crucial role in the activation of intraepithelial cytotoxic CD8+ T cells, leading to epithelial damage and villous atrophy [172]. In a clinical trial evaluating AMG 714, a mAb directed against IL-15, patients with CeD demonstrated significant improvement in clinical symptoms, particularly diarrhea, underscoring the potential of cytokine-targeted therapies [173].
Inflammation is a shared pathophysiological hallmark of CeD, MetS, and MASLD, despite their differing primary etiologies and target tissues. Emerging biomarkers, such as the systemic immune-inflammation index and uric-acid-to-creatinine ratio, reflect this inflammatory burden in MASLD and MetS, respectively [174, 175]. This shared inflammatory process may partly explain their clinical convergence and supports further investigation into common immunometabolic pathways. In addition, the interaction between the GM and sIgA has been shown to modulate intestinal inflammation, with shifts in GM composition potentially playing a role in reducing inflammatory responses [176]. Thus, the convergence of immunometabolic dysfunction in these conditions suggests that targeting inflammatory pathways may offer therapeutic benefits across disease contexts, requiring integrated treatment strategies.
Within the context of this discussion, it is also important to highlight that CeD is associated with a range of psychiatric manifestations, including depression, anxiety, eating disorders, autism spectrum disorder, attention deficit/hyperactivity disorder, bipolar disorder, schizophrenia, and mood disorders [177]. Nutritional psychiatry is an emerging field that employs rigorous scientific methods to evaluate the efficacy and define appropriate therapeutic applications of dietary supplements and nutraceuticals in individuals with and without mental health conditions [178]. This approach addresses safety concerns and side effects commonly associated with pharmacological treatments, such as dyslipidemia, altered glucose metabolism, extrapyramidal symptoms, sexual dysfunction, weight gain, MetS, and T2DM [178]. Consequently, nutritional psychiatry may play a pivotal role in CeD management, as personalized dietary interventions could not only alleviate disease-specific symptoms but also improve comorbid psychiatric conditions. Moreover, nutritional psychiatry encompasses the use of psychobiotics, which are a novel class of psychotropic agents that include live microorganisms and bioactive compounds demonstrated to be effective in treating stress, anxiety, and depression [179]. Thus, these therapeutic strategies hold significant potential as adjunctive treatments in CeD.
As with all narrative reviews, the present study is subject to several inherent limitations. First, considerable heterogeneity was observed across the reviewed studies, stemming from differences in sequencing technologies, experimental protocols, analytical pipelines, and sample types. These methodological discrepancies complicate cross-study comparisons and hinder the identification of consistent microbial biomarkers for tracking disease progression. Furthermore, differences in sample collection procedures and storage conditions may introduce additional variability, impacting the reproducibility and reliability of findings. This underscores the need for standardized methodologies and reporting guidelines to improve study comparisons and facilitate meta-analyses. Second, short-term studies typically highlight the immediate alleviation of gastrointestinal symptoms and inflammatory markers, but they often fail to capture the long-term impact on metabolic dysfunctions commonly associated with CeD. These metabolic complications may persist even in patients adhering to a GFD, and their long-term consequences are still not fully understood. Longitudinal studies are crucial to determine whether microbial interventions can provide sustained benefits in modifying the GM in a way that addresses not only the acute inflammatory responses but also the chronic metabolic imbalances of CeD. Moreover, the dynamic nature of the GM and its complex interactions with diet, lifestyle, and disease progression require longer intervention periods to assess the potential of microbial therapies in preventing or mitigating metabolic dysfunctions over time. Third, the diagnosis of CeD remains challenging due to the broad spectrum and non-specific nature of clinical manifestations. A substantial proportion of individuals with CeD remain undiagnosed, with an average diagnostic delay of approximately 12 years [180]. In this context, predictive models aimed at estimating CeD risk based on symptomatology and clinical risk factors offer potential utility. However, these models often demonstrate limited efficacy when relying solely on clinical data [181]. To address these challenges, future studies should consider the following recommendations: (i) the incorporation of microbial biomarkers derived from stool samples, which offer a non-invasive and accessible diagnostic modality, may significantly enhance the predictive performance of current risk models; (ii) the integration of microbial signatures with clinical markers of mucosal integrity could provide a more robust framework for disease monitoring and prognosis. Notably, evidence suggests that the GM composition differs among CeD patient subgroups with varying clinical phenotypes, indicating a potential role of GM in the persistence of symptoms despite adherence to a GFD [182].
CeD constitutes a heterogeneous condition with diverse clinical presentations and underlying mechanisms, which complicates the development of a universal treatment strategy. The emergence of personalized medicine is likely to become increasingly important, as it offers the potential for customizing therapeutic interventions to the individual’s genetic, immunological, microbial, and metabolic profiles. Consequently, this personalized approach may enhance treatment efficacy and minimize adverse outcomes. Moreover, it is pivotal to educate patients about the potential risks associated with CeD and its management, particularly those related to metabolic complications and dietary imbalances. Encouraging the adoption of a healthy lifestyle mainly consisting in a nutritionally plant-based balanced diet and regular physical activity should be an integral component of comprehensive care for individuals with CeD.
A synthesis of existing studies underscores the potential of several protective factors and targeted interventions for future research aimed at preventing CeD. Among these, the adoption of a health-promoting dietary pattern within the first two years of life, such as the Mediterranean or vegetarian diet, has demonstrated a protective role in reducing CeD risk. These diets are rich in dietary fiber and phytochemicals, which foster the growth of beneficial commensal gut microbiota and support the production of SCFAs. SCFAs, in turn, play a critical role in maintaining intestinal barrier integrity by enhancing mucus production and upregulating the expression of tight junction proteins. Furthermore, SCFAs exert immunomodulatory effects, promoting immune tolerance through the induction of Tregs, which secrete IL-10 and mitigate pro-inflammatory Th1 responses and autoantibody production. These findings highlight the relevance of early-life nutritional strategies as a foundation for future preventive approaches in CeD.
Precision probiotics have demonstrated multifaceted therapeutic potential in the context of CeD. These probiotics exert their effects through several key mechanisms. They attenuate inflammatory responses associated with CeD by disrupting the activity of pathogenic and pro-inflammatory microbial species and restoring eubiotic populations that produce SCFAs. In addition, certain microbial strains synthesize peptidases capable of degrading immunogenic gliadin peptides, thereby mitigating antigenic stimulation. Precision probiotics also contribute to immune homeostasis by enhancing Treg activity and modulating intestinal barrier integrity through the regulation of tight junction proteins. Moreover, they are capable of producing AhR ligands, which are associated with increased IL-22 production, enhanced intestinal stem cell proliferation, and the repair of mucosal injury. Complementing these effects, postbiotics have been shown to further support gut barrier function by reinforcing tight junctions and preventing gliadin-induced inflammatory effects. All these promising outcomes highlight the potential of precision probiotics and postbiotics. However, further well-designed clinical studies are needed to confirm their proven efficacy.
Therefore, the integration of clinical models with microbial biomarkers holds considerable promise for enhancing both the diagnosis and longitudinal monitoring of CeD. This combined approach would substantially improve current clinical practice by enabling more accurate risk stratification, earlier detection, and personalized management strategies that reflect the heterogeneous nature of the disease.
AhR: aryl hydrocarbon receptor
ASVs: amplicon sequence variants
BMI: body mass index
CeD: celiac disease
CVD: cardiovascular disease
E. coli: Escherichia coli
GFD: gluten-free diet
GM: gut microbiome
GRDs: gluten-related disorders
HDL: high-density lipoprotein
IFN-γ: interferon-gamma
IgA: immunoglobulin A
mAb: monoclonal antibody
MASLD: metabolic dysfunction-associated steatotic liver disease
MetS: metabolic syndrome
mTG: microbial transglutaminase
NCGS: non-celiac gluten sensitivity
NGS: next-generation sequencing
PPIs: proton pump inhibitors
SCFAs: short-chain fatty acids
sIgA: secretory immunoglobulin A
T2DM: type 2 diabetes mellitus
tCD: treated celiac disease
tCD-TG−: negative transglutaminase serology
tCD-TG+: positive transglutaminase serology
TNF-α: tumor necrosis factor alpha
tTG: tissue transglutaminase
ABR: Conceptualization, Investigation, Writing—original draft, Writing—review & editing. JJB: Conceptualization, Investigation, Writing—original draft, Supervision. Both authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 4464
Download: 41
Times Cited: 0
Natalia Baryshnikova ... Valeria Novikova
