Affiliation:
Department of Radiology, Juntendo University Graduate School of Medicine, Tokyo 113-8421, Japan
Email: yuya.saito.fwd@gmail.com
ORCID: https://orcid.org/0000-0002-4033-1424
Explor Neuroprot Ther. 2023;3:186–206 DOI: https://doi.org/10.37349/ent.2023.00047
Received: December 07, 2022 Accepted: May 18, 2023 Published: August 25, 2023
Academic Editor: Christopher A. Shaw, The University of British Columbia, Canada
Amyotrophic lateral sclerosis (ALS) is the most prevalent type of motor neuron disease (MND) and is diagnosed with a delay from the first appearance of symptoms. Surrogate markers that may be used to detect pathological changes before a significant neuronal loss occurs and allow for early intervention with disease-modifying therapy techniques are desperately needed. Using water molecules that diffuse within the tissue and experience displacement on the micron scale, diffusion magnetic resonance imaging (MRI) is a promising technique that can be used to infer microstructural characteristics of the brain, such as microstructural integrity and complexity, axonal density, order, and myelination. Diffusion tensor imaging (DTI) is the primary diffusion MRI technique used to evaluate the pathogenesis of ALS. Neurite orientation dispersion and density imaging (NODDI), diffusion kurtosis imaging (DKI), and free water elimination DTI (FWE-DTI) are only a few of the approaches that have been developed to overcome the shortcomings of the diffusion tensor technique. This article provides a summary of these methods and their potential as surrogate markers for detecting the onset of ALS at an early stage.
Amyotrophic lateral sclerosis (ALS) is the most prevalent type of motor neuron disease (MND), a neurodegenerative illness that causes muscle weakness [1]. The incidence of sporadic ALS shows little variation in Western countries, ranging from 1 to 2 per 100,000 person-years [2–6], with an estimated lifetime risk of 1 in 400 [7]. MND is caused by upper motor neuron (UMN) or lower motor neuron (LMN) lesion weakness as shown in Table 1 [8–10]. ALS is characterized by the progressive loss of UMNs in the cerebral primary motor cortex (PMC) and corticospinal tract (CST), together with degeneration of LMNs in the brainstem and spinal anterior horns. This both progressive death of UMNs and LMNs leads to respiratory failure owing to diaphragm weakness and consequently, death. The median survival duration is 3–4 years from symptom onset, while it is also reported that a survival duration is at least 10 years in over 10% of patients. Thus, ALS has a wide range of survival duration. Most ALS is a sporadic disorder (around 70–80%) and hardly distinguished from a family history case (around 10–20%) [2, 11–15]. The most common familial ALS cases include mutation of the chromosome 9 open reading frame 72 (C9orf72, present in 7.7% of ALS cases), followed by Cu/Zn-superoxide dismutase 1 (SOD1, 2.0%), never in mitosis A (NIMA)-related kinase 1 (NEK1, 1.8%), TAR DNA binding protein (TARDBP) (1.4%), and kinesin family member 5A (KIF5A, 0.8%) [11]. For risk factor and prognostic variants, compared to the above gene mutations, ataxin-2 gene (ATXN2) intermediate trinucleotide expansion in 3.9% of ALS subjects and increased ALS risk [11, 16, 17]. In spite of the fact that the disease was first described almost 200 years ago [18] and the simultaneous involvement of both the UMN and LMN has been identified as the disease’s distinctive hallmark, the diagnosis of ALS is still mostly a clinical one. Over the course of the last 30 years, the average diagnostic delay from the first appearance of symptoms has remained close to a year and a half [19]. This is a significant share of the average amount of time people live. It is quite probable that the most significant barrier that stands in the way of reducing this amount of time is a basic lack of understanding about the greater group of individuals who are at risk of developing ALS. It is possible that a higher understanding among primary care doctors and other medical professionals who are not neurologists might be of some value in lowering the amount of time it takes to diagnose [20–22]. This diagnosis delay highlights the lack of clearly identifiable surrogate markers that are currently available in ALS. In spite of the fact that there are several candidates derived from cerebrospinal fluid (CSF) and blood [1], none of them have been validated for usage in clinical settings as of yet. Despite a variety of adverse prognostic variables, such as greater age at start and bulbar rather than limb-onset symptoms, individual survival often surpasses these criteria. Age of onset and bulbar versus limb-onset symptoms [23]. It is thus of the utmost importance to develop more strong and trustworthy prognostic surrogate markers. Emerging as a possible accurate surrogate marker for ALS is neuroimaging that does not need any kind of invasive procedure, such as magnetic resonance imaging (MRI).
Differentiating features of UMN and LMN disease
| Feature | Upper Motor Neuron Lesion | Lower Motor Neuron Lesion |
|---|---|---|
| Site of the lesion | Cerebral hemispheres, cerebellum, brainstem, spinal cord | Anterior horn cell, nerve roots, peripheral nerves, neuromuscular junction, muscles |
| Muscle weakness | Quadriplegia, hemiplegia, diplegia, paraplegia | Proximal (myopathy) Distal (neuropathy) |
| Muscle tone | Spasticity, rigidity | Hypotonia |
| Fasiculations | Absent | Present (particularly tongue) |
| Tendon reflexes | Hyperreflexia | Hypo/areflexia |
Abdominal reflexes | Absent (depending on the involved spinal level) | Present |
| Sensory loss | Cortical sensations | Peripheral sensations |
Electromyography (EMG) | Normal nerve conduction Decreased interference pattern and firing rate | Abnormal nerve conduction Large motor units Fasciculations and fibrillations |
Note. Reprinted from “Concise outline of the nervous system examination for the generalist,” by Jan MM, Al-Buhairi AR, Baeesa SS. Neurosciences (Riyadh). 2001;6:16–22 (https://nsj.org.sa/content/6/1/16). CC BY-NC.
Different tissue subtypes’ water content creates contrast for MRI. MRI, a non-invasive and ubiquitous investigative technology, may provide surrogate markers for future therapy monitoring [1]. MRI’s diagnostic and prognostic significance rivals known and new neurophysiological modalities, and it may provide surrogate markers for future treatment monitoring.
When combined with clinical involvement of the UMN and LMN, as well as growing weakness in the context of preserved feeling, there are very few perceptible differential diagnoses that can be made, particularly when the sickness originates in the bulbar region (which accounts for less than 25% of all cases). Despite the fact that comprehensive lists of putative “mimics” continue to emerge in the literature, this condition is difficult to identify because of the combination of these symptoms. The new El Escorial (EE) criteria for ALS are predicated on the presentation of simultaneous UMN and LMN signs [24]. These criteria were developed by Airlie House.
Insidious development of symptoms is characteristic of ALS, and it is predicted that ~30% of the cells in the anterior horn must die off before weakness becomes noticeable [25]. The subclinical diagnosis of LMN involvement by EMG is not required but may be useful in raising diagnostic confidence by revealing involvement of numerous areas. However, this detection method is not required. Despite the neuropathological evidence that many of these individuals obviously exhibit CST involvement [26] and are recognized by the majority of physicians as part of the ALS spectrum, the EE criteria, which were established mainly as a research tool, do not take into consideration the ~20% of patients who may first present with simply LMN indications on clinical examination, which is frequently labeled progressive muscular atrophy (PMA). This is because the EE criteria were developed as a research tool in the first place. Multifocal motor neuropathy with conduction block (MFMNCB) is one of PMA’s few imitators. In those instances in which an EMG test was unable to confirm a conduction block, the evidence of obvious sub-clinical UMN involvement would make it possible to rule out the possibility of its presence more quickly. MFMNCB is a rare, treatable autoimmune disease that solely affects LMNs.
In addition, the same EE criteria exclude the lesser number of patients with UMN-only signals during the first four years of their illness history, which is called primary lateral sclerosis (PLS). This proportion is less than 5%. Patients diagnosed with PLS may be identified by their continuously lengthy life expectancy, which ranges anywhere from 10 to 20 years on average [27]. However, there is data that suggests a common neuropathological signature between ALS and this disorder, and these people generally develop LMN symptoms later in the course of the illness. It is possible that PLS is misdiagnosed as hereditary spastic paraparesis or primary progressive multiple sclerosis, two very rare disorders that affect only UMN patients.
There is little question that MRI has had its most significant influence to date via the elimination of various types of diseases [28]. Individuals with a limb-onset of symptoms should be evaluated for cervical radiculomyelopathy and lumbosacral radiculopathy, two possible mimics of ALS. About 70% of all patients fall into this category. However, MRI is now positioned to play a larger part in the diagnosis process because of the widespread acceptance of ALS as a brain neurodegenerative condition. This is because it may identify subclinical involvement of the UMN compartment, in addition to the cerebral motor and extra-motor neuron involvement, in patients with apparent LMN-only results on examination. Due to its potential to identify subclinical involvement, MRI has recently been positioned to play a larger part in the diagnostic process. MRI is set up in such a way that it may include the whole range of phenotypes that are grouped together under the “umbrella” term of MND. Recent developments in diffusion MRI, which reflects image contrast based on the diffusion of water molecules in brain tissue, have allowed for the detection of microstructural abnormalities, and a variety of diffusion MRI techniques have been used to assess diagnosis and prognosis in ALS. The diffusion MRI technique is expected to early detection and prognosis for ALS pathology and symptom and could contribute to the exploration of neuroprotective therapy.
It is expected that surrogate markers could identify pathological alterations prior to the development of severe neuronal loss and, as a result, promote early intervention with disease-modifying treatment methods. MRI, one of the many potential surrogate markers that have been presented, is an outstanding surrogate markers candidate because it offers a strong method that permits noninvasive in vivo brain examination. This makes it stand out from the other candidate surrogate markers. Diffusion MRI in particular holds great promise due to its ability to infer brain microstructural features. Microstructural white matter (WM) complexity and integrity, axonal density and organization, and myelination are all examples of these features. To do this, we make use of water molecules that displace one another inside the tissue on a micron scale as they disperse [29]. The fact that these features can be inferred by diffusion MRI makes it a potential tool. The most common diffusion MRI technique for evaluating neurodegenerative disease pathophysiology is diffusion tensor imaging (DTI). However, DTI has a number of drawbacks that have prompted the development of new technologies like diffusion kurtosis imaging (DKI), free water elimination DTI (FWE-DTI), and neurite orientation dispersion and density imaging (NODDI). This analysis provides a summary of these tools and their potential as surrogate markers for identifying and halting the progression of prevalent neurodegenerative diseases.
Brownian motion, commonly known as random thermal motion, describes the movement of water molecules as they diffuse across space in three dimensions. When applied to water molecules, isotropy happens when the diffusion of water is completely unfettered. Water diffusion in the brain can be categorized into isotropic diffusion, in which water diffuses equally in all directions [e.g., in CSF and gray matter (GM)]. When water diffusion takes on a particular pattern, we say that the flow of water is anisotropic since it is not purely at random. Anisotropy, or anisotropic diffusion (like water moving down the straws in a glass), represents WM is a phenomenon in which the direction of diffusion changes. If there is a larger degree of anisotropy, then the diffusion of water molecules will be more directed and linear. These principles enable clinically meaningful imaging to take place, notably the DTI. Water molecules will diffuse across space in a manner that is distinct from one another based on the kind of tissue, its components, its structure, its architecture, and its integrity. This latter method, which is compared to the straws in a glass of water, determines the amount of water that moves down the axons.
The gathering of data is done statistically by the use of volume elements, also known as voxels, using diffusion MRI. When a voxel includes scalar values that constitute a vector, we refer to that particular voxel as a tensor. This is where DTI got its name, and it also explains the extra information that can be obtained via using DTI [30]. DTI settings are able to measure the diffusion of water down an axon in a variety of directions, including 6, 9, 33, and 90 directions. These number of directions are the most common parameters employed, and anything over 33 directions increases confidence in the accuracy. Because obtaining a scan in ninety directions normally takes anywhere from ten to twenty minutes longer in the MRI scanner, this technique may not be appropriate for everyday clinical practice. DTI will, in essence, give an indirect technique of examining the structure of neuroanatomy on a microscopic level by measuring the degree of anisotropy and structural orientation of water molecules inside a voxel. This will be accomplished via the use of DTI. Because of this, the primary use for DTI is in the imaging of WM, which allows for the orientation, location, and anisotropy of the tracts to be quantified and analyzed. The organization of the axons into parallel bundles and the myelin sheaths that surround them both work together to make it easier for water molecules to diffuse along the main path of the axons.
DTI may be used to construct a variety of different metrics, each of which has the potential to give quantitative power, such as fractional anisotropy (FA) for the summative direction of the diffusion, mean diffusivity (MD) for the rate of molecular diffusion, axial diffusivity (AD) for the rate of diffusion parallel to the main vector, and radial diffusivity (RD) for the rate of diffusion perpendicular to the main vector.
FA measures the directionality of diffusivity in a summative way and is very sensitive to changes in microstructure. Despite its great sensitivity, FA may be unspecific to the origin of changes in microstructure. A rise in MD is indicative of disease processes such as edema or necrosis, although MD itself provides a quantitative measure of cellular and membrane density. Demyelination is accompanied by a rise in RD, which is used to quantify myelin neuropathology. Quantification of axonal deterioration is accomplished using AD, which rises as the brain matures [31, 32].
The FA values reflect the overall health of the axon. Abnormal FA readings suggest axonal injury. It is possible to determine FA values by using the region of interest (ROI) approach. The whole-brain analysis such as voxel-wise analysis is also becoming increasingly common as a result of advancements in automation and the capability to examine a greater number of tracts. The ROI approach, in which the areas that are to be evaluated are first traced by a technician and then analyzed by a computer, is trustworthy and easy to reproduce [33–36]. The segmented corpus callosal values are one of the most used and well-accepted ways of calculating ROI [37–39]. Because it is the greatest axonal tract in the brain, the damage that occurs to the corpus callosum as a result of head trauma and other disorders has been well characterized [40].
The values of FA may change depending on which of the aforementioned three techniques of analysis is employed, in addition to other aspects such as the MRI technique and the kind of post-processing that is carried out [41]. The FA values obtained via the use of a standardized procedure are very repeatable; they are not reliant on the expertise of the technician; they may be subjectively assessed by a radiologist; and they can be approximately compared to chosen values found in the literature.
Normal values for children are often lower than those established for adults. On the other hand, the majority of changes take place before the age of 5, and the corpus callosum reaches 90% of its adult FA values by the age of 11 years old [42]. After reaching maturity, FA levels have a tendency to drop in a linear fashion with increasing age.
However, its applicability in clinical settings is limited by a number of drawbacks. First, although some water molecules in certain components such as the cell membrane and the myelin sheath, have non-Gaussian diffusion properties which result in biological restrictions; DTI cannot reflect the non-Gaussian diffusion properties, because DTI model hypothesizes the Gaussian diffusion properties [43, 44]. As a result, DTI is unable to identify microstructural changes in GM since this tissue is mostly made up of neuronal cell bodies and displays greater isotropic water diffusion [43, 44]. Second, DTI makes the assumption that each voxel contains a single tissue compartment. It causes a partial volume effect owing to the extracellular free water such as the CSF [32] and DTI measurement might not be estimated accurately at the GM/WM boundary [45, 46] and that of the GM voxels contaminated by the CSF [47, 48]. In the third place, the parameters of the DTI do not offer any disease-specific or pathological information [29]. For instance, the answer to the issue of whether a drop in FA reflects a reduction in the axon-bundle cross-section or axon density is still unknown, and the interpretation of DTI parameters is a contentious topic [49–52]. Last but not least, the DTI model is an example of an oversimplification of the brain’s architecture. Although WM voxels include crossing or kissing fibers and make ~90% of all WM voxels in the healthy adult brain [53, 54], DTI expresses only a single major direction; as a result, FA diminishes in such voxels even in normal brain tissue [55].
DKI approximates the diffusion-weighted signal attenuation more accurately with respect to DTI, by quantifying the degree of non-Gaussian diffusion. Kurtosis is a dimensionless measurement that determines how much of a non-Gaussian distribution there is in water diffusion in a voxel [43, 44]. Because of the intricacy of the components that make up brain tissue, such as the cell membrane and the myelin sheath, the DKI is able to detect the limitation of water transport that results from this complexity [56]. When the diffusion kurtosis is larger, the water molecule diffusion deviates more from the Gaussian distribution. This shows that the environment has more restrictions on the diffusion of water molecules. On the other hand, a lower diffusion kurtosis suggests less limited diffusion, which is similar to what happens when neuronal death is occurred [57].
For a more sophisticated model, DKI needs at least three b-values and 15 diffusion gradient directions, while DTI only requires at least two b-values and six directions [58]. Mean kurtosis (MK), axial kurtosis (AK), and radial kurtosis (RK) are the three different types of kurtosis metrics that are used by DKI to characterize the condition of brain tissue. DKI has been used in the evaluation of neurodegeneration in WM with complex architectures, including voxels that include crossing fibers [59–62]. In addition, although DTI parameters imply no constraint on water diffusion, DKI parameters account for it both in anisotropic and isotropic situations. Thus, it has been shown that DKI may be used to assess microstructural alterations in GM, which is mostly made up of neuronal cell bodies and has isotropic water transport [44, 52, 63, 64].
In conclusion, despite the fact that it is better than DTI for evaluating pathological alterations in the brain, DKI suffers from a number of drawbacks. First, the acquisition time for DKI is much greater than that of DTI (about ten minutes) [43, 65], which restricts its applicability in clinical practice. This is because a more sophisticated DKI model needs more parameters than the DTI model does. Because neither DKI nor DTI incorporates any biophysical assumptions [43, 44], neither model can adequately describe disease-specific and pathological alterations. Some examples of these kinds of changes are the density, dispersion, and cross-section of axons or dendrites in neurons.
DTI can accurately estimate indices that reflect tissue-specific microstructures only in voxels containing a single type of brain tissue but cannot quantify tissue-specific indices in voxels contaminated by extracellular free water, such as the CSF, which is contained inside the ventricles and surrounding the brain parenchyma [32, 45–48]. Vasogenic edema is the accumulation of free water in the brain parenchyma’s extracellular space caused by disruptions in the blood-brain barrier due to pathologies such as tumors, brain trauma, or hemorrhage. This may lead to an increase in the volume of the brain [66–68]. DTI allows for the detection of free water since it demonstrates isotropic diffusion with MD of about 3 × 10–3 mm2/s at 37℃. This is roughly four times bigger than the average MD values of the brain parenchyma [69].
If each voxel in an image only includes one kind of tissue, then the DTI indices may be considered tissue-specific. DTI indices are normally regarded as identifiers for a single tissue, although this is no longer the case when partial volume of different diffusion compartments occurs. The reason for this is that the DTI indices are representative of the whole system [70]. CSF contamination occurs in voxels that are shared by CSF and brain tissue and may be seen along the ventricle’s contour lines and the brain parenchyma’s periphery. When the CSF barrier ruptures, this may occur [71, 72]. As a direct consequence of this, the MD values of voxels that are polluted with CSF are much higher than their FA counterparts. A WM voxel that has been tainted by free water will almost certainly be given a diffusion tensor that has a reasonably isotropic shape, which will prevent it from being classified as WM. CSF contamination might impair the delineation of fibers that pass close to the ventricles, including the fornix, the cingulum, and parts of the corpus callosum. This is a problem for both voxel-based and histogram-based comparisons of DTI-related variables [71, 73, 74]. Edema has an impact that is comparable to that of CSF contamination; however, its location and spread are determined by the specific pathology that generated it in the first place. In addition to this, edema often infiltrates the brain tissue, contaminating vast regions with partial volume effects that make it hard to detect the state of the infiltrated tissue and to carry out an investigation such as tractography [29, 75].
The fluid-attenuated inversion recovery (FLAIR)-diffusion-weighted imaging (DWI) sequence, which blocks the CSF signal, is often used in proposed approaches for removing CSF contamination [71, 73, 74]. FLAIR-DWI does not enable gating according to the cardiac cycle, which is important to eliminate pulsation artifacts and misalignment. It also suffers from a poor signal-to-noise ratio (SNR), which lengthens the scan duration and raises the specific absorption rate (SAR) [74]. Pierpaoli and Jones [76] presented a method that is based on models for removing CSF contamination from traditional diffusion pictures. In both cases, free water predominates the signal attenuation, which nullifies the specificity of the derived DTI indices for the underlying tissue, such as WM and GM. They hypothesized that vasogenic edema has diffusion properties similar to those of free water and causes partial volume effects similar to those caused by CSF contamination. Therefore, it was postulated that lowering pollution levels in the CSF might also mitigate the effect of edema on signal attenuation. In their study, Pierpaoli and Jones [76] used a bi-tensor model that has two compartments: a free water compartment that is characterized by an isotropic tensor with the diffusivity of free water, and a tissue compartment that is modeled by a diffusion tensor. Both of these compartments are separated by a membrane. Therefore, FWE-DTI has the ability to increase the accuracy of single-tensor DTI indices and particularly analyzes brain tissue microstructures after free water has been removed from the equation. In addition, the free-water map is regarded as a potential surrogate marker because it may differentiate between neuronal degeneration and the buildup of free water in the extracellular space that is linked with neuronal disorders such as neuroinflammation [77–80]. Because FWE-DTI is able to quantitatively estimate the degree of edema and atrophy, in addition to neuroinflammation, it has the potential to provide a better understanding of the pathology that is behind neurodegenerative diseases such as Parkinson’s disease [79], and psychiatric disorders such as schizophrenia [81, 82], and depression [83]. FWE-DTI may be estimated from clinically common single-shell diffusion data by using the same methodology that is used for DTI [78], and it has shown accuracy that is equivalent to that of FWE-DTI generated from multi-shell data [84].
The NODDI method was designed to offer a more accurate assessment of the microstructures of brain tissue compared to the above models such as DTI, DKI, and FWE-DTI. NODDI estimates the three different types of brain tissue compartments. NODDI [85] allows for the estimation of three important aspects of neuronal tissue for each voxel: neurite density index (NDI), which quantifies the packing density of axons or dendrites; orientation dispersion index (ODI), which assesses the orientational coherence of neurites; and isotropic volume fraction (ISOVF), which estimates the extent of CSF contamination. NODDI histological validation studies have shown that ODI is consistent with its histological equivalent [86–90], and NDI has shown the same level of consistency [88, 91]. Changes in ISOVF are compatible with the development of an inflammatory response, as shown by results from a transgenic Alzheimer’s disease model tracking the progression of diffusion-derived markers [92].
NODDI is a member of the multicompartment model-based family of diffusion MRI methods [93]. The voxel-level MRI signals are interpreted by multicompartment models as the total of the contributions from the various compartments. In comparison to traditional DTI and cutting-edge DKI, our interpretation of voxel-wise MRI data offers significant benefits [44]. DTI and DKI only provide a partial picture of the many different contributions in a voxel via their metrics. Therefore, as indicated above, a change in DTI-derived FA may be produced by a variety of underlying alterations to the contributing compartments. Multicompartment models, on the other hand, attempt to separate the effect of each compartment so that it may be characterized separately.
The number of presumed compartments inside a voxel is a fundamental differentiator between the NODDI model and other multicompartment models. Many models built about the same time as NODDI use a two-part model of neural tissue, with one portion reflecting the signal contribution from water contained inside the intraneurite space and the other part capturing the contribution from water located outside of the neurites. The possible contribution from CSF contamination is non-negligible for cortical GM, periventricular subcortical GM, and WM, however, this method does not take this into consideration. In order to solve this problem, NODDI was designed with a third compartment to specifically account for CSF contamination.
Direct inference on microstructural characteristics from indirect MRI scans is made possible by model-based techniques like NODDI. An important aspect of this strategy is the use of modeling assumptions. Various multicompartment models make different trade-offs in the microstructural characteristics they may predict due to the low information content available from standard diffusion MRI data. NODDI has the benefit over other contemporary models in a number of respects, including its ability to quantify free water contamination. Unfortunately, in order to make this decision, it is required to forsake the possibility of estimating diffusivity characteristics like din. Thus, NODDI must make an a priori guess at this value. This value was computed from the corpus callosum in the first version of NODDI and was thought to be appropriate for both WM and GM. In any case, it was eventually shown that the correct number for GM is somewhat different from the typical [94]. In addition, whereas the NODDI application for people of all ages (> 10 years old) is quite consistent, the value for newborns (1 year old) is somewhat different. Consequently, NODDI predicts a decrease in the intrinsic parallel diffusivity as a result of the crowded conditions in the extraneurite region, where water molecules disperse. It has been shown by Lampinen et al. [95] that a new type of diffusion MRI data obtained using b-tensor encoding is incompatible with NODDI, and it has been hypothesized that this limitation is to blame. But further research by Guerreri et al. [96] showed that this conclusion was unwarranted, and that an improperly selected din was the major cause of the incompatibility, necessitating tissue-specific tuning. In spite of these caveats, NODDI is one of the few multicompartment models that has undergone thorough histological validation [86–90], and NDI has shown the same level of consistency [88, 91].
DTI is the diffusion MRI technique that has been subjected to the most study and is now the method of choice for determining the presence of WM structural changes in the brain and spinal cord of ALS patients. FA was shown to be consistently reduced in the CST, the posterior limb of the internal capsule, and the cingulate gyrus in individuals who had ALS, according to the findings of two voxel-based meta-analyses [97, 98]. FA decrease was also found in the corpus callosum, and this shift extended to main motor cortices [98–106]. In addition, a decrease in FA in the left corona radiata was shown to have a positive correlation with the ALS Functional Rating Scale-Revised (ALSFRS-R) score [98, 107–109]. The ALSFRS-R is a standard tool for evaluating the quality of life of people with ALS. In a whole-brain spatial statistical analysis of people with ALS caused by C9ORF72 mutations, researchers found that FA was significantly reduced along the CST, with projections to the frontal and hippocampal areas [110]. In addition, research that compared individuals with ALS who had C9ORF72 mutations to healthy controls reported significant FA reductions in the corticorubral and corticopontine tracts, in addition to the corticostriatal pathway and the proximal perforant route [111, 112]. These studies show that ALS-related neurodegeneration affects not only motor but also non-motor regions of the brain [110–112]. However, a single-tensor diffusion model was utilized in these researches, which severely constrained our ability to characterize complex microstructures within brain tissues and to depict key clinical aspects of ALS. This suggests that DTI is a promising tool for revealing WM microstructural alterations.
In contrast to DTI, DKI allows for simultaneous microstructural assessment of both GM and WM without regard to the spatial orientation of structures. This is due to the fact that DKI doesn’t care which way buildings face in space. Therefore, DKI may be useful in providing more data on the microstructure of the whole brain in ALS. Decreased MK and RK in WM, which comprises the bilateral precentral gyri, bilateral corona radiata, bilateral middle corpus callosum, the left occipital lobe, and the right superior parietal lobule, have been seen in ALS patients using voxel-based analysis. This is in comparison to controls, who exhibit normal levels of MK and RK in these regions. Patients with ALS had less myelin kinase in the left anterior cingulate gyrus, bilateral precentral gyri, and paracentral lobules of their GM areas (Figure 1). Additionally, numerous WM areas display reduced FA as well as increased MD/RD; however, the geographic breadth of these changes is lower in comparison to the changes that were identified when utilizing the decreased DKI measurements.
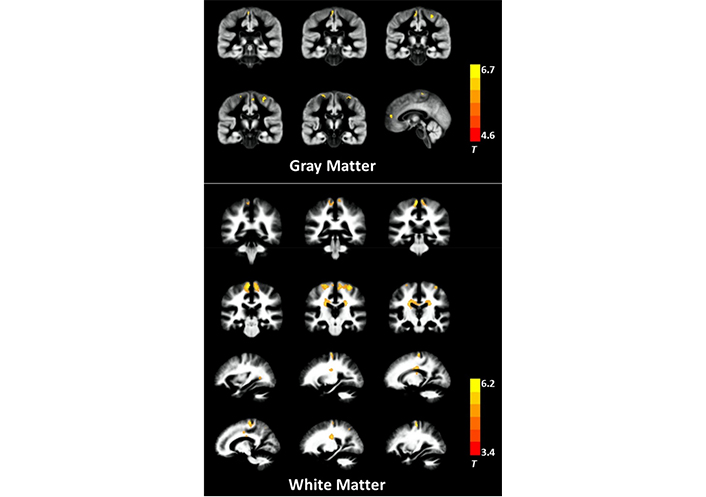
GM and WM areas in ALS patients exhibiting dramatically lower levels of MK. The presented pictures have been superimposed above the aggregated GM and WM maps obtained from all of the individuals. The T-values that are shown by the color bar illustrate the degree to which patients with ALS and controls vary in terms of their MK
Note. Reprinted from “Abnormal cerebral microstructures revealed by diffusion kurtosis imaging in amyotrophic lateral sclerosis,” by Huang NX, Zou ZY, Xue YJ, Chen HJ. J Magn Reson Imaging. 2020;51:554–62 (https://doi.org/10.1002/jmri.26843). CC BY.
MK and RK in the left precentral gyrus were shown to have a negative correlation with illness duration in one research [113]. On the other hand, right precentral gyrus MK and left caudate body RK were found to have a positive correlation with the ALSFRS-R score. In separate research [114], the ROI analysis was utilized to establish that there were substantial changes in the motor cortex between the groups of people with ALS and controls. MK, AK, and RK were all considerably lower in the ALS group, although the standard DTI measurements were not statistically different between the two groups. In addition, RK was shown to have a favorable correlation with the ALSFRS-R score [114]. In individuals diagnosed with ALS, both MK and RK levels were shown to have decreased, as was the case in both trials. A decrease in RK, which is a reflection of myelin changes in the brain, may thus be an indicator of myelin degradation in people with ALS.
To the best of our knowledge, there is no previously published study that has looked at the usefulness of FWE-DTI in the treatment of ALS.
NODDI may be more sensitive than DTI, despite the fact that it was just recently included in the process of assessing people with ALS. In fact, the ALS group showed significantly lower NDI across large swaths of the CST, corpus callosum, and right precentral gyrus, lower ODI across the right anterior internal capsule and right precentral gyrus, and higher ISOVF across the right lateral ventricle in a whole-brain voxelwise analysis using NODDI (Figure 2) [115]. The ALS group also showed considerably lower ODI throughout a large portion of the right. Additionally, patients with both limb and bulbar involvement showed a significantly lower NDI in the corona radiata and subcortical WM of the right hemisphere compared to those with limb involvement alone. It was shown that a decline in ODI in the precentral gyri, precuneus, and dorsolateral prefrontal cortex was associated with disease progression. Whole-brain DTI analysis revealed reduced FA within the CST in the ALS group [116]. However, the impacted regions were more restricted when compared to those that were detected by NDI.
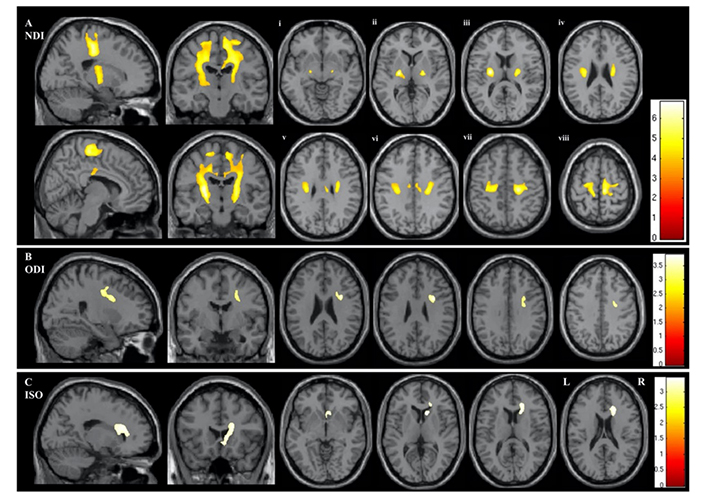
Regions demonstrating statistically significant differences between the ALS and control groups in the whole-brain NODDI parameters of NDI (A), ODI (B), and ISOVF (C). The results are given based on P < 0.05 after family-wise error correction at the cluster level, with each cluster’s P-value being 0.001. Panels Ai–viii indicate statistically significant differences in NDI on axial sections commencing at the posterior limb of the internal capsule (vi) and extending rostrally into the subcortical WM of the precentral gyrus (viii). This research employed the acronyms ALS, ISOVF, NDI, NODDI, ODI, and WM
Note. Reprinted from “Neurite orientation and dispersion density imaging (NODDI) detects cortical and corticospinal tract degeneration in ALS,” by Broad RJ, Gabel MC, Dowell NG, Schwartzman DJ, Seth AK, Zhang H, et al. J Neurol Neurosurg Psychiatry. 2019;90:404–11 (https://jnnp.bmj.com/content/90/4/404). CC BY-NC.
Research involving ROI analysis of NODDI and DTI in premanifest C9ORF72 mutation carriers found that these individuals showed NDI-WM abnormalities in 10 tracts, including the CST, based on NDI, but in only five tracts using DTI metrics (increased AD, RD, and MD rather than decreased FA; Figure 3). Two distinct tracts showed significantly larger NDI effect sizes compared to DTI measures. ISOVF was increased in 13 sites, while volumetric atrophy was seen in 11 sites in C9ORF72 mutant carriers. These results provide support to the idea that the loss of axonal fibers, rather than an increase in the complexity or dispersion of the tracts themselves, is responsible for the decreased FA (or increased diffusivity) seen in the CST and corpus callosum [117].
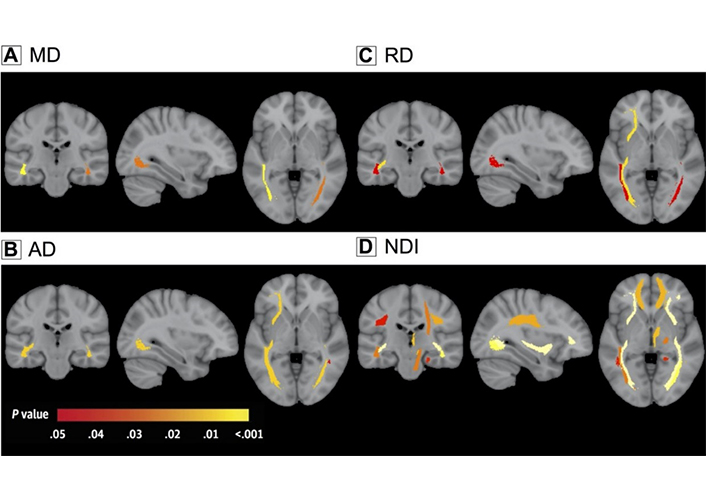
Changes in the WM have been seen in C9ORF72 mutant carriers. A depiction in color coding of the P-values that correlate to the connection of the C9ORF72 mutation with the integrity of the WM, after multiple comparisons have been corrected for. (A) MD; (B) AD; (C) RD; (D) NDI
Note. Reprinted with permission from “Neurite density is reduced in the presymptomatic phase of C9orf72 disease,” by Wen J, Zhang H, Alexander DC, Durrleman S, Routier A, Rinaldi D, et al.; Predict to Prevent Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis (PREV-DEMALS) Study Group. J Neurol Neurosurg Psychiatry. 2019;90:387–94 (https://jnnp.bmj.com/content/90/4/387). © Author(s) [or their employer(s)] 2018.
Furthermore, both presymptomatic and symptomatic animals exhibited reduced NDI and increased ODI compared to control mice in research that paired NODDI with the histological evaluation of the manually segmented WM anterolateral area in the spinal cord of a rat model of ALS [115]. Animals who showed no outward signs of illness were regarded to be in the earliest stages of the disease, but a reduction in the total axonal area was seen histologically in presymptomatic mice at the same period of the experiment. It was also observed that the extra-axonal compartment had greatly expanded in the early stage of the sickness, and that the extra-axonal compartment had further expanded and the axonal areas had decreased in later stages of the illness. Histological data suggested that less myelin and axonal area corresponded to a lower NDI. In sum, these studies show that NODDI can detect WM abnormalities brought on by the axonal degeneration typical of ALS [115–117].
The intricacies of neurodegenerative conditions have been better understood as a result of the use of more recent diffusion MRI methods such as FWE-DTI, DKI, and NODDI. These techniques provide fresh insights into the microstructures of the brain. However, owing to the lack of clinical data on their efficacy, these more sophisticated approaches have not yet been used in clinical settings. One example of this is the regular use of DWI to evaluate myocardial infarction. The cost of time consumption is higher in advanced diffusion MRI-based surrogate markers than in the measurement of hippocampal volume. The high related cost is another barrier that prevents clinical studies from being carried out. As a result, the collection of clinical data is hampered by the effectiveness of sophisticated diffusion MRI methods despite their cheap cost.
In order to reach the practical application of sophisticated diffusion MRI methods for the diagnosis of neurodegenerative illnesses, there are a few restrictions that need to be solved first. To begin, there is a lack of clarity about the connection between the pathogenic alterations seen in neurodegenerative disorders and the advanced diffusion MRI measures. The use of diffusion MRI is essential for the success of DKI, FWE-DTI, and NODDI in modeling and forecasting brain microstructures. However, the extent to which these models are able to accurately represent and explain specific neurodegenerative diseases remains unclear. For this reason, further studies are needed on neurodegenerative illnesses in postmortem human tissues or animal models to clarify the relationship between pathological findings and the advanced diffusion MRI metrics of FWE-DTI, DKI, and NODDI. Modern diffusion MRI could have these measurements. Second, the results of studies using cutting-edge diffusion MRI techniques are very unreliable and difficult to replicate. This is mostly because of the low statistical power that results from using such a tiny sample. Therefore, in order to demonstrate the usefulness of FWE-DTI, DKI, and NODDI as the surrogate marker for ALS, solid data from multi-site research with bigger sample numbers for the purpose of improving the statistical power of these studies should be gathered. Despite the fact that various large-scale multi-site studies are now being conducted, MRI scanners and acquisition settings vary greatly, depending not only on the imaging location but also on the individual scanner [118]. Because of these discrepancies across research locations, the repeatability and reliability of advanced MRI investigations of diffusion may be compromised. For instance, Andica et al. [119] examined the scan-rescan and inter-vendor repeatability of DTI and NODDI by using two 3-T MRI scanners that were manufactured by two different companies. Both scanners produced scan-rescan coefficients of variation (CoVs) in the NODDI measures that were equivalent to those of the DTI metrics (around 4%). Nevertheless, the scan-rescan CoV for NODDI measures ranged from 2.3% to 14%. The inter-vendor CoV was greater. In addition, the inter-sequence variability of DTI measures was measured using three distinct sequences, and the results indicated that the CoVs for FA and MD were, respectively, 5.45–7.34% and 1.72–5.56% [120]. In addition, Kamagata et al. [121] investigated the inter-site dependability of DTI measurements using identical 3-T MRI scanners at two distinct locations and acquisition conditions. This was done in order to determine how consistent the DTI metrics were between the two locations. According to the authors’ findings, the CoV of DTI varied from 0.6% to 5.6% [121]. Therefore, poor reproducibility and reliability in multi-site studies using sophisticated diffusion MRI methods may be the consequence of variances in diffusion MRI metrics induced by site differences such as MRI scanners and acquisition settings [118]. Since the changes in diffusion MRI metrics in neurocognitive and psychiatric disorders are mild (~5–6%) compared to healthy controls and on the same order as that of site difference, it is challenging for a multi-site investigation to identify pathological alterations in patients with these conditions [122–124]. Therefore, it is necessary to reduce inter-site variability in diffusion MRI metrics by standardizing MRI methods, including MRI scanners and acquisition parameters, and harmonizing multi-site diffusion MRI data. This will be accomplished by reducing the inter-site variability in diffusion MRI metrics. Several harmonization methods for diffusion MRI, such as a combined association test called ComBat [125], linear regression based on rotation invariant spherical harmonics [126], and the deep learning approach [127], have been proposed, for instance, in order to reduce the variability that can occur between MRI scanners and protocols. Empirical Bayes inference is used by ComBat for the purpose of data harmonization of diffusion MRI measurements. This is accomplished by the regression of variables. The linear rotation invariant spherical harmonics method utilizes features for the purpose of harmonizing diffusion MRI signal and mapping diffusion MRI data from a target location to a reference site. During the learning stage of the deep learning harmonization approach, the neural network parameters are optimized with the help of diffusion MRI signals acquired at target and reference sites. Afterwards, the trained neural network is used to harmonize diffusion MRI data. Not only can these harmonization strategies cut down on undesirable changes in DTI measurements that are induced by variances in locations, but they also keep the biological variability that is produced by age and sex.
A further shortcoming is the lack of clarity on the connection between the pathological changes brought on by ALS and the more sophisticated diffusion MRI measurements. DKI, FWE-DTI, and NODDI can only predict and forecast brain microstructures using diffusion MRI. It’s unclear how well these models reflect and explain neurodegenerative diseases. To explain the association between pathology findings and DKI, FWE-DTI, and NODDI, further study on neurodegenerative disorders in postmortem human tissues or animal models is needed. MRI measures these parameters. Resolution of these limits could enable clinical application of advanced diffusion MRI techniques as surrogate markers for neurodegenerative diseases.
Recently, frameworks that integrate diffusion tensor MRI and relaxometry have been created to increase their specificity for quantifying myelin and axonal characteristics independent of fiber organization inside the voxel, even with crossing fibers [128, 129]. These frameworks quantify myelin and axonal characteristics independently of fiber complexity. Increasing evidence suggests that iron buildup may contribute to neurodegenerative diseases. Quantitative susceptibility mapping imaging offers the potential for studying brain iron distribution [130]. By studying the features of different kinds of biological tissue, the diffusion tensor-relaxometry framework or the combination of advanced diffusion MRI techniques and quantitative susceptibility mapping might provide a fuller picture of neurodegenerative diseases.
ALS is the most prevalent type of MND and is diagnosed with a delay from the first appearance of symptoms. Surrogate markers are needed to detect pathological changes before neuronal death and allow early disease-modifying therapy. Diffusion MRI can be used to infer microstructural characteristics of the brain, including microstructural integrity and complexity, as well as axonal density, order, and myelination, by using water molecules that are diffused within the tissue, with micron-scale displacement. DTI is the most used MRI approach for evaluating ALS. NODDI, DKI, and FWE-DTI bypass DTI’s restrictions. This article describes these technologies and their potential as ALS surrogate markers.
AD: axial diffusivity
ALS: amyotrophic lateral sclerosis
ALSFRS-R: amyotrophic lateral sclerosis Functional Rating Scale-Revised
C9orf72: chromosome 9 open reading frame 72
CoVs: coefficients of variation
CSF: cerebrospinal fluid
CST: corticospinal tract
DKI: diffusion kurtosis imaging
DTI: diffusion tensor imaging
DWI: diffusion-weighted imaging
EE: El Escorial
EMG: electromyography
FA: fractional anisotropy
FWE-DTI: free water elimination diffusion tensor imaging
GM: gray matter
ISOVF: isotropic volume fraction
LMN: lower motor neuron
MD: mean diffusivity
MK: mean kurtosis
MND: motor neuron disease
MRI: magnetic resonance imaging
NDI: neurite density index
NODDI: neurite orientation dispersion and density imaging
ODI: orientation dispersion index
PLS: primary lateral sclerosis
RD: radial diffusivity
RK: radial kurtosis
ROI: region of interest
UMN: upper motor neuron
WM: white matter
I extend sincere gratitude to my supervisor for their unwavering support, invaluable guidance, and scientific insight, which significantly improved this paper. Their constructive feedback enriched my research skills. I’m also deeply thankful to my boss for their leadership and patience. Their belief in my capabilities encouraged me to overcome challenges and focus on my research goals. Their invaluable contributions were key to the success of this research. Any recognition this paper receives is a testament to their dedication and guidance.
YS: Conceptualization, Investigation, Writing—original draft.
The author declares that he has no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
This study was partially supported by the Juntendo Research Branding Project, JSPS KAKENHI I grant no. [21K07690, 21K12153, 22H04926]; a Grant-in-Aid for Special Research in Subsidies for ordinary expenses of private schools from The Promotion and Mutual Aid Corporation for Private Schools of Japan; the Brain/MINDS Beyond program grant no. [JP19dm0307101] of the Japan Agency for Medical Research and Development (AMED), under grant no. [JP21wm0425006]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
