Affiliation:
1DIITET Department, National Research Council (CNR), Institute for Systems Analysis and Computer Science, 00185 Rome, Italy
2Experimental Neuroscience and Neurological Disease Models, Fondazione Santa Lucia, 00143 Rome, Italy
Email: cinzia.volonte@cnr.it; c.volonte@hsantalucia.it
ORCID: https://orcid.org/0000-0001-7362-8307
Affiliation:
3Molecular Neurobiology Laboratory, Department of Biochemistry and Molecular Biomedicine, Faculty of Biology, Universitat de Barcelona, 08028 Barcelona, Spain
4CiberNed, Network Center for Neurodegenerative Diseases, National Spanish Health Institute Carlos III, 28029 Madrid, Spain
5School of Chemistry, Universitat de Barcelona, 08028 Barcelona, Spain
ORCID: https://orcid.org/0000-0003-2549-4919
Explor Neuroprot Ther. 2023;3:429–434 DOI: https://doi.org/10.37349/ent.2023.00059
Received: July 13, 2023 Accepted: September 19, 2023 Published: November 29, 2023
Academic Editor: Shile Huang, Louisiana State University Health Science Center, USA
Purinergic P2 and P1 receptors respond to the most prevalent and ancestral chemical messengers existing in the animal and plant kingdoms, the natural nucleotide ligands adenosine triphosphate (ATP), adenosine diphosphate (ADP), guanosine triphosphate (GTP), cytidine triphosphate (CTP), uridine 5’-triphosphate (UTP), uridine diphosphate (UDP), inosine triphosphate (ITP), and the purine and pyrimidine nucleosides adenosine, guanosine, inosine, etc. These molecules are basically present in all cells because they participate in the energy metabolism and in the anabolism/catabolism of nucleic acids. Evolution has however granted an extra value to nucleotides and nucleosides. They are present also in the extracellular space where they act as specific ligands for cell surface receptors, the purinergic P2 and P1 receptors respectively for nucleotides and nucleosides, which are similarly ancestral and widely expressed not only in eukaryotes, but also in prokaryotes. Perhaps because of this wide-ranging distribution, the purinergic receptors are characterized by a vast diversification of features at the molecular and functional level.
P2 receptors are categorized into ion channels (P2X) and G protein-coupled proteins (P2Y). Seven ionotropic P2X (P2X1–7) and eight metabotropic P2Y types (P2Y1, P2Y2, P2Y4, P2Y6, P2Y11, P2Y12, P2Y13, and P2Y14) have been cloned so far from mammalian species. In addition, four adenosine P1 receptors have been identified (A1, A2A, A2B, A3), all belonging to the G protein-coupled receptor family. Each purinergic receptor possesses unique molecular and pharmacological properties and this diversification and heterogeneity extends also to their functions. Indeed, both P1 and P2 receptors are directly or indirectly involved in a plethora of distinct biological events and moreover participate in several pathological conditions [1–3]. Despite thriving research over the years on purinergic signaling, their sophistication is however staggering and intriguing even today, more than 50 years after the seminal discovery by Geoffrey Burnstock that ATP is a neurotransmitter, particularly the neurotransmitter for non-adrenergic inhibitory nerves in the gut [4]. Regardless of fervent research on purinergic receptors, several issues remain unanswered so far, and the two most essential are i) the reason why there are several different receptors for purines and pyrimidines expressed in each cell phenotype, and ii) why there are several functionalities that are associated with the activation of these receptors.
Focusing particularly on these unknowns, the combinatorial receptor web model was formulated a while ago with the purpose of categorizing such a large complexity into an intelligible paradigm [5]. The combinatorial receptor web model was based on the fact that multiple P2X, P2Y, and P1 receptors can actually be co-expressed on the same cell [6–10], and combine to form unique complexes that correlate with unique cellular functions [5]. A simple combinatorial calculation indeed demonstrated that combinations of P2 and/or P1 receptors, instead of single receptors, may be associated with a larger variety of cellular actions, thus amplifying and diversifying the response to extracellular purine/pyrimidine nucleotides. According to the combinatorial receptor model: i) multiple purinergic subtypes are enrolled for performing an increased number of unique functions; ii) the expression of P2 and P1 receptors is a dynamic event that is regulated, among other, by the extracellular environment; iii) P2 and P1 proteins combined in complexes are more than the sum of their single entities, thus satisfying features of molecular cooperation and receptor cross-talk [5].
Based on the “chunking theory”, which is about a collection of elements that have strong associations with each other but weaker links to items within other collections, the combinatorial receptor web model has then naturally evolved into the “chunking model” with the intent of further explaining the grouping of purinergic receptors [11]. The notion of “chunk” was introduced back in 1956 in the psychology field by Miller [12] to describe how information is processed, and to distinguish between “units of information” and “chunks of information” (containing various units), in other words recognizing “the importance of grouping or organizing the input sequence into units or chunks”. In the field of receptor pharmacology, and particularly in the field of purinergic receptors [11], the chunking consists of clustering receptors that are simultaneously expressed on a given cell at a given time, in order to participate in a function. In this way, specific purinergic chunks can generate novel collective receptor features that are different from those of single/individual receptors. In this way, further diversification can be reached, for instance in agonist and/or antagonist selectivity, channel and/or desensitization properties, signal transduction mechanisms, all leading to further functional divergence and versatility. The chunking theory finds also a correlate in the receptor mosaic hypothesis of the engram previously proposed for G protein-coupled receptors [13], which establishes that “aggregates” of different receptor species (mosaics) may form for instance in neuronal membranes (typically synapses) and constitute a memory trace (engram) of their activity [14].
Based on these mosaics of receptors [15], or chunks [11], occurring within membrane compartments [16, 17], one could explain why a given receptor, for instance P2Y1, may stimulate nitric oxide synthesis in hypothalamus, amygdala, hippocampus [18], promote excitatory conductance in hippocampal interneurons [19], inhibit ion fluxes in glutamatergic cortical neurons [20], modulate extra-synaptic cross-depolarization in dorsal root ganglia [21], induce cell death in prostate cancer cells [22], and improve vasodilatory capacity in arteries [23]. However, plastic changes may also occur with a given chunk. For instance, the chunk containing P2Y1 may change in response to the environment to lead to a different P2Y1-containing chunk, which in turn may lead to another cell response when different purines/pyrimidines are present in the extracellular environment.
A way to explain this functional variance is that P2Y1 is comprised in different chunks within each different cellular context, and acts in synergy (or antagonism) with different P2/P1 receptors while responding to extracellular nucleotides. It is important to note that the simple belonging to a chunk can impact not only on functional responsiveness, as just described, but also on ligand binding and signal transduction mechanisms. For instance, all four adenosine receptors are known to interact with each other to form homodimers and/or heterotetramers, and may be part of receptor chunks. In particular, when A2A and A3 receptors form a complex in cortical neurons, the A3 functionality and signaling becomes negligible, but selective A2A antagonists then cause A3 activation [24]. Moreover, the A2A can form complexes, directly interact, and display cross-antagonism also with N-methyl-D-aspartic acid (NMDA) receptors in microglia, a mechanism useful to prevent exacerbation of NMDA activity by A2A antagonists [25]. A close interrelationship is recently demonstrated in microglia also between A2A and CB2 receptors that are able to physically interact and affect the signaling of each other, likely by conformational changes within the A2A-CB2 receptor heteromer. Remarkably, blockade of A2A results in increased CB2-mediated signaling, thus suggesting that A2A antagonists may potentiate the neuroprotective action of endocannabinoids [26].
What reported so far has highlighted the high presence of purinergic receptors in various cell phenotypes, their evident cell phenotype-specificity, and strong cooperativity. Among the most intriguing feature, there is the co-expression within the same cell type of both ionotropic and metabotropic receptors, which are moreover coupled to different signal transduction mechanisms, for instance to stimulatory G protein-coupled receptors (Gs) or inhibitory G protein-coupled receptors (Gi), respectively activating or inhibiting the adenylate cyclase pathway. As a consequence of this grouping, the chunk can be interpreted as a unique functional unit, a concentration-sensing device, a relay system with the role of integrating and modulating the activation of single receptors into a cumulative global response. The presence of these “chunks” for instance in nerve terminals may lead to a particular regulation of neurotransmitter release. If only one type of receptor is expressed, only one type of regulation is allowed. By the co-expression and cross-interaction for instance of A1 and A2A receptors, adenosine may coordinate and fine-tune, either inhibit or enhance, neurotransmitter release, exactly depending on the local and time-dependent concentration of the nucleoside present in the synaptic cleft [27].
In this way, for each given cellular condition, the chunk would comprise different receptor units and take on a particular dynamic shape, as for instance exemplified by the different arrangements of the units, or seeds, that macroscopically constitute a dandelion (Figure 1A). Considering for instance four different interacting purinergic receptors on the cell surface, there could be two levels of organization, the first one due to direct interactions (heteromers) that must comply with structural restrictions (Figure 1B); the second one due to the disposition in chunks of these heteromers.
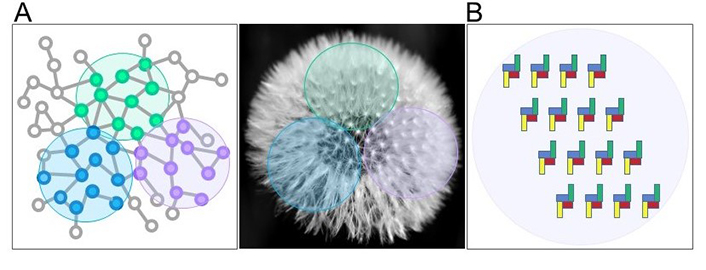
Chunk in biological contexts. (A) Units having strong associations (vicinities) with each other (blue, versus green, versus purple little dots) are clustered within single chunks (colored circle areas), as the units (the seeds) contained in the chunks (colored circle areas) of a dandelion; (B) purinergic receptors are arranged as chunks in a given cell in a particular condition: In this example the chunk is made up of heteromers formed by 4 directly interacting receptors (blue, yellow, red, and green)
In conclusion, further arguments in favor of different purinergic receptor combinations, or purinergic chunks, characterizing each cell phenotype and instructing specific and diversified cellular responses, are provided here. As such, we do suggest that purinergic receptor expression may be included in that unique regulatory gene expression signature that delineates the identity, the origin, and evolution of each cell phenotype [28]. However, a phenotype-specific purinergic signature is not immutable and can vary in response to the modulation of the extracellular environment, and of the physiological or pathological conditions. It can change with cell development, aging, metabolism, replication, differentiation, and most importantly, pharmacological treatments. While the relationship between receptor expression and function, and the identity of purinergic signatures both require further validation, this information will surely increase our knowledge about physiological and pathological processes at the molecular level. Most importantly, it will increase our ability to distinguish among different cell phenotypes and predict or refine their classification toward more functional relevance. Indeed, a cell state is a transient property that characterizes the cell in a specific surrounding context. Similarly, the expression of purinergic receptors, chunks, and signatures is a dynamic process within a mutable extracellular environment. Understanding how purinergic receptors “chunk together” into the cell surface to respond to extracellular nucleotides/nucleosides will help draw clearer boundaries among cell phenotypes and distinguish different cell states. These new insights will surely contribute to filling up the recently proposed “periodic table of cell types” [29]. However, further research is required to evaluate for instance whether quantitative associations might also exist within receptor chunks characterizing cell phenotypes and signatures, and whether synergistic or antagonistic effects might occur among different receptors. Investigating the upstream and downstream signaling networks that govern receptor expression, and understanding the functional roles of different purinergic signatures or chunks is a challenging task and provocative project that will require multidisciplinary approaches combining in vitro and in vivo studies, novel techniques, and ultimately, a higher level of image-driven in silico analysis on the expression of P2 and P1 receptors. This commentary shows the tip of the iceberg, and aims to motivate deeper knowledge about the grouping and participation of purinergic receptors in the classification of cell phenotypes and signatures.
CV: Conceptualization, Writing—original draft, Writing—review & editing. RF: Writing—original draft, Writing—review & editing. Both authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
This work was supported by FATALSDRUG Project (Progetti di Ricerca@CNR [SAC.AD002.173.058]) from National Research Council, Italy, to CV; and grant [PID2021-126600OB-I00] by Spanish [MCIN/AEI/10.13039/501100011033]; grant “ERDF A way of making Europe” by the “European Union Next Generation EU/PRTR”, to RF. The research group of the University of Barcelona is considered of excellence (grup consolidat #2021 SGR 00304) by the Regional Catalonian Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Authors 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 2101
Download: 22
Times Cited: 0
