Affiliation:
1Department of Medicine, University of Padova, 35123 Padova, Italy
ORCID: https://orcid.org/0000-0003-2077-9202
Affiliation:
2Department of Clinical and Biological Sciences, Unit of Experimental Medicine and Clinical Pathology, School of Medicine, University of Torino, 10125 Torino, Italy
Email: maurizio.parola@unito.it
ORCID: https://orcid.org/0000-0001-7720-5141
Explor Dig Dis. 2024;3:22–41 DOI: https://doi.org/10.37349/edd.2024.00038
Received: September 07, 2023 Accepted: November 16, 2023 Published: February 28, 2024
Academic Editor: Natalia Nieto, University of Illinois at Chicago, USA
The article belongs to the special issue Fibrosis and Hepatobiliary Cancer
Chronic liver diseases (CLDs), which are typically characterized by fibrogenic progression towards liver cirrhosis and related complications eventually leading to organ failure and can also lead to the development of primary liver cancers, represent a major burden for human health on a worldwide basis. Although the present knowledge on the pathogenesis of CLDs progression and primary liver cancers development has remarkably increased in the last decades, critical molecular mediators remain incompletely understood, and approved antifibrotic therapies to efficiently counteract CLDs progression and liver cancer are lacking. In the present review, this study will specifically analyse the putative contribution of SERPINB3, a member of the superfamily of serine-protease inhibitors (SERPINs), which has been shown to exert significant pro-inflammatory and pro-fibrogenic roles in progressive CLDs as well as to be involved in the development of primary liver cancers, including hepatocellular carcinoma (HCC), cholangiocarcinoma, and hepatoblastoma.
Chronic liver diseases (CLDs) and, particularly, liver cirrhosis, the end stage of fibrogenic progression, represent a major burden for human health on a worldwide basis. Several studies have reported that liver cirrhosis alone can affect approximately 1–2% of the global population, with an impact which is significantly raised in the last 30 years and is actually estimated to cause as much as 1 million of deaths per year worldwide [1–4]. Moreover, a recent analysis indicated that cirrhosis was associated with 2.4% of global deaths in 2019 [5]. Liver cirrhosis can lead to hepatic decompensation and failure, with affected patients developing ascites, hepatic encephalopathy, hepatorenal syndrome, and variceal bleeding [2, 6, 7]. Moreover, liver cirrhosis is also considered as the most relevant risk factor for developing hepatocellular carcinoma (HCC), the most common primary liver cancer. Indeed, irrespective of aetiology, approximately 90% of HCC occur in cirrhotic patients [8–10] although in 25–30% of non-alcoholic steatohepatitis (NASH) patients HCC may occur in the absence of a diagnosis of cirrhosis [10]. Along these lines, HCC is a very aggressive tumor which represents the sixth most common human cancer and the fourth leading cause of cancer mortality worldwide [3, 4, 8–10].
Whether fibrogenic progression of CLDs is concerned, liver fibrosis involves an excess deposition of extracellular matrix (ECM) components associated with concurrent matrix degradation and remodelling. This scenario is a common pathological outcome of a sequela of tissue events including chronic parenchymal injury, persistent and sustained activation of inflammatory response as well as liver fibrogenesis (i.e., the process leading to fibrosis), and then chronic wound healing [11–17].
From an epidemiological point of view, CLDs progression can develop as a consequence of the following etiological conditions [2, 18–22]: i) chronic infection by either hepatitis-B or -C virus (HBV, HCV); ii) metabolic-related injury, mainly detected in obese and/or type 2 diabetes patients, under the definition of non-alcoholic fatty liver disease (NAFLD) and the related progressive form of NASH, which are now emerging as the leading causes of CLDs for the next decades; the reader should be aware that a worldwide consensus has been very recently reached to replace NAFLD with the more appropriated definition of “metabolic dysfunction-associated steatotic liver disease (MASLD)” [22]; for the sake of clarity and in order to refer to current and past literature, in this review, we will still use the previous definition of NAFLD and NASH; iii) chronic toxic injury, mainly related to excess alcohol consumption; iv) autoimmune injury directed versus either hepatocytes, as in autoimmune hepatitis (AIH), or cholangiocytes, as in primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC) [23]; v) different chronic cholangiopathies [24, 25]; vi) hereditary diseases such as mainly hereditary hemochromatosis, Wilson’s disease, and α1-antitrypsin deficiency [18, 19].
At present, our knowledge on the pathogenesis of CLDs progression and HCC development has remarkably increased in the last decades by a very impressive number of experimental, translational, and clinical studies. These studies have started to reveal the complexity of the so-called “pro-fibrogenic microenvironment” which involves complex, dynamic, and interrelated interactions (i.e., in terms of molecular mediators and signalling pathways) between different hepatic and extrahepatic cell populations. These studies, mostly dedicated in the last decade to NAFLD/NASH, have pointed out the major role of cells of the innate and adaptive immunity as well as of fully activated liver myofibroblasts (MFs) [13, 15, 16, 26–30].
However, we still lack approved antifibrotic therapies to efficiently counteract CLDs progression and HCC development and there are several unmet needs that should represent the future target of basic, translational, and clinical studies, as recently pointed out [17]. In particular, we still suffer from incomplete knowledge of critical molecular mediators able to modulate either CLDs progression as well as the development of HCC as well as of other primary liver cancers, including cholangiocarcinoma and hepatoblastoma. In the present review, we will specifically analyse the putative contribution of SERPINB3, a member of the superfamily of serine-protease inhibitors (SERPINs), which has been shown to exert significant pro-inflammatory and pro-fibrogenic roles in progressive CLDs as well as to be involved in the development of primary liver cancers.
SERPINs constitute a large superfamily of proteins and accurate analyses of the human genome have identified as much as 36 related protein-coding genes and 8 pseudogenes [31, 32]. SERPINs exhibit different peculiar biological activities which have been extensively investigated particularly in cardiovascular diseases, metabolic disturbances, and cancer [33, 34]. The majority of SERPINs have a true serine-protease inhibitory activity and exhibit, for example, antifibrinolytic and anticoagulant activities; this is true for two of the best characterized proteins of this superfamily like antithrombin (AT or SERPINC1) and plasminogen activator inhibitor-1 (PAI-1 or SERPINE1). SERPINs are also known to operate as anti-inflammatory or immune-modulatory proteins and AT has been also reported to act as a potent cardioprotective molecule by activating the AMP-activated protein kinase pathways [35, 36]. Along these lines, α1-antitrypsin (SERPINA1) is best known for its ability to inhibit neutrophil elastase (NE), being able to prevent NE-related tissue injury [37], as for example in lung diseases characterized by chronic injury and inflammation. SERPING1, also known as C1 esterase inhibitor (C1-INH), is a regulator of the classic complement cascade by either inhibiting C1s/C1r complex or the mannan binding lectin (MBL) pathway through the inhibition of MBL-associated serine protease (MASP)-1/MASP-2 tetrameric complex [38]. It should be noted that there are SERPINs that can also exhibit an inhibitory activity versus other proteases like caspases or papain-like cysteine proteases [31]. By contrast, some SERPINs do not exert inhibitory functions at all but operate as hormonal transporters and molecular chaperones [39].
From a phylogenetical point of view, human SERPINs have been subdivided into nine clades (i.e., from A to I clades). Along these lines, 13 ovalbumin SERPINs (ov-SERPINs) have been classified as clade B SERPINs that, differently from the majority of SERPINs, which are proteins secreted extracellularly, lack a critical signaling peptide necessary for secretion and are then prevalently intracellular proteins [40]. Three of clade B genes are located at chromosome 6p25 (SERPINB1, SERPINB6, and SERPINB9) whereas all the other genes, including SERPINB3, are clustered at chromosome 18q21.3 [40, 41].
The history of SERPINB3 started in 1977 when Kato and Torigoe isolated what they originally proposed as a tumor-specific antigen identified in squamous cell carcinoma (SCC) of the uterine cervix [42]. For this reason, the protein was first defined as tumor antigen 4 (TA4) [43] and later referred to as SCC antigen (SCCA) [44]. In particular, in the original study, SCCA was detected in 27 out of 35 women carrying SCC, with all 27 SCCA-positive women at an advanced stage of the disease [42]. Interestingly, it was later recognized that the original SCCA was not a single protein but, rather, a mix of two different protein isoforms of 390 amino acids, sharing an identical apparent molecular weight of 45 kDa [43]. Using isoelectric focusing the two isoforms were separated into a neutral isoform, called SCCA1, and an acidic isoform or SCCA2. These two isoforms were later classified as SERPINB3 and SERPINB4, respectively, which shared a 98% identity at nucleotide level and 92% identity at amino acid sequence. Moreover, it was discovered, through genomic analysis, which the two genes were encoded from independent loci arising from a tandem gene duplication [44–46]. Soon after the first studies, the serum levels of SCCA (SERPINB3/SERPINB4, as detected by an antibody recognizing both isoforms) were found to be elevated in the human SCC [47, 48] and proposed as a putative diagnostic and prognostic biomarker for that tumor [49]. Starting from these original studies related to SCC, SCCA and then SERPINB3 and -B4 were progressively reported to be involved in other cancer types as well as in other pathological conditions [39, 40, 50], then stimulating the interest of the scientific community.
While the present review is primarily dedicated to SERPINB3, it is necessary to introduce data also on SERPINB4 since these two proteins, sharing a very high structural homology, exhibit distinct biochemical features, substrate protease specificity and are often co-expressed in both normal and diseased tissues. As mentioned before, SERPINB3 and -B4 belong to the group of ov-SERPINs and this means that they have a so-called “ovalbumin-like domain” which represents the main body of the protein. From a structural point of view, these proteins possess nine α-helices and three antiparallel β-sheets as well as a hydrophobic c-terminal reactive center loop (RCL), also referred to as reactive site loop (RSL), which is essential for binding and inhibiting the target protease [51–55]. The interested reader may find more structural and functional details on these two proteins in dedicated and more comprehensive reviews or articles [50, 53–55] and just a number of relevant issues, including tissue distribution in physiological and pathological conditions (liver excluded) can be synthetized as follows:
Human SERPINB3 in vitro mostly recognizes as target proteases papain-like cysteine proteases, including cathepsins L, S, and K, as well as papain, whereas SERPINB4 inhibits chymotrypsin-like serine proteases, including chymase and cathepsin G [53–55];
Both SERPINB3 and -B4, like other SERPINs, form a sodium dodecyl sulfate (SDS)-resistant complex with the target protease through an acyl-oxyester bond in order to inhibit the proteolytic activity [50];
The genes encoding SERPINB3 and -B4 are frequently co-expressed in different tissues, including lung, trachea, prostate, uterine cervix, and testis, with the exception for bladder and thymus where only SERPINB3 is expressed [56, 57];
In other tissues such as, pertinent to this review, the liver, SERPINB3 is physiologically undetectable but its levels (see later) were found to rise in pathological conditions, including conditions of CLDs as well as in primary liver cancers [58–60];
According to the previous issue, both SERPINs have been detected in conditions of chronic inflammation including several respiratory diseases (tuberculosis, asthma, and other chronic obstructive pulmonary diseases) [61–64] and certain skin diseases (atopic dermatitis and psoriasis) [65, 66]; in some of these chronic diseases, both SERPINs were found to be up-regulated in response to cytokines released by Th2 lymphocytes such as interleukin (IL)-4 and IL-13 [61, 63, 65].
As mentioned in the previous section, SERPINB3 as well as the structurally related SERPINB4 are undetectable in normal human liver. However, their expression has been reported to progressively increase in samples from human conditions of CLDs, likely as a part of the chronic and multi-faceted response to persistent liver injury [58–60, 67, 68]. Moreover, as it will be recalled later in this review, the expression of the two SERPINs was earlier found to be increased in HCC cells as well as in cells of highly dysplastic nodules and in hepatocytes of peritumoral cirrhotic tissue, leading to the suggestion that the overexpression of SERPINB3 and -B4 may also represent a relatively early event in liver carcinogenesis [58–60, 67–69].
The first study suggesting the involvement of SERPINB3 and -B4 in CLD progression emerged by analyses showing that circulating immune complexes (IC) composed by SCCA and immunoglobulin M (SCCA-IgM), undetectable in the serum of healthy controls, were detectable in a cohort of 188 chronic hepatitis patients in relation to disease outcome [59]. Authors observed a significant increase of SCCA-IgM serum levels over time only in patients with histopathological detectable fibrosis progression. These data were the first to hypothesize a relationship between SERPINB3 and -B4 and fibrosis progression, also suggesting that monitoring serum SCCA-IgM levels over time may be useful to identify chronic hepatitis patients at higher risk to develop cirrhosis [59]. This study opened the way to start to investigate and characterize the biological action of SERPINB3 in relation to CLD progression. In particular, the focus has been on the hypothesis that this protein may not simply serve as a possible biomarker for diagnostic and/or prognostic purposes but also have a role as a mediator involved in the crucial processes underlying CLDs progression (i.e., inflammation, fibrogenesis, and carcinogenesis). These studies have taken advantage of the availability of mice genetically manipulated in order to either overexpress SERPINB3 in hepatocytes or to carry a hepatocyte-specific deletion of the related gene as well as of human recombinant SERPINB3 and of tools able to discriminate in vivo and in vitro between SERPINB3 and -B4.
The first consistent hypothesis of an involvement of SERPINB3 as a mediator of critical processes involved in the progression of a fibrogenic chronic disease was raised in a study investigating the role of SCCA in metaplastic epithelial cells in idiopathic pulmonary fibrosis (IPF), a chronic progressive and fibrogenic lung disorder with a poor prognosis [70]. This study, in addition to providing for the first time the notion that SCCA was significantly over-expressed in IPF patients versus control individuals, also outlined a positive correlation between SCCA expression, the expression of the critical pro-fibrogenic mediator transforming growth factor β1 (TGFβ1) and the extension of fibroblastic foci. Moreover, exposure of cultured lung epithelial A549 cells to SCCA led to up-regulation of TGFβ1, suggesting for the first time that SCCA produced by metaplastic lung epithelial cells (a consequence of chronic injury and death of normal epithelial cells) may induce, in a paracrine way, proliferation and activation of lung fibroblasts, then favoring the development of lung fibrosis [70]. The same laboratory published two years later a study dedicated this time to investigate the relationships among SERPINB3, TGFβ1, and liver fibrosis in CLDs by evaluating these two proteins in liver biopsies obtained from 94 CLD patients [58]. SERPINB3 and TGFβ1 at protein and mRNA level significantly correlated in liver biopsies and both proteins correlated to the extent of liver fibrosis. Moreover, primary human hepatocytes responded to human recombinant SERPINB3 (hrSERPINB3) by up-regulating TGFβ1 expression [58]. Similarly, when HepG2 and HuH7 cells were transfected to overexpress the intact SERPINB3 human gene, all transfected cells showed increased TGFβ1 mRNA and protein production; the integrity of the RSL of the SERPIN was critical to achieve this effect [58]. This study generated the hypothesis that chronically damaged hepatocytes can release both SERPINB3 and TGFβ1, with an active role of SERPINB3 in mediating TGFβ1 expression and release. Of interest, it was also proposed that up-regulation of SERPINB3 expression, in addition to being related to chronic hepatocyte injury, was also elicited by exposure of hepatocytes to hypoxic conditions. In particular, by employing human hepatocyte-derived cell lines exposed to controlled conditions of hypoxia it was shown that this was followed by up-regulation of SERPINB3 expression. This effect was related to hypoxia-induced stabilization of hypoxia-inducible factor-2α (HIF-2α) and very selective since HIF-2α was found, by chromatin immunoprecipitation (ChIP) analysis, to directly bind the SERPINB3 promoter leading to increased transcription of the gene [71]. This is potentially relevant since accumulating evidence has indicated a direct relationship among hypoxia, HIFs, and fibrogenic progression of CLDs, as extensively reviewed elsewhere [15, 72–76].
The putative pro-fibrogenic action of SERPINB3 has been further investigated using transgenic mice manipulated to overexpress SERPINB3 in hepatocytes and employed into two distinct murine protocols resulting in liver fibrosis like chronic administration of carbon tetrachloride (CCl4) or the methionine and choline deficient (MCD) dietary NASH-related protocol [77]. In both experimental protocols, the overexpression of SERPINB3 significantly worsened experimental liver fibrosis that was strictly related to a significant increase in the transcript levels of pro-fibrogenic genes as well as of collagen deposition and the number of α-smooth muscle actin (αSMA)-positive MFs, as compared to wild-type mice. Further in vitro experiments provided evidence that hrSERPINB3 strongly and directly upregulated the expression of the same critical pro-fibrogenic genes, including TGFβ1, αSMA, collagen 1A1 (col1A1), tissue inhibitor of metalloprotease 1 (TIMP1), platelet-derived growth factor B (PDGF-B) and the related receptor β (PDGFRβ) in either human HSC/MFs or human LX2 cells. Moreover, in the same MF-like cells hrSERPINB3 also promoted their oriented migration in a reactive oxygen species (ROS)—dependent manner through the activation of Akt and c-Jun-aminoterminal kinases (JNKs) [77].
More recently, evidence has emerged that indicates SERPINB3 as a novel pro-inflammatory mediator in NAFLD/NASH progression by means of in vivo and in vitro experiments [78]. For these experiments were used the previously mentioned mice either genetically manipulated to overexpress SERPINB3 in hepatocytes (TG/SB3 mice) or to carry a deletion in the RSL of SERPINB3A gene [SB/knock out (KO) mice]. These mice were fed on two different dietary protocols [MCD diet or the choline-deficient, amino acid-refined (CDAA) diet] to induce experimental NASH. These experiments revealed that the dietary induction of NASH in TG/SB3 mice led to an impressive increase in macrophages infiltrating the chronically injured liver, forming the characteristic crown-like aggregates described also by others, as well as to an up-regulation of hepatic transcript levels of pro-inflammatory cytokines [including tumor necrosis factor-α (TNF-α) and IL-1β] as well as of chemokine ligand 2 (CCL2). Of relevance, all these parameters and the extent of liver injury were significantly blunted in KO/SB3 mice. The opposite changes in liver macrophage activation observed in TG/SB3 or KO/SB3 mice following dietary NASH induction were associated with a parallel modulation in the expression of triggering receptor expressed on myeloid cells-2 (TREM2), cluster of differentiation 9 (CD9), and galectin-3 markers, recently detected in NASH-associated macrophages. Additional in vitro experiments employed phorbol-myristate acetate-differentiated human THP-1 macrophages that were exposed to hrSERPINB3. These experiments on cultured THP1 macrophages confirmed that hrSERPINB3 was able to stimulate macrophages to produce M1-cytokines such as TNF-α and IL-1β, as well as ROS, TGFβ1, and vascular endothelial growth factor (VEGF) through the activation of the nuclear factor-kappa B (NF-κB) transcription factor. These results outlined that SERPINB3, as released by chronically activated/injured hepatocytes, may operate both as a pro-inflammatory and pro-fibrogenic mediator in NASH, then contributing to disease progression (Figure 1).
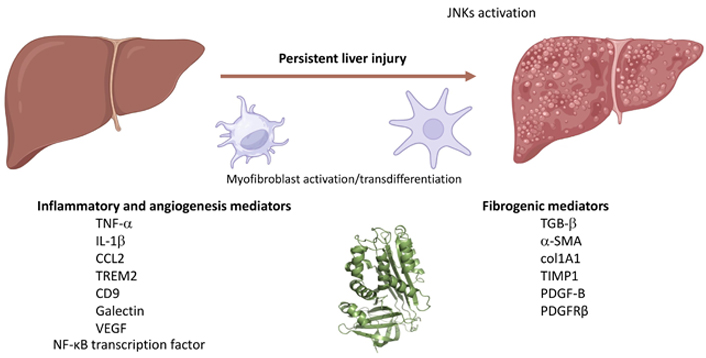
SERPINB3 can contribute to the progression of CLDs by inducing the release of pro-inflammatory and pro-fibrogenic mediators or ECM components by macrophages or MFs. TGB-β: transforming growth factor-β. Created with BioRender.com
Several laboratories have proposed a pro-carcinogenic role for SERPINB3 and -B4 which are indeed significantly up-regulated in the sera and in tumor tissue of a number of human squamous carcinomas developed in those tissues in which they are also physiologically expressed, including lung, esophagus, and uterine cervix [57, 79, 80]. Whether SCC and squamous esophageal carcinoma are concerned, the levels of these SERPINs correlate with tumor infiltration and lymph node metastasis [81–83] and have been proposed as biomarkers to predict disease stage and response to therapy [84–86]. An increased expression of both SERPINs has been also detected in the adenocarcinoma of lung, pancreas, and breast, as detailed later in this review, in HCC and other primary liver cancers [67, 87–90].
The pro-tumorigenic role of these SERPINs in earlier studies, mostly related to extrahepatic cancer, has been attributed to several biological effects; the most relevant ones are represented by: i) the induction of epithelial to mesenchymal transition (EMT), a process favoring invasiveness and then metastasis in cancers of epithelial origin [91–93]; ii) the ability to inhibit cell death, particularly by preventing cancer cell apoptosis induced by several conditions or chemotherapeutic agents [94], by either inhibiting JNK or p38 mitogen-activated protein kinase (MAPK) and/or suppressing mitochondrial ROS generation [95–97]. In particular, SERPINB3 has been suggested to be able to contribute to tumor cell resistance to anti-neoplastic drugs as a consequence of its localization at the inner mitochondrial compartment. In this location, SERPINB3 can bind to the respiratory complex I and for this reason, SERPINB3 has been suggested to protect cells from the toxicity of pro-oxidant chemotherapeutic agents such as doxorubicin and cisplatin [97]. Specifically, SERPINB3, by inhibiting ROS generation induced by the two chemotherapeutics, can prevent or reduce the opening of the mitochondrial permeability transition pore (MPTP) and the induction of apoptotic cell death.
As an additional pro-carcinogenic mechanism, SERPINB3 was also proposed to induce constitutive and chronic activation of the endoplasmic reticulum (ER) stress-related unfolded protein response (UPR), possibly through the inhibition of lysosomal proteases [92].
Historically, the earlier data related to HCC were published in 2004 in a study describing the detection of SCCA (as it was still named) in a series of 65 biopsies from CLD patients of different etiology carrying HCC and compared to normal livers [67]. SCCA, undetectable in the control liver, was observed by immunohistochemistry in 85% (55 out of 65) of the HCC biopsies, with positivity mostly in the cytoplasm of HCC cells. Further analyses performed on a total of 18 HCC available surgical samples from the same cohort of patients showed that 14 on 18 of these samples were expressing SCCA mRNA but never in nontumoral tissue. Sequence alignment outlined that SCCA1 variant was present in six patients and SCCA2 in three patients but also revealed the expression of a novel SCCA1 variant (G351 to A) in five cases. This earlier study then indicated that SCCA variants were overexpressed in HCC cells independently of tumor etiology but not in normal liver. These earlier data were confirmed and further supported by two independent studies that analyzed cirrhotic patients and cirrhotics carrying HCC. A first study [98] confirmed that SCCA expression was much higher in HCC mass than in the peritumoral tissue. Moreover, SCCA serum levels were significantly higher in samples from HCC patients than in cirrhotic samples. In the same year, a second study [68] showed that circulating IC composed by SCCA-IgM IC, undetectable in the serum of healthy controls, were detected in 35 on 50 patients carrying HCC. Moreover, the SCCA-IgM IC was detected in the serum of only 15 out of 50 patients with cirrhosis but at levels significantly lower than those evaluated in HCC patients. Both studies [68, 98] suggested that SCCA expression or the SCCA-IgM IC may be useful for either screening of serum in patients at risk to develop HCC and/or to increase the sensitivity for HCC diagnosis. A later study, performed on two series of patients with cirrhosis that in a period of follow up of 4 years either developed HCC or not, demonstrated a progressive increase of SCCA-IgM IC over the time and that such a progressive increase was strongly associated with HCC development, suggesting that SCCA-IgM IC might be useful to identify cirrhotic patients at higher risk of HCC development [99]. Finally, by still monitoring serum SCCA-IgM levels, an additional study revealed that the relative balance between SERPINB3 and -B4 isoforms was altered in HCC patients being characterized by a lower SERPINB4-IgM/SERPINB3-IgM ratio, determined by lower SERPINB4 levels [60].
The initial evidence of the role of SERPINB3 in the impairment of the immune system dates back to 2001, when Suminami et al. [100] described that SCCA1, but not SCCA2, was able to suppress in vitro natural killer (NK) cell migration induced by monocyte-chemoattractant protein-1 (MCP1, now renamed as C-C motif chemokine ligand 2 or CCL2). This inhibitory effect was lost by mutation of the RSL of SCCA1, highlighting the crucial role of its anti-protease activity in the induction of this effect.
More recent evidence has confirmed that SERPINB3 plays a crucial role in the impairment of immune surveillance through its cross-talk with the tumor immune microenvironment, favoring cancer progression [101]. In esophageal cancer, the presence of this serpin in tumoral tissue was indeed inversely correlated with the extent of expression of antigen-presenting cells and the presence in serum of both free and IgM bound SERPINB3 was inversely correlated with activation markers of both innate and adaptive immunity within the tumor [102]. In addition, SERPINB3 and programmed death-ligand 1 (PD-L1) showed a positive correlation within the tumor tissue, while this serpin determined a paracrine induction of this check point regulator in cultured mononuclear cells [102]. Another study in human primary cervix tumors documented that SERPINB3 positive tumors secreted high levels of chemokines, like C-X-C motif chemokine ligand (CXCL) 1/8 and S100A8/A9, which attracted myeloid cells with potent immunosuppressive activity and inhibited T cell activation, leading to an environment resistant to radiotherapy [103]. It is interesting to note that this effect was mediated via signal transducer and activator of transcription (STAT)-3 signaling activation [103], a transcription factor that has been previously identified as a required element for the continuous activation of SERPINB3 and SERPINB4 genes [104]. In addition, TGFβ1, a cytokine induced by SERPINB3 and implicated in the pro-fibrogenic process, as described above, is also one of the major cytokines involved in impaired immune response, since it orchestrates T lymphocyte regulation by limiting effector T cell functions and by promoting regulatory T cell development and functions [105]. This assumption has been also supported by the fact that in a murine model of lung fibrosis, despite mice transgenic for SERPINB3 showed more collagen deposition, they presented lower inflammation and lower peribronchiolar and interstitial follicular inflammatory infiltration [106]. In line with these findings, mice that were genetically modified and expressed a SERPINB3 form deleted in its RSL (SERPINB3/KO), showed not only lower TGFβ1 expression, but also a markedly reduced macrophage infiltrate in the liver [78]. These findings are relevant since tumor-associated macrophages (TAMs) represent the most relevant leukocyte population in solid tumors and with their anti-inflammatory and pro-tumorigenic features they modulate the local microenvironment to facilitate tumor growth and metastasis [107].
One of the main mechanisms through which SERPINB3 determines the aggressive profile of cancer concerns its impact on stem cell state. Stemness features confer indeed tumor growth advantage and increased therapeutic resistance, due to their increased ability of DNA repair [108] and drug efflux pumps [109]. Among the stemness-related signaling, the Wnt/β-catenin has been identified as one of the more important signaling pathways [110] and, as already described, SERPINB3 is able to upregulate this pathway through the activation of membrane low-density lipoprotein receptor-related proteins (LRPs) [111]. It should be considered that one of the most important downstream target genes of β-catenin is Myc oncogene, which was identified as one of four genes, including SRY-box transcription factor 2 (also known as SOX2), Octamer-4, and Krüppel-like factor 4 (KLF4), which could reprogram cells to a pluripotent stem cell state [112]. Therefore, it is not surprising that different studies have described the association of SERPINB3 with a stemness profile, not only in the different forms of primary liver cancers, including hepatoblastoma [113], HCC [114], and cholangiocarcinoma [115], but also in glioblastoma, where SERPINB3 knockdown reduced cancer stem cell signaling [116].
β-catenin signaling is another critical regulator of cancer aggressiveness, not only by the induction of EMT and invasiveness, but also because activated β-catenin impairs immune surveillance and promotes immune escape. Two different studies have indeed documented that in HCC β-catenin activation correlates with T-cell exclusion [117] and resistance to anti-programmed cell death 1 (PD-1) therapy [118]. These results were also confirmed in a genetically-engineered mouse model of HCC where the expression of β-catenin promoted immune escape, which involved defective recruitment of dendritic cells and consequently, impaired T cell activity. In addition, β-catenin-driven tumors were resistant to anti-PD-1 treatment [119]. In this scenario, SERPINB3 was found as an effective inducer of β-catenin, since its overexpression, obtained in transfected hepatoma cells, determined a remarkable up-regulation of both cytoplasmic and nuclear β-catenin levels, together with EMT features and increased cell proliferation, compared to cells transfected with the empty plasmid vector [93]. In addition, it has been recently reported that SERPINB3 is able to upregulate Wnt signaling and β-catenin expression also in monocytic cells by interacting with the membrane-associated LRP family members, including LRP-1, LRP-5, and LRP-6, a feature which is shared with other SERPINs, at least for LRP-1 [111]. The results obtained in both primary human monocytes and in the THP-1 cell line have demonstrated that the exogenous addition of hrSERPINB3 significantly increased the lifespan and the proliferation of both cell types. In particular, the addition of hrSERPINB3 resulted in a significant increase in the replication of primary monocytes, starting on the fifth day of culture. The positive effect of this SERPIN was confirmed in THP-1 cells, in which increased proliferation was achieved starting on the second day of culture. It is worth noting that increased monocyte proliferation promoted by hrSERPINB3 was associated with an increase in its endogenous expression, leading to a paracrine positive loop induction.
Among the SERPINB3 and -B4 isoforms, SERPINB3 has been more closely associated with cancer progression. Despite these two polymorphic variants share a 92% identity at amino acid sequence, their degree of homology drops to 54% at the catalytic site RCL, accounting for the different targeted proteases [120]. While previous findings indicate that the integrity of RCL is essential for TGFβ1 induction [58], other studies indicate that SERPINB3 specifically inhibits cathepsin L [50] and this protease, released from the lysosomes and other acidic compartments, has been shown to cause caspase activation and apoptotic cell death [121, 122].
Another remarkable effect of SERPINB3 in tumorigenesis consists in its ability to up-regulate Myc oncogene [113]. Among transcription factors involved in liver carcinogenesis, Myc overexpression is indeed detectable in up to 70% of viral and alcohol-related HCCs, often in correlation with the more advanced and aggressive tumor forms [123, 124]. At mechanistic level, it has been found that this oncogene is up-regulated by SERPINB3 through calpain and Hippo-dependent molecular mechanisms both in human liver cancers and in transgenic mice and hepatoma cells overexpressing SERPINB3 [125]. This SERPIN was able to inhibit the activity of calpain in vitro, likely reducing its ability to cleave Myc in its not oncogenic Myc-nic cytoplasmic form. The hypothetical proposed model assumes that the lack of retention of Myc into the cytoplasm, due to the block of calpain-induced cleavage of Myc, allows in turn its nuclear translocation, where it could exert its pro-oncogenic transcriptional activity. Myc transcription is also upregulated by SERPINB3 through the induction of Yes-associated protein (YAP) and Hippo pathways, although the precise signal cascade has not been yet elucidated [125].
Strategies by which SERPINB3 leads to the development of primary liver cancers are multiple and are supported by the previously described findings, including apoptosis resistance, increased proliferation and EMT induction, impaired immune response, and induction of stemness features. While these properties confer cell protection in acute damage conditions [126–128], they favor malignant transformation in case of chronic injuries. It is worth noting that SERPINB3 has been detected in the more aggressive forms of all the types of primary liver cancers, including HCC, cholangiocarcinoma, and hepatoblastoma and this is not surprising, considering that this SERPIN is expressed in the hepatic stem cells compartment, consisting in the epithelial cell adhesion molecule (EpCAM) positive fraction, sorted from both foetal and adult cirrhotic livers [129]. Recapitulation of the major known pro-carcinogenic actions of SERPINB3 is illustrated in Figure 2.
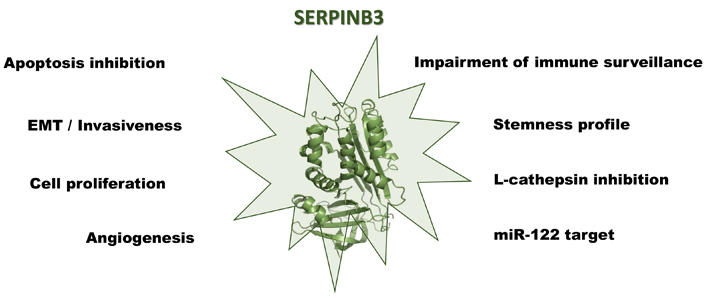
The multiple roles of SERPINB3 in the development and progression of primary liver cancers. Created with BioRender.com
One of the initial properties described for SERPINB3 consists in its ability to determine EMT, a feature that facilitates cell invasion and metastasis formation [93]. Hepatoma cell clones transfected to overexpress SERPINB3 showed indeed morphological changes, characterized by clusters of loosely connected cells with elongated shape and ultrastructural analysis confirmed the decrease of desmosomal junctions and widening of intercellular spaces. These alterations were associated with a reduction of E-cadherin and an increase of β-catenin, associated with a parallel increase in cell proliferation. The autocrine effect of this serpin was maintained also at paracrine level, since recombinant SERPINB3 induced significant cell scattering, migration, and invasiveness in untransfected cells.
Concerning the resistance to chemotherapeutic drugs, beside apoptosis resistance, a computational analysis has identified SERPINB3 as a miR-122 hypothetical target gene [114]. miR-122 is the most abundant miRNA in the liver and is crucial for the normal liver function [130, 131], while it is down-regulated in preneoplastic nodules and in HCC and inversely associated with metastasis formation and poor prognosis, although the underlying mechanisms are still unclear [132]. Using a dual-luciferase assay, miR-122 overexpression determined a decrease of the reporter gene activity in pGL3-SERPINB3 co-transfected HepG2 cells, while an opposite result was obtained in miR-122-silenced cells, where an upregulation of SERPINB3 at both mRNA and protein levels was observed [114]. The interplay of miR-122 and SERPINB3 was confirmed in a well-characterized chemically induced diethyl-nitrosamine (DEN)-HCC rat model and the results showed an up-regulation of SERPINB3 in HCC tissues and a down-regulation of miR-122. In addition, higher miR-122 levels were associated with lower SERPINB3 expression, suggesting a possible regulation of SERPINB3 by miR-122 in this animal model. An inverse correlation between miR-122 and SERPINB3 was also found in two cohorts of patients with HCC, both al mRNA and protein level [114]. In addition, while miR-122 overexpression determined sensitization to Sorafenib in different cell lines, the presence of SERPINB3 overexpression determined resistance and increased cell viability, compared to controls, emphasizing the importance of patient-tailored stratification to optimize therapeutic strategies.
A recent study has described that specific deletion of HIF-2α in a mice model of NASH-related liver carcinogenesis determines a significant reduction of the number and volume of liver tumors, compared to controls [133]. These effects, not involving HIF-1α, were associated with a reduction of the cell proliferation marker Ki67 at nuclear level. It is worth noting that, both in liver experimental NASH and in human liver tumors of metabolic origin, HIF-2α levels were closely correlated with SERPINB3 expression. Accordingly, a positive correlation between the transcripts of HIF-2α, YAP, and its downstream target Myc in tumor specimens was observed, while HIF-2α deletion determined down-modulation of Myc and YAP expression. Since SERPINB3 is up-regulated by HIF-2α [71] and in turn it is able to enhance HIF-2α stabilization through the conjugation with the neural precursor cell expressed developmentally down-regulated-8 (NEDD8) [134], these results support the relevant role of these two molecules in the modulation of the YAP/Myc pathway in liver carcinogenesis during NASH progression and may be considered as novel putative therapeutic targets.
Hepatoblastoma is the most common liver malignancy in early childhood and activating mutations in the β-catenin gene, associated with cytoplasmic and nuclear accumulation of β-catenin and increased levels of Myc and cyclin D1 have been frequently reported, especially in proliferative and poorly differentiated forms [135, 136]. An international collaborative study has documented that SERPINB3 is detectable in the majority of the hepatoblastoma cases, with different extents of expression in individual cases [113]. It is interesting to note that the highest levels of this serpin were detectable in the more immature embrional cell compartment and in the small-cell undifferentiated (SCUD) pattern, typical features of the most aggressive hepatoblastoma subtypes [137]. Also, in this type of primary liver tumor SERPINB3 up-regulation was significantly correlated with Myc expression. This positive relationship was further corroborated by the results obtained in HepG2 cells stably transfected to over-express SERPINB3, confirming the involvement of this serpin in Myc up-regulation. This effect was independent of the presence of the serpin reactive loop, suggesting that the antiprotease activity of the protein is not required [113]. Since Myc is a downstream gene of β-catenin, these data are in line with recent results indicating that the site of interaction of SERPINB3 with the surface receptor LRP-1, which determines up-regulation of the Wnt pathway, is located downstream of the RSL [111].
Cholangiocarcinoma (CCA) is a primary liver cancer deriving from biliary epithelial cells and represents the second most common primary liver tumor, after HCC, characterized by a dismal prognosis, with a 5 years overall survival less than 10% [138]. This pathological condition and the resultant health problems are of particular relevance since the incidence of CCA is constantly increasing and its mortality has been rising in the last decades [139]. Several studies have identified cancer stem cells (CSCs) as a driving force for CCA initiation, dissemination, and drug resistance [140, 141]. A recent study has identified SERPINB3 as a novel and pivotal modulator of the stemness features of CCA [115]. Several lines of evidence supported this conclusion, deriving from in vitro experiments, in vivo tumor xenograft models, and from different cohorts of patients with CCA. Cultured cells experiments provided evidence that SERPINB3 expression was markedly up-regulated in the subset of stem-like cells of CCA that formed 3D spheres (SPH) and these findings were in line with previous studies, reporting a higher tumorigenic potential and stemness features of this subset, able to activate macrophages toward a TAM phenotype [142, 143]. In addition, the presence of SERPINB3 determined up-regulation of gene expression of stem-like markers [CD24, CD44, CD90, CD133, c-MYC, NOTCH1, STAT3, YAP, BMI1, octamer 4 or OCT4, SOX2], EMT (β-catenin, snail family zinc finger 2 or SLUG, SNAIL), and ECM remodelling-related genes like matrix metalloproteinases (MMP), including MMP1, MMP7, and MMP9, a disintegrin and metalloproteinase (ADAM) isoforms like ADAM9, ADAM10, and ADAM17, as well as integrin beta-3 (ITGB3) [115]. It is worth noting that these changes in gene expression were associated with activation of key molecular pathways, such mitogen-activated protein kinases like extracellular regulated kinases (ERK) 1/2, p38, JNK-1, phosphorylation of p65 subunit of NF-κB transcription factor as well as up-regulation of c-Myc, NOTCH, MMP9, and β-catenin [115]. In agreement with these findings, CCA cells transfected to overexpress SERPINB3, when injected in immune-deficient mice, determined increased tumor formation and the neoplastic mass presented higher weight and volume, compared to the findings observed in controls. The potential clinical relevance of these results was confirmed in human intrahepatic CCA (iCCA) specimens. Using a published dataset of 104 patients with iCCA [144], the presence of high levels of SERPINB3 was associated with lower survival and shorter time to recurrence and these results were validated in another cohort of patients with CCA [145]. Immunohistochemistry results, obtained in iCCA specimens from additional 38 patients, revealed that patients with high SERPINB3 scores had a three-fold lower time to recurrence compared to patients with low SERPINB3 expression [115]. These findings, indicating a role of SERPINB3 in mediating malignant phenotype of iCCA, are in line with preliminary results described in extrahepatic CCA (eCCA), where the presence of SERPINB3 in the bile compartment was associated with a higher frequency of portal invasion and a higher rate of tumor recurrence after surgery [146].
As described in the present review, evidence from experimental, translational, and clinical studies has outlined that SERPINB3, in addition to its canonical role as protease inhibitor, may serve as a putative biomarker of CLDs progression and liver cancer development. Even more relevant, SERPINB3 has been reported to act as a molecular mediator that can exert a critical role in modulating CLDs progression (i.e., by affecting both inflammatory response and fibrogenesis) as well as the processes leading to the development of all the primary liver cancers, including HCC, hepatoblastoma, and both iCCA and eCCA. This bulk of knowledge has opened the way to research, currently in progress, devoted to designing and testing therapeutic strategies able to counteract its pro-inflammatory, pro-fibrogenic, and pro-carcinogenic actions.
ADAM: a disintegrin and metalloproteinase
CCA: cholangiocarcinoma
CCL2: chemokine ligand 2
CD9: cluster of differentiation 9
CLDs: chronic liver diseases
EMT: mesenchymal transition
HCC: hepatocellular carcinoma
HIF-2α: hypoxia-inducible factor-2α
hrSERPINB3: human recombinant serpin family B member 3
IC: immune complexes
iCCA: intrahepatic cholangiocarcinoma
IL: interleukin
JNKs: c-Jun-aminoterminal kinases
KO: knock out
LRPs: low-density lipoprotein receptor-related proteins
MFs: myofibroblasts
MMP: matrix metalloproteinases
NAFLD: non-alcoholic fatty liver disease
NASH: non-alcoholic steatohepatitis
NF-κB: nuclear factor-kappa B
PD-1: programmed cell death 1
RCL: reactive center loop
ROS: reactive oxygen species
RSL: reactive site loop
SCC: squamous cell carcinoma
SCCA: squamous cell carcinoma antigen
SCCA-IgM: squamous cell carcinoma antigen and immunoglobulin M
SERPINB3: serpin family B member 3
TGFβ1: transforming growth factor β1
TNF-α: tumor necrosis factor-α
YAP: Yes-associated protein
αSMA: α-smooth muscle actin
PP and MP: Conceptualization, Writing—original draft, Writing—review & editing. Both authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Siva Bala Subramaniyan, Balasubramaniyan Vairappan
Anna Fichera, Mirella Fraquelli
Lorenzo Mainardi ... Chiara Raggi
